Abstract
The branching structures of neurons are a long-standing focus of neuroscience. Axonal and dendritic morphology affect synaptic signaling, integration, and connectivity, and their diversity reflects the computational specialization of neural circuits. Altered neuronal morphology accompanies functional changes during development, experience, aging, and disease. Technological improvements continuously accelerate high-throughput tissue processing, image acquisition, and morphological reconstruction. Digital reconstructions of neuronal morphologies allow for complex quantitative analyses that are unattainable from raw images or 2D tracings. Furthermore, digitized morphologies enable computational modeling of biophysically realistic neuronal dynamics. Additionally, reconstructions generated to address specific scientific questions have the potential for continued investigations beyond the original reason for their acquisition. Facilitating multiple re-use are repositories like NeuroMorpho.Org, which ease the sharing of reconstructions. Here, we review selected scientific literature reporting the reconstruction of axonal or dendritic morphology with diverse goals including establishment of neuronal identity, examination of physiological properties, and quantification of developmental or pathological changes. These reconstructions, deposited in NeuroMorpho.Org, have since been used by other investigators in additional research, of which we highlight representative examples. This cycle of data generation, analysis, sharing, and re-use reveals the vast potential of digital reconstructions in quantitative investigations of neuronal morphology.
Axonal and dendritic morphology
The branching structures of neurons - dendrites and axons - form the substrate for network connectivity, synaptic communication, and signal integration. The stunning diversity of neuronal morphology (Figure 1) has thus profound implications for information processing in the nervous system. The importance of axonal and dendritic wiring in neural function had already been recognized by Santiago Ramón y Cajal, whose pioneering pencil-on-paper drawings beautifully immortalized the complexity of brain circuitry more than a century ago. Today much of neuroscience still focuses on the relationship between neuronal structure, activity, and function, but the techniques for data acquisition and analysis have advanced greatly. Specifically, over the past three decades axonal and dendritic arborizations have been increasingly reconstructed from optical microscopy in digital format (Halavi and others 2012). This transition has transformed the investigation of neural architecture, as detailed quantification of branching morphology is only possible via digitized data (Meijering 2010).
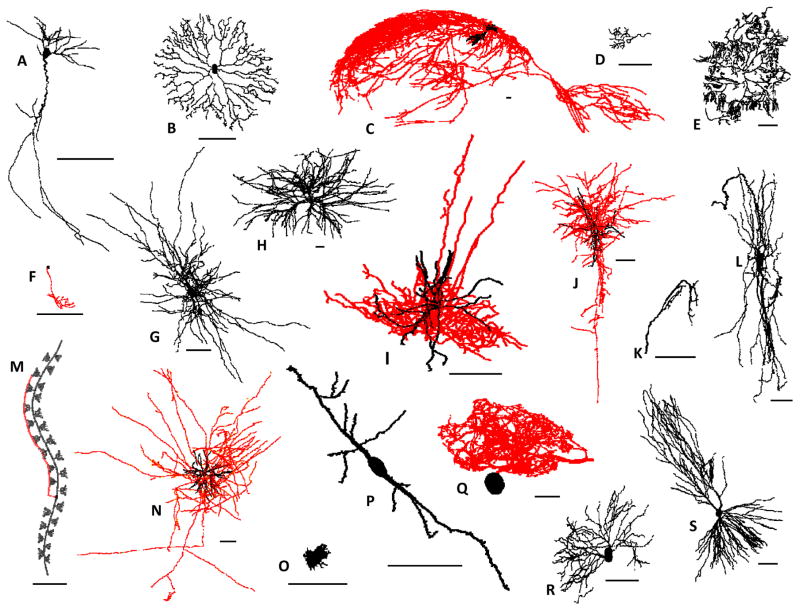
Figure 1. Structural diversity in dendrites (black) and axons (red).
The species, brain region, and reported neuron type of representative reconstructions from NeuroMorpho.Org (numbers in parenthesis indicate the corresponding unique identifier “NMO_ID” in the database); scale bars: 100 μm. A) Proechimys, hippocampus, pyramidal-like (7254); B) Rabbit, retina, amacrine (765); C) Rat, hippocampus, von Economo (930); D) Goldfish, optic nerve, sensory (6897); E) Drosophila, peripheral nervous system, pyramidal (7046); F) Zebrafish, spinal cord, motoneuron (9367); G) Elephant, neocortex, motoneuron (6239); H) Frog, spinal cord, motoneuron (7289); I) Mouse, neocortex, parvalbumin containing (8508); J) Monkey, neocortex, sensory interneuron (1869); K) Cricket, cercal sensory system, neurogliaform (4594); L) Turtle, spinal cord, motoneuron (7291); M) C. elegans, somatic nervous system, sensory (9857); N) Cat, neocortex, pyramidal (856); O) Chicken, brainstem, bipolar (8909); P) Human, neocortex, basket (1078); Q) Spiny lobster, stomatogastric ganglion, retinotectal (6634); R) Salamander, retina, ganglion (770); S) Guinea-pig, hippocampus, pyramidal (7904).
The rapid growth of digitized morphological reconstructions in neurobiology is due in large part to the powerful synergy of state-of-the-art molecular labeling, microscopic imaging, neuroinformatics tools, and computational simulations of biophysical activity (Parekh and Ascoli 2013). Yet most individual labs typically only interact with specific aspects of this collective pipeline. Although applications of digital reconstructions are numerous, most datasets are acquired to address a specific scientific question. Investigators may not always realize that, once collected and utilized to pursue an original research goal, those same reconstructed morphologies, if shared, are suitable to produce additional knowledge. Digital reconstructions are easier to share than the original image stacks or tissue preparations. Moreover, once shared, these data can be (and typically are) re-used multiple times, therefore augmenting the impact of the original research multiplicatively rather than additively. Leveraging shared neuronal reconstructions in independent novel re-analyses or in computational models of biophysically realistic neuronal activity avoids the substantial expense of time and labor involved in acquiring new data (Ascoli 2007). Furthermore, these follow-up applications often require and therefore catalyze collaborative interactions between researchers with different expertise.
The scientific literature on the development, structure, and function of neuronal arbors in physiological and pathological conditions spans the gamut of cellular neuroscience. This review focuses on the quantitative studies based on digital reconstructions of axonal and dendritic morphologies. Specifically, we highlight representative investigations in which neurons were digitally reconstructed, leading to primary discoveries, and then shared with the research community. We then analyze the growth and utilization of the main open repository for publicly sharing neuronal reconstructions. Lastly, we provide examples of secondary discoveries from the re-use of shared reconstructions.
Digital morphological reconstructions - primary discoveries
The process of generating digital reconstructions begins with labeling neurons, followed by visualizing and then tracing their morphology. Several alternatives exist at each step with complementary advantages and disadvantages, depending on experimental conditions, available instruments, and scientific goals. Earlier digital reconstructions of neurons were generated for quantitative and structural description of dendritic and axonal morphologies in species as diverse as frog (Bregman and Cruce 1980), lobster (Beltz and Kravitz 1987), mudpuppy (Arkin and Miller 1988), salamander (Toris and others 1995), and cat (Oliver and others 1991). These efforts continued at the turn of the century in macaques (Burman and others 2000), squirrel monkeys (M. Parent and A. Parent 2005), guinea-pigs (Palmer and others 2003), agoutis (Santiago and others 2010), elephants and humans (Jacobs and others 2011), and dragonflies (Gonzalez-Bellido and others 2013). Most recently, digital reconstructions of a variety of cerebellar neurons were generated from a number of large-brained mammals including chimpanzee, human, manatee, humpback whale, clouded leopard, giraffe, and Siberian tiger (Jacobs and others 2014). Detailed comparative morphometric analyses conducted with these reconstructions indicated significant variations in dendritic measures among all species.
Digital reconstructions have been instrumental in quantifying morphological aberrations in pathological conditions. For example, simplified dendritic architecture and decreased spine density were observed in cortical pyramidal cells with increasing seizure duration in people with partial epilepsy (Multani and others 1994). In contrast, pyramidal neurons in the prefrontal cortex of patients with Huntington’s disease revealed increases in dendritic length and complexity (Sotrel and others 1993). A decrease in spine density in hippocampal CA1 pyramidal neurons was reported in two transgenic mouse models of Alzheimer’s disease (Perez-Cruz and others 2011), and in basal ganglia spiny neurons in a rat model of Parkinson’s disease (Stephens and others 2005). Digital reconstructions enabled the extensive quantification of the damaging developmental effects of alcohol (Hoffman and others 2008) and of in utero exposure to cocaine (Lloyd and others 2003) on the morphology of cortical neurons. Similarly, stress was found to cause experience-dependent reduction of apical dendritic arbors in medial prefrontal cortex (Liston and others 2006) and time-sensitive alterations followed paternal deprivation (Helmeke and others 2009).
Changes in dendritic morphology are not the sole purview of pathologic conditions, but also occur during development (Nunez-Abades and others 1994) and in adulthood (Lee and others 2006). Digital reconstructions have shed light on the developmental roles of specific molecules such as membrane cytoskeletal proteins in the development of Purkinje cell dendrites (Gao and others 2011), nicotinic α5 receptor subunits in layer VI neurons of the prefrontal cortex (Bailey and others 2012), and nicotinic α7 receptors in axonal outgrowth (Nordman and Kabbani 2012). Similar approaches have elucidated the influence of growth factors, including brain-derived neurotrophic factor (Finsterwald and others 2010) and insulin-like growth factor I (Niblock and others 2000), on dendritic development.
Axonal and dendritic arbors play a critical role in establishing and maintaining connectivity between and within brain regions. Well before the word “connectomics” was coined, tract-tracing demonstrated connection specificity between sensory axons and motor neurons in the bullfrog spinal cord (Lichtman and others 1984) and of callosal axons between cortical areas 17 and 18 of the cat (Houzel and others 1994). More recent advances towards the generation of comprehensive connectivity maps in the nervous systems of moth (Kvello and others 2009), cockroach (Wei and others 2010), honeybee (Rybak and others 2010), and fruit fly (Chiang and others 2011) have been accomplished by virtually embedding three-dimensional tracings of neurons into template atlases of the brain. Connectivity maps in rodents have started to yield useful knowledge on cell-specific pathways underlying selected behaviors (Oberlaender and others 2011). Even individual reconstruction can have considerable impact towards quantitative analysis of neural circuitry, as exemplified by the detailed characterization of the 3D spatial distribution of dendritic synapses in a single CA1 pyramidal neuron from the rat hippocampus (Megias and others 2001).
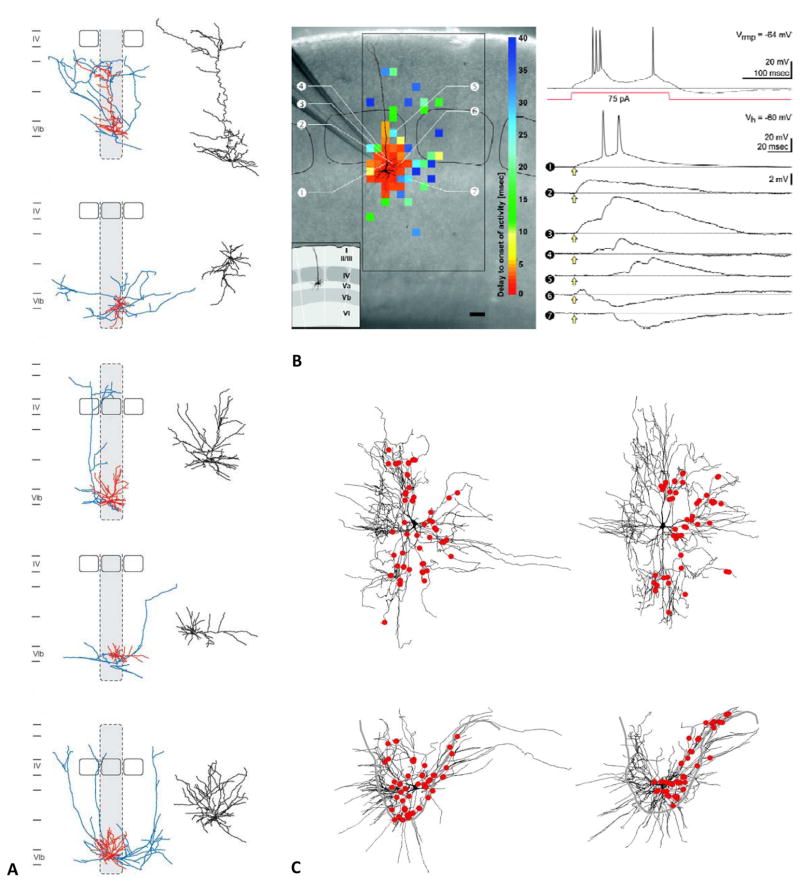
Figure 2 highlights three case studies in which digitally reconstructed morphologies aided in neuron identification, functional classification, quantitative morphometry, and analysis of circuit connectivity at both macroscopic and microscopic levels. In the first example, detailed 3D reconstructions of 46 electrophysiological identified cells helped classify excitatory neurons in previously unexplored layer 6b of the rat barrel cortex (Marx and Feldmeyer 2012). Specifically, both dendritic and axonal arborizations were digitally traced after whole-cell patch-clamp recording, biocytin filling, and subsequent histological processing (Figure 2A). The combined structural and functional characterization in the same neurons demonstrated the existence of distinct groups of cells with specific input, output, and biophysics. The reconstructions were shared via NeuroMorpho.Org (Feldmeyer archive) and downloaded more than 3500 times in less than a year.
Figure 2. Examples of primary discoveries made with digital reconstructions.
A) Diversity of excitatory neurons in layer 6b of the rat barrel cortex from the Feldmeyer archive of NeuroMorpho.Org (adapted from Marx and Feldmeyer 2013). B) Morphological and electrophysiological characterization of newly identified pyramidal neurons from the Staiger archive (adapted from Schubert and others 2006). The left photomicrograph of the native coronal slice is superimposed to the somatodendritic reconstruction of the recorded neuron. The color-coded topographic map represents the delay between glutamate uncaging and activity onset in the recorded cell, separating likely direct and indirect connections. The larger rectangular and smaller rounded black frames mark the extent of the investigated cortical area and the layer IV barrels, respectively (scale bar: 100 μm). The inset illustrates the laminar and columnar organization. The top right voltage recording corresponds to the firing pattern upon depolarizing current injection of the same neuron at the resting potential (Vrmp). The traces below are membrane potential recordings at the given holding potential (Vh) obtained after glutamate uncaging (yellow arrows) at the positions indicated by the circled numbers. C) Distribution of vestibulospinal synapses on ipsilateral splenius motoneurons in an adult cat spinal cord from the Rose archive (adapted from Grande and others 2010).
In a conceptually similar line of research, Schubert and others (2006) reconstructed the morphologies of pyramidal neurons in layer Va, which had been largely ignored until then. In addition to the detailed description of the axonal and dendritic arbors, these neurons were also characterized physiologically and by functional connectivity based on patch-clamp recordings and caged glutamate photolysis, respectively (Figure 2B). These combined techniques clearly identified a distinct pyramidal cell population in a previously ill-defined neocortical layer. This lab deposited 68 neuronal reconstructions in NeuroMorpho.Org (Staiger archive), which were downloaded over 36,000 over seven-and-a-half years.
In the third case study, the spatial distribution of serotonin boutons on the dendrites of splenius motoneurons in the cat spinal cord was measured along the reconstructed neuron (Figure 2C), allowing for pathway-specific neural connectivity inferences (Grande and others 2010). Moreover, mapping the excitatory and inhibitory contacts on the dendritic reconstructions enabled the determination of the influence of synapse location on the output of motoneurons by computational modeling. The 11 motoneurons uploaded to NeuroMorpho.Org by this lab (Rose archive) were downloaded 1100 times in the first two years of sharing.
NeuroMorpho.Org – Online repository for data sharing
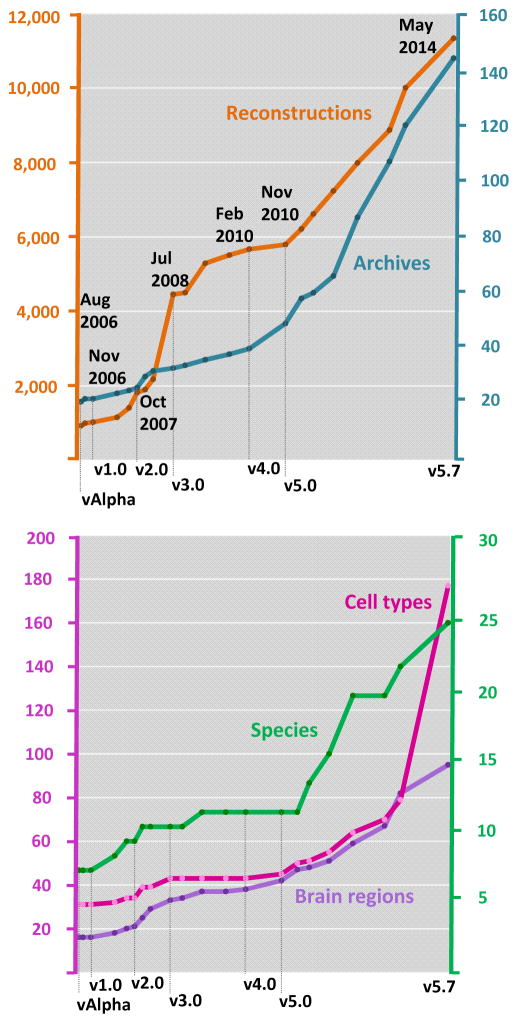
Much like gene bank for genetic sequences, digital reconstructions of neuronal morphology can be deposited in and downloaded from the public repository NeuroMorpho.Org. Each shared reconstruction is centrally inspected, standardized, measured, imaged, and annotated with relevant metadata with minimal burden to the contributing labs (Halavi and others 2008). Processed data are regularly uploaded to the repository and made available to researchers worldwide at periodic version releases (Figure 3).
Figure 3. Availability of reconstructed neuronal morphology in NeuroMorpho.Org.
A) At the introductory release in August 2006, the repository contained ~1,000 reconstructions and 21 archives. Subsequent version releases show a steady increase in the content of the repository. At the May 2014 release (v5.7), the total number of reconstructions is 11,335 from 144 archives. B) With increased data content, the numbers of represented species, brain regions, and cell types have also steadily grown since over the years.
The introductory alpha-version of NeuroMorpho.Org, released in August 2006, included just shy of 1,000 reconstructions (Figure 3A). In May 2013 (v5.6), the repository crossed a milestone bringing the total number of available, fully curated reconstructions over 10,000. The number of contributing laboratories (“archives”) in the database has grown steadily along with the reconstructions, suggesting a progressively increased willingness of the community to share data. For example, since v5.0 (released in 2010), the number of new archives has nearly tripled, reflecting successful outreach efforts to raise awareness on the importance of data sharing. The number of species, brain regions, and cell types represented in the repository show a similar growth trend with each release (Figure 3B). As the experimental and computational pipeline for generating reconstructions becomes more efficient, new cell types and brain regions become accessible from previously challenging species. For example, digital neuron morphologies from eight new species, ranging from C. elegans to elephant, have been made available since June 2011. Additional new species currently in the processing pipeline include giraffe, humpback whale, clouded leopard, manatee, and Siberian tiger (Jacobs and others 2014).
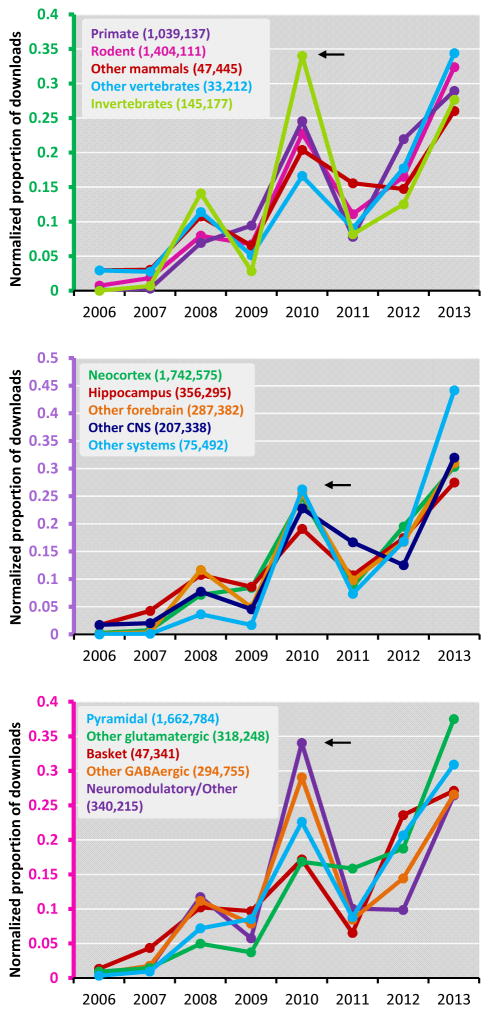
Without any sign-in requirements, NeuroMorpho.Org visitors can freely browse, search, analyze, inspect, and download the processed reconstruction files, the original files contributed by the authors, detailed log reports of the changes made in the standardization procedure, and all metadata. The processed reconstructions are available in the standard SWC format, but files can also be downloaded in NEURON, Genesis, and NeuroML formats, allowing users to import the reconstructions files directly into their preferred simulation environments or other intended applications. To explore download trends, we analyzed activity across species, brain regions, and cell types (Figure 4). For clarity of analysis, we grouped species by taxonomy, brain regions by anatomical hierarchy, and cell types according to their classification in NeuroMorpho.Org. Hence, 21 different species were divided into five groups, namely primate, other mammals, rodent, other vertebrates, and invertebrates. Similarly, 25 brain regions were grouped into neocortex, hippocampus, other forebrain, other central nervous systems, and other systems; and 79 cell types were grouped into pyramidal, other glutamatergic, basket cell, other GABAergic, and neuromodulatory/others.
Figure 4. Data download from NeuroMorpho.Org.
Reconstructions downloaded from NeuroMorpho.Org across three categories: A) species, B) brain regions, and C) cell types. The abscissa refers to the time of download, thus 2006 only counts for its last 5 months since the first public (pre-) release in August. Within every entry of each category, downloads in one year are normalized by the total number of downloads for that same entry over the entire 2006–2014 period. The absolute numbers of total downloads are reported for each entry in parentheses.
A general observation about download activity is that the rate of data access is a direct consequence of the amount of available data. Furthermore, users tend to download entire sets of reconstructions belonging to a specific metadata category (data not shown). Downloads of specific data increase (as expected) over time, as datasets are available online for a longer period. However, larger-than-expected amounts of data were downloaded from the website in September 2010 (arrows in Figure 4). This anomaly corresponded to the selection of NeuroMorpho.Org as one of the data sources for an annual national modeling contest (shumo.com/home – in Chinese) held by the South China University of Technology in Guangzhou, China. Nearly 2,000 teams that included more than 300 PhD students from 230 universities took part in this contest. Participating graduate students (599 teams) chose a specific problem from available options. The challenge given for NeuroMorpho.Org reconstruction data was to generate algorithms for classifying neurons based on their morphology. Files accessed over this three-day period pushed the total number of downloads from the repository in excess of 1.5 million.
Beside the peculiarity of the Chinese contest, in which relatively more reconstructions from invertebrate than vertebrate species were downloaded (Figure 4A), the general emerging trend is a steady increase in data accessed from the repository across all three categories (species, brain region and cell type). More specifically, rodents constitute the largest downloaded species group closely followed by primates (human and monkey), and separated by an order of magnitude from all other species. Neocortical neurons are the most downloaded brain regions, nearly twice as much as all others combined and with a five-fold difference from the second largest downloaded region, the hippocampus (Figure 4B). However, as neuroscience research diversifies, “other” regions and nervous systems are clearly attracting more and more interest. While neocortex tallied over 20 times more total downloads than these “other systems”, on a relative scale this Cinderella group grew by a factor of 26 since 2009 against the more modest 3.5x growth of the lead group. Similarly, pyramidal cells have always been the most commonly studied and downloaded cell type (Figure 4C), but adjusting for total number of downloads reveals a pattern of increased activity of non-glutamatergic cells.
Digital neuron morphology - secondary discoveries
Download activity from the repository clearly shows an overall trend of increasing awareness of the research potential of shared data. Digitized morphologies of axonal and dendritic arbors are amenable to measure comprehensive quantitative morphometrics, to simulate anatomically and biophysically realistic neural activity, to determine potential network connectivity, and to study the developmental principles underlying neuronal growth. By employing experimental measures as constraints, computational modeling has a synergistic relation with traditional “wet lab” research. Both electrophysiological and morphological simulations can help explain and interpret empirical observations by rigorously implementing mechanisms that are technologically inaccessible to direct experimental verification. In this way, computer simulations can validate, quantify, and assess proposed or leading theories. Furthermore, computer modeling allows the rapid exploration of parameters affecting neuronal function. This exercise often results in quantitative and experimentally testable predictions, thus aiding development of new hypotheses as well as design of novel experiments, which in turn can be invaluable to disprove or improve the underlying theory.
For example, dendritic growth can be modeled by global algorithms implementing a sequence of random branching events (van Pelt and Verwer 1983). The underlying production rules reflect biological constraints such as trophic gradients and metabolic resources. Complementary approaches implement local recursive rules corresponding to intrinsic influences (Eberhard and others 2006). In the extreme, morphological models can be based on compartmental dynamics corresponding to individual growth cones and related molecular cascades (van Ooyen 2011). Individual neurons can be distributed in three-dimensional space to generate virtual networks by activity dependent selection of synaptic contacts among the pairs of sufficiently close axonal and dendritic branches (Koene and others 2009). An alternative approach to modeling anatomically realistic networks involves simulation of developmental processes like inside-out neurogenesis, physical interactions of migrating neurons, and diffusible guidance molecules (Zubler and Douglas 2009).
The increasing production and availability of digital reconstructions of dendritic morphology has progressively enabled a tighter and tighter integration of these computational simulations with experimental data. For example, a simple modeling approach demonstrated that anatomically plausible arbors can be virtually synthesized by optimally balancing the minimization of wiring cost and conduction time (Cuntz and others 2010). This model required measuring the cell type-specific spatial distribution of synapses (a key additional parameter) from real neuronal reconstructions. To test the broad applicability of this growth algorithm, this study employed a variety of digital morphologies from NeuroMorpho.Org that had originally been acquired for very different scientific purposes. These include data generated to identify, characterize, classify and describe neurons in the somatosensory cortex (Wang and others 2002) and perirhinal cortex (Furtak and others 2007), to establish a relation between structure and function in retinal ganglion cells (Bloomfield and Miller 1986), and to quantify changes in structure during postnatal development of dentate gyrus granule cells (Rihn and Claiborne 1990). The tool used to implement this simulation is now available as an open-access software package (Cuntz and others, 2011), thus underscoring the research potential of community built resources.
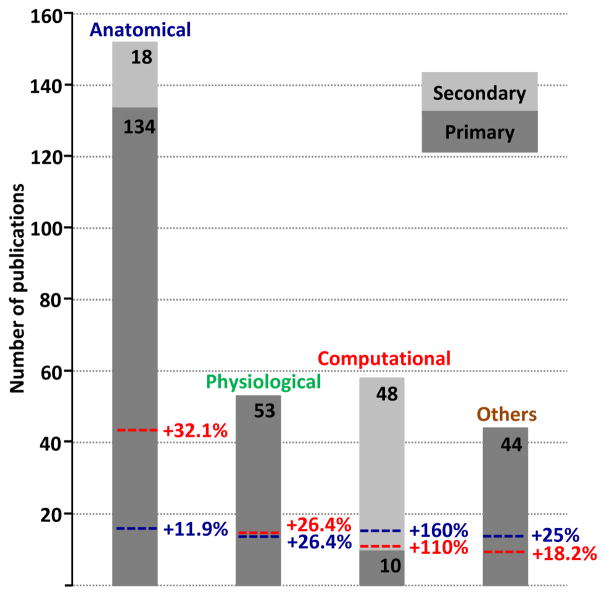
The above example is far from unique. The number of scientific reports published using morphological reconstructions downloaded from NeuroMorpho.Org is constantly growing. One and the same dataset, if shared, can yield both the primary discovery for which it was intended and many additional secondary discoveries that would not have been otherwise possible. To quantify the impact of shared morphological reconstructions beyond the realm of their original intent, we examined the literature database of NeuroMorpho.Org. We identified 241 “primary” publications reporting digital reconstructions of dendritic or axonal morphology that were contributed to NeuroMorpho.Org and 66 “secondary” publications that utilized data from the same repository. Each of these 307 publications was categorized according to their main scientific or technical focus (Figure 5). Publications primarily identifying and describing neuronal morphologies were categorized as anatomical (134 primary, 18 secondary). This category also included quantitative approaches to determine connectivity or to measure structural changes. Experimental studies that detailed the electrophysiological properties of neurons, such as passive and active membrane properties, or factors affecting neuronal dynamics, were classified as physiological (53 primary). Reports relying on computer simulations or describing tools and resources related to reconstructions, such as neuron tracing and data curation, were designated as computational (25 primary, 49 secondary). Articles quantifying changes in neuronal morphologies under pathological conditions or during development, together with those focusing on molecular markers or gene expression, were categorized as others (82 primary). Although the research content of most publications typically addressed more than one topic, each article was only assigned to its primary categories. For example, if a study prominently described the dendritic arborization of a given cell type, complementing it with data on protein expression and membrane properties, it would only be listed as anatomical rather than also as physiological or others.
Figure 5. Categories of publications that contributed to (primary) and utilized data from (secondary) NeuroMorpho.Org.
A total of 307 publications, divided in primary (dark grey) and secondary (light grey) discovery, were categorized based on their primary focus as anatomical, physiological, computational or others. The colored dashed lines reflect the “added impact” or percentage scientific gain expressed as the equivalent number of secondary publications enabled by data shared from a given category.
While all publications categorized as physiological or others describe primary discoveries, secondary discoveries contribute more than 10% of the anatomical papers and (not surprisingly) the majority of the computational articles. We also assessed the added value of primary publications in each category by tallying the number of secondary publications in which the corresponding shared data were used. This analysis is illustrated in Figure 5 as the “additional percentage” effect of the primary publications. For example, as many as 43 of the 48 secondary publications in the computational category used data shared from primary anatomical publications. Therefore the original 134 anatomical publications virtually enabled an additional 43 publications corresponding to an added value of +32.1%. An analogous computation for the secondary anatomical publications (16 out of 18 utilized primary anatomical data) adds a further +11.9% to the added value of this category, summing up to a total scientific gain of 44%. In other words, the 134 primary anatomical publications had an effective impact of 193 publications. Each of the four primary publication categories contributed substantially to both anatomical and computational secondary discoveries. Thus, the research potential of shared data is independent of the scientific goal of the original study. Moreover, because most secondary publications use a large number of primary publications (in the extreme case, database-wide analyses use all available primary data), the vast majority of shared data from primary publications are re-used at least once and more typically multiple times.
Selected examples of secondary discoveries are illustrated in Figure 6. The first use-case leveraged the power of pooling together massive amounts of data to investigate the relationship between branching patterns and spatial sampling in dendritic arbors (Teeter and Stevens 2011). This study quantified arbor territory (the space occupied by the dendritic trees) and branch density (an index of dendritic complexity) from numerous reconstructions shared from independent laboratories. Analysis of reconstructions that invade a mostly flat surface, such as retinal ganglion cells and cerebellar Purkinje cells, as well as of morphologies occupying 3D space (pyramidal and non-pyramidal cells), suggested an inverse relationship between arbor territory and branch density that holds true across species, brain regions, and cell types. Hence, less dense arbors tend to sample greater territories (Figure 6A). While not the only or first scaling law ever discovered in neuroanatomy, this finding is representative of the scientific added value of data sharing. Given the scale of the investigation, these results could not have been obtained without re-using data that had not been specifically created for this intent.
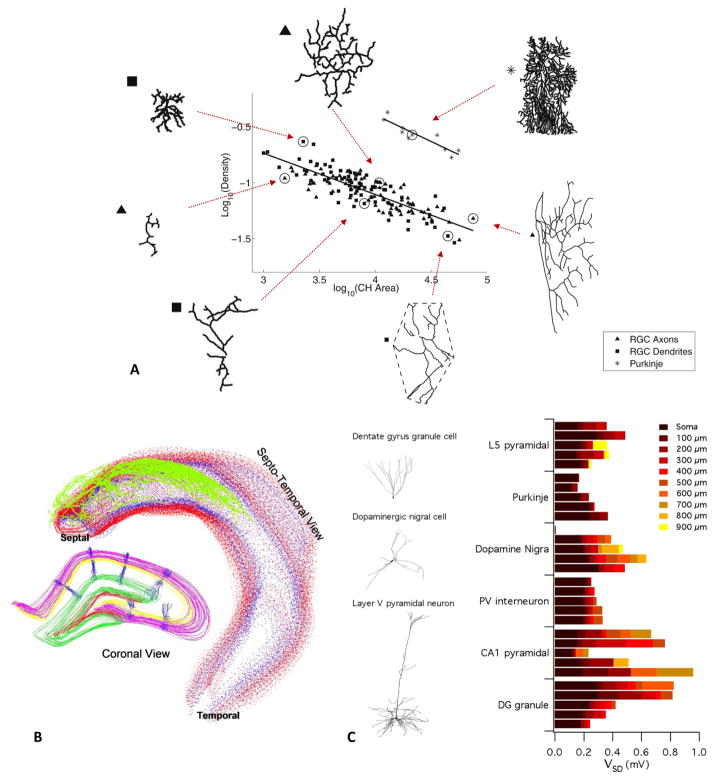
Figure 6. Examples of secondary discoveries made with reconstructions downloaded from NeuroMorpho.Org.
A) Structural design principle describing an inverse relationship between branching complexity and total arbor area in all neurons (adapted from Teeter and Stevens 2011). These data illustrate the case of relatively flat trees from retinal ganglion cells (RGC) and Purkinje cells. B) Space occupancy, dendritic overlap, and orientation of pyramidal cells in the rat hippocampus (Ropireddy and others 2012). The fully reconstructed axon of a single pyramidal cell (green) is embedded in the volumetric rendering of a digital reconstruction of the hippocampal tissue. In the left inset, the reconstructed dendritic trees of pyramidal cells from NeuroMorpho.Org are embedded in the same space. C) Neuron models with stochastically gated, identically distributed ion channels in distinct morphologies, exemplified on the left. On the right, the average standard deviations of the simulated membrane potentials are plotted for each morphological type as a function of increasing distance along the dendrite from the soma (adapted from Cannon and others 2010).
In particular, data downloaded from NeuroMorpho.Org for this study were shared from original publications cutting across the anatomical, physiological, and computational categories. These included morphological characterizations of functionally identified neurons to shed light on the role of layer IV spiny neurons in intracortical processing of sensory information (Staiger and others 2004), in addition to one of the studies described in more detail above under primary discoveries (Schubert and others 2006). Similarly, reconstructions re-used in this application also came from a study that distinguished several diverse sub-classes of basket cells in layers II-IV in the rat somatosensory cortex based on their functional, anatomical, and molecular characterization (Wang and others 2002). Another part of the re-used dataset had been collected for mapping synaptic projections to layer II/III neurons in the rat barrel cortex to determine their functional organization (Shepherd and Svoboda 2005). Other re-used morphologies had previously addressed age-related (Bergstrom and others 2007) and nicotine-induced dendritic remodeling (Ehlinger and others 2012), as well as species-specific neuronal identities, such as von Economo neurons in the human anterior cingulate cortex (Watson and others 2006).
The claimed “universality” of the scaling law was corroborated with neuronal arbors from less represented species, such as those originally reconstructed to investigate projection neurons in the macaque prefrontal cortex (Duan and others 2002), the patterns of axonal terminations of cells in layers II and III of the cat visual cortex (Kisvarday and Eysel 1992), and the in vivo synaptic physiology of the same type of neurons (Hirsch and others 2002). Several datasets reused in this first example of data sharing application had been originally generated for computational modeling, such as the pyramidal neurons in layers V and VI of the temporal sulcus initially employed to investigate the role of local inhibition on synchronized thalamic inputs (Contreras and others 1997). Likewise, other reconstructions re-used in the above study had been previously employed to simulate the effect of dendritic morphology on action potential propagation (Vetter and others 2001) and to constrain biophysical models of reconstructed neurons with their own firing properties recorded in vitro (Hay and others 2011).
As a second example of scientific discoveries enabled by sharing of neuronal morphology, we selected our own lab’s mapping of potential synaptic connectivity in the hippocampus (Ropireddy and Ascoli 2011). This study is particular because it combined shared digital reconstructions available in NeuroMorpho.Org from both the same authors and from several other independent investigators, further integrating them with additional experimental data that were specifically collected for the investigation. The goal of this research was to quantify the potential for synaptic connectivity by estimating the spatial overlap between axonal and dendritic arbors. Most connectivity studies focus either on the electron microscopy synaptic scale or on the light microscopy tract-tracing scale. The probabilistic approach undertaken here falls in between these two scales by quantifying the close spatial appositions of dendrites and axons. Such research strategy requires virtually embedding axonal and dendritic arbors into the appropriate 3D anatomical space of corresponding sub-regional and layer boundaries of the hippocampus (Figure 6B). While NeuroMorpho.Org contains an abundance of dendritic reconstructions from a variety of neuron types in the hippocampus, complete axonal arbors of projection neurons are extremely challenging to collect, and thus much rarer. Furthermore, a detailed 3D reconstruction of the hippocampal tissue with its internal histological demarcations was not yet available.
The full axonal arbors of three CA3 pyramidal neurons from the hippocampus were first meticulously traced and semi-automatically digitized from an in vivo dye-filling preparation. Quantitative analysis of the axonal morphologies revealed differences in length and branching complexity among the neurons depending on their somatic location within area CA3 (Ropireddy and others 2011). A 3D model of the hippocampus was then separately reconstructed from thin Nissl-stained rat brain sections, and the three axonal reconstructions were embedded in it, enabling the measurement of their spatial spread over different layers and sub-regions through the longitudinal extent of the hippocampus. Subsequent quantitative re-analysis of the whole hippocampus reconstruction enabled the 3D stereological characterization of all major anatomical subdivisions (Ropireddy and others 2012). The underlying raw images and corresponding digitized data were freely shared online (http://krasnow1.gmu.edu/cn3/hippocampus3d). Lastly, hundreds of reconstructed pyramidal cells and interneurons downloaded from NeuroMorpho.Org were embedded into the same 3D hippocampus model, enabling neuron-type specific potential circuit analysis.
The shared reconstructions utilized in this application came from multiple primary publications spanning a broad range of topics. Several papers reported morphological alterations due to aging (Pyapali and Turner 1996), toxic lesions (Pyapali and Turner 1994), and postnatal development (Rihn and Claiborne 1990; also used in the growth model discussed at the beginning of this section); in all of these cases only the control groups were re-used. Other articles employed reconstructions to quantify the spatial distribution of inhibitory and excitatory synapses on pyramidal neurons (Megias and others 2001) and the laminar branching patterns of dendrites (Ishizuka and others 1995). Additional reports studied dendritic voltage attenuation in vitro (Golding and others 2005) or firing patterns in vivo (Turner and others 1995). A detailed analysis quantitatively compared the morphologies reconstructed from in vivo vs. in vitro labeling (Pyapali and others 1998).
Two primary publications contributing to this shared dataset described digital reconstructions in a neuroinformatics context: to test and demonstrate the then-new ImageJ digital tracing plug-in Neuron_Morpho (Brown and others 2005), and to introduce a pioneering online repository of neuronal morphology in the rat hippocampus (Cannon and others 1998). Many of the primary discoveries enabled by the digital morphologies later downloaded to map the potential connectivity in the hippocampal circuit were in the computational domain. Two studies simulated the passive properties of reconstructed CA3 interneurons (Chitwood and others 1999) and pyramidal cells (Henze and others 1996) to determine the efficacy of signal transfer from dendrites to soma. Similar approaches compared the electrical signaling of a large number of neurons via a novel algorithm transforming the neuromorphological space into electrotonic space (Carnevale and others 1997), and investigated the combined effect of biophysical properties and dendritic structure on individual post-synaptic potentials (Jaffe and Carnevale 1999).
In fact, the majority of the secondary discoveries relying on data downloaded from NeuroMorpho.Org also employ computational approaches that simulate biophysically realistic membrane dynamics, often revealing emergent properties that can be further tested. We therefore selected the third example of applications of shared neuronal morphologies from this realm. This study elucidated the functional consequences of discrete transitions between open and closed states of ion channels on membrane voltage in a biophysically and anatomically realistic computational model (Cannon and others 2010). With stochastic ion channels distributed in a variety of arbor morphologies from different brain regions obtained through NeuroMorpho.Org, simulations demonstrated that dendritic spike initiation may be a probabilistic (rather than deterministic) function of patterns of synaptic input. This effect differed both among neuron types and within the dendritic arbor, as the voltage fluctuation increased with the distance from the soma (Figure 6C). The shared reconstructions downloaded for this study had been previously collected to quantify excitatory and inhibitory inputs onto specific cell types (Gulyas and others 1999), investigate voltage attenuation in dendrites (Stuart and Spruston 1998) or demonstrate an online database on registered neuronal microscopic images and reconstructions (Martone and others 2003). Many of the datasets re-used in this work have already been described above (Rihn and Claiborne 1990, Pyapali and others 1998, Megias and others 2001, Brown and others 2005, Golding and others 2005, Shepherd and Svoboda 2005), as they had also been downloaded for different examples of secondary discovery. This many-to-many relationship between a primary publication contributing to several secondary discoveries, and each in turn utilizing multiple primary sources, is the norm rather than the exception, once more highlighting the potentially combinatorial benefit of data sharing.
Concluding remarks and implications for future big data analyses
The quantitative investigations of axonal and dendritic arbors reviewed here yielded primary discoveries in a wide range of topics, including development, structure, function, and pathology. Similarly, the few highlighted examples of secondary discoveries spanned broadly from the biophysics of signal integration and propagation to spatial occupancy and potential circuit connectivity; and from the effect of structural diversity on neuronal excitability to the basic principles that could explain axonal and dendritic growth. The interest on neuronal architecture pervades most of the sub-disciplines and research communities in cellular and systems neuroscience. Digital reconstructions transformed the characterization of neuronal morphology from largely qualitative and descriptive to rigorously quantitative and simulation-ready. Moreover, these data are exquisitely amenable to sharing, and many new and important results have already been obtained by re-using shared reconstructions. Both primary use and secondary re-use of neuronal reconstructions typically involve quantitative measures of dendritic or axonal arbors. Nevertheless, the selected studies reviewed here illustrate the general observation that the original use of the reconstructions typically differs considerably from the goals upon re-use. Moreover, the extent of data re-use appears to be limited by the amount of available data rather than its first intended purpose. Thus, independent of the origin or initial aim for digital reconstruction, if the resulting neuronal morphologies are publicly shared, their scientific impact is likely to continuously grow for many years.
Although some research designs require custom-reconstructions of axonal or dendritic arbors in specific conditions, many investigations can be carried out seamlessly with morphologies digitally reconstructed by one’s own lab or re-use existing available data from previous studies. While not often recognized, there are also certain large-scale or comparative analyses that are impractical or impossible to perform without community-wide sharing, because they require a substantial critical mass that is beyond the means of individual researchers (Costa and others 2010). At the same time, rapid technical advancements in all the components of the pipeline to generate digital reconstructions (labeling, imaging, and tracing) are broadly expected to trigger an imminent deluge of information. Data curation and management will then become even more necessary to maximize the opportunity for scientific discovery.
The culture of data sharing is not yet as strong in neuroscience as in more mature disciplines such as molecular biology (gene expression, protein structure, etc.) and physics (e.g. high-energy particles and astronomy). Nevertheless, other neuroscience domains in addition to neuronal morphology are also advancing in the same direction, as evident with gene expression patterns (Wan and Pavlidis 2007) and non-invasive brain imaging (Poldrack and others 2013). In addition to facilitating free access to large datasets, centrally stored and curated repositories such as NeuroMorpho.Org also help formulate standards for metadata annotation, information reporting, consistent terminologies, and ontology engineering for smarter searches. As more evidence for the powerful research applications of shared reconstructions continues to accumulate, the scientific recognition of primary studies will progressively shift from their original reported discoveries to the expanded impact of the potentially many more secondary discoveries subsequently enabled by the same data.
Acknowledgments
The authors are grateful to Dr. Deepak Ropireddy for Figure 6B, and to Hoang Nguyen, Juhi Saxena and Apoorva Veerareddy for assistance in literature mining. This work is supported by NIH R01 NS39600 from NINDS to GAA, ONR MURI 14101-0198, Keck NAKFI IB1 , and Burroughs-Wellcome.
References
- Arkin MS, Miller RF. Mudpuppy retinal ganglion cell morphology revealed by an HRP impregnation technique which provides Golgi-like staining. Journal of Comparative Neurology. 1988;270:185–208. doi: 10.1002/cne.902700204. [DOI] [PubMed] [Google Scholar]
- Ascoli GA. Successes and rewards in sharing digital reconstructions of neuronal morphology. Neuroinformatics. 2007;5:154–160. doi: 10.1007/s12021-007-0010-7. [DOI] [PubMed] [Google Scholar]
- Bailey CDC, Alves NC, Nashmi R, De Biasi M, Lambe EK. Nicotinic α5 subunits drive developmental changes in the activation and morphology of prefrontal cortex layer VI neurons. Biological Psychiatry. 2012;71:120–128. doi: 10.1016/j.biopsych.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltz BSB, Kravitz EAE. Physiological identification, morphological analysis, and development of identified serotonin-proctolin containing neurons in the lobster ventral nerve cord. Journal of Neuroscience. 1987;7:533–546. doi: 10.1523/JNEUROSCI.07-02-00533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom HC, McDonald CG, French HT, Smith RF. Continuous nicotine administration produces selective, age-dependent structural alteration of pyramidal neurons from prelimbic cortex. Synapse. 2007;62:31–39. doi: 10.1002/syn.20467. [DOI] [PubMed] [Google Scholar]
- Bloomfield SA, Miller RF. A functional organization of ON and OFF pathways in the rabbit retina. Journal of Neuroscience. 1986;6:1–13. doi: 10.1523/JNEUROSCI.06-01-00001.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman BSB, Cruce WLW. Normal dendritic morphology of frog spinal motoneurons: a Golgi study. Journal of Comparative Neurology. 1980;193:1035–1045. doi: 10.1002/cne.901930415. [DOI] [PubMed] [Google Scholar]
- Brown KM, Donohue DE, D'Alessandro G, Ascoli GA. A cross-platform freeware tool for digital reconstruction of neuronal arborizations from image stacks. Neuroinformatics. 2005;3:343–360. doi: 10.1385/NI:3:4:343. [DOI] [PubMed] [Google Scholar]
- Burman K, Darian-Smith C, Darian-Smith I. Geometry of rubrospinal, rubroolivary, and local circuit neurons in the macaque red nucleus. Journal of Comparative Neurology. 2000;423:197–219. [PubMed] [Google Scholar]
- Cannon RC, O'Donnell C, Nolan MF. Stochastic ion channel gating in dendritic neurons: morphology dependence and probabilistic synaptic activation of dendritic spikes. PLoS Computational Biology. 2010;6(8):e1000886. doi: 10.1371/journal.pcbi.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon RC, Turner DA, Pyapali GK, Wheal HV. An on-line archive of reconstructed hippocampal neurons. Journal of Neuroscience Methods. 1998;84:49–54. doi: 10.1016/s0165-0270(98)00091-0. [DOI] [PubMed] [Google Scholar]
- Carnevale NT, Tsai KY, Claiborne BJ, Brown TH. Comparative electrotonic analysis of three classes of rat hippocampal neurons. Journal of Neurophysiology. 1997;78:703–720. doi: 10.1152/jn.1997.78.2.703. [DOI] [PubMed] [Google Scholar]
- Chiang A-S, Lin C-Y, Chuang C-C, Chang H-M, Hsieh C-H, Yeh C-W, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Current Biology. 2011;21:1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- Chitwood RA, Hubbard A, Jaffe DB. Passive electrotonic properties of rat hippocampal CA3 interneurones. The Journal of Physiology. 1999;515(Pt 3):743–756. doi: 10.1111/j.1469-7793.1999.743ab.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras D, Destexhe A, Steriade M. Intracellular and computational characterization of the intracortical inhibitory control of synchronized thalamic inputs in vivo. Journal of Neurophysiology. 1997;78:335–350. doi: 10.1152/jn.1997.78.1.335. [DOI] [PubMed] [Google Scholar]
- Costa LDF, Zawadzki K, Miazaki M, Viana MP, Taraskin SN. Unveiling the neuromorphological space. Frontiers in Computational Neuroscience. 2010;4:1–13. doi: 10.3389/fncom.2010.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Borst A, Hausser M. One rule to grow them all: a general theory of neuronal branching and its practical application. PLoS Computational Biology. 2010;6(8):e1000877. doi: 10.1371/journal.pcbi.1000877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuntz H, Forstner F, Borst A, Hausser M. The TREES toolbox--probing the basis of axonal and dendritic branching. Neuroinformatics. 2011;9(1):91–6. doi: 10.1007/s12021-010-9093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Wearne SL, Morrison JH, Hof PR. Quantitative analysis of the dendritic morphology of corticocortical projection neurons in the macaque monkey association cortex. Neuroscience. 2002;114:349–359. doi: 10.1016/s0306-4522(02)00305-6. [DOI] [PubMed] [Google Scholar]
- Eberhard J, Wanner A, Wittum G. NeuGen: A tool for the generation of realistic morphology of cortical neurons and neural networks in 3D. Neurocomputing. 2006;70:1327–1342. [Google Scholar]
- Ehlinger DG, Bergstrom HC, McDonald CG, Smith RF. Nicotine-induced dendritic remodeling in the insular cortex. Neuroscience Letters. 2012;516:89–93. doi: 10.1016/j.neulet.2012.03.064. [DOI] [PubMed] [Google Scholar]
- Finsterwald C, Fiumelli H, Cardinaux J-R, Martin J-L. Regulation of dendritic development by BDNF requires activation of CRTC1 by glutamate. Journal of Biological Chemistry. 2010;285:28587–28595. doi: 10.1074/jbc.M110.125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtak SC, Moyer JRJ, Brown TH. Morphology and ontogeny of rat perirhinal cortical neurons. Journal of Comparative Neurology. 2007;505:493–510. doi: 10.1002/cne.21516. [DOI] [PubMed] [Google Scholar]
- Gao Y, Perkins EM, Clarkson YL, Tobia S, Lyndon AR, Jackson M, et al. β-III spectrin is critical for development of purkinje cell dendritic tree and spine morphogenesis. Journal of Neuroscience. 2011;31:16581–16590. doi: 10.1523/JNEUROSCI.3332-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding NL, Mickus TJ, Katz Y, Kath WL, Spruston N. Factors mediating powerful voltage attenuation along CA1 pyramidal neuron dendrites. The Journal of Physiology. 2005;568:69–82. doi: 10.1113/jphysiol.2005.086793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Bellido PT, Peng H, Yang J, Georgopoulos AP, Olberg RM. Eight pairs of descending visual neurons in the dragonfly give wing motor centers accurate population vector of prey direction. Proceedings of the National Academy of Sciences. 2013;110:696–701. doi: 10.1073/pnas.1210489109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande G, Bui TV, Rose PK. Distribution of vestibulospinal contacts on the dendrites of ipsilateral splenius motoneurons: an anatomical substrate for push-pull interactions during vestibulocollic reflexes. Brain Research. 2010;1333:9–27. doi: 10.1016/j.brainres.2010.03.065. [DOI] [PubMed] [Google Scholar]
- Gulyas AI, Megias M, Emri Z, Freund TF. Total number and ratio of excitatory and inhibitory synapses converging onto single interneurons of different types in the CA1 area of the rat hippocampus. Journal of Neuroscience. 1999;19:10082–10097. doi: 10.1523/JNEUROSCI.19-22-10082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halavi M, Hamilton KA, Parekh R, Ascoli GA. Digital Reconstructions of Neuronal Morphology: Three Decades of Research Trends. Frontiers in Neuroscience. 2012;6:1–11. doi: 10.3389/fnins.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halavi M, Polavaram S, Donohue DE, Hamilton G, Hoyt J, Smith KP, et al. NeuroMorpho.Org implementation of digital neuroscience: dense coverage and integration with the NIF. Neuroinformatics. 2008;6:241–252. doi: 10.1007/s12021-008-9030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay E, Hill S, Schürmann F, Markram H, Segev I. Models of neocortical layer 5b pyramidal cells capturing a wide range of dendritic and perisomatic active properties. PLoS Computational Biology. 2011;7:e1002107. doi: 10.1371/journal.pcbi.1002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmeke C, Seidel K, Poeggel G, Bredy TW, Abraham A, Braun K. Paternal deprivation during infancy results in dendrite- and time-specific changes of dendritic development and spine formation in the orbitofrontal cortex of the biparental rodent Octodon degus. Neuroscience. 2009;163:790–798. doi: 10.1016/j.neuroscience.2009.07.008. [DOI] [PubMed] [Google Scholar]
- Henze DA, Cameron WE, Barrionuevo G. Dendritic morphology and its effects on the amplitude and rise-time of synaptic signals in hippocampal CA3 pyramidal cells. Journal of Comparative Neurology. 1996;369:331–344. doi: 10.1002/(SICI)1096-9861(19960603)369:3<331::AID-CNE1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hirsch JA, Martinez LM, Alonso J-M, Desai K, Pillai C, Pierre C. Synaptic physiology of the flow of information in the cat's visual cortex in vivo. The Journal of Physiology. 2002;540:335–350. doi: 10.1113/jphysiol.2001.012777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman EJ, Mintz CD, Wang S, McNickle DG, Salton SRJ, Benson DL. Effects of ethanol on axon outgrowth and branching in developing rat cortical neurons. Neuroscience. 2008;157:556–565. doi: 10.1016/j.neuroscience.2008.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houzel JC, Milleret C, Innocenti G. Morphology of callosal axons interconnecting areas 17 and 18 of the cat. European Journal of Neuroscience. 1994;6:898–917. doi: 10.1111/j.1460-9568.1994.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Ishizuka N, Cowan WM, Amaral DG. A quantitative analysis of the dendritic organization of pyramidal cells in the rat hippocampus. Journal of Comparative Neurology. 1995;362:17–45. doi: 10.1002/cne.903620103. [DOI] [PubMed] [Google Scholar]
- Jacobs B, Johnson NL, Wahl D, Schall M, Maseko BC, Lewandowski A, Raghanti MA, Wicinski B, Butti C, Hopkins WD, Bertelsen MF, Walsh T, Roberts JR, Reep RL, Hof PR, Sherwood CC, Manger PR. Comparative neuronal morphology of the cerebellar cortex in afrotherians, carnivores, cetartiodactyls, and primates. Frontiers in Neuroanatomy. 2014;8(24) doi: 10.3389/fnana.2014.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs B, Lubs J, Hannan M, Anderson K, Butti C, Sherwood CC, et al. Neuronal morphology in the African elephant (Loxodonta africana) neocortex. Brain Struct Funct. 2011;215:273–298. doi: 10.1007/s00429-010-0288-3. [DOI] [PubMed] [Google Scholar]
- Jaffe DB, Carnevale NT. Passive normalization of synaptic integration influenced by dendritic architecture. Journal of Neurophysiology. 1999;82:3268–3285. doi: 10.1152/jn.1999.82.6.3268. [DOI] [PubMed] [Google Scholar]
- Kisvarday ZF, Eysel UT. Cellular organization of reciprocal patchy networks in layer III of cat visual cortex (area 17) Neuroscience. 1992;46:275–286. doi: 10.1016/0306-4522(92)90050-c. [DOI] [PubMed] [Google Scholar]
- Koene RA, Tijms B, van Hees P, Postma F, de Ridder A, Ramakers GJA, et al. NETMORPH: a framework for the stochastic generation of large scale neuronal networks with realistic neuron morphologies. Neuroinformatics. 2009;7:195–210. doi: 10.1007/s12021-009-9052-3. [DOI] [PubMed] [Google Scholar]
- Kvello P, Løfaldli BB, Rybak J, Menzel R, Mustaparta H. Digital, Three-dimensional Average Shaped Atlas of the Heliothis Virescens Brain with Integrated Gustatory and Olfactory Neurons. Frontiers in Systems Neuroscience. 2009;3:14. doi: 10.3389/neuro.06.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-CA, Huang H, Feng G, Sanes JR, Brown EN, So PT, et al. Dynamic remodeling of dendritic arbors in GABAergic interneurons of adult visual cortex. PloS Biology. 2006;4:e29. doi: 10.1371/journal.pbio.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Jhaveri S, Frank E. Anatomical basis of specific connections between sensory axons and motor neurons in the brachial spinal cord of the bullfrog. Journal of Neuroscience. 1984;4:1754–1763. doi: 10.1523/JNEUROSCI.04-07-01754.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston C, Miller MM, Goldwater DS, Radley JJ, Rocher AB, Hof PR, et al. Stress-induced alterations in prefrontal cortical dendritic morphology predict selective impairments in perceptual attentional set-shifting. Journal of Neuroscience. 2006;26:7870–7874. doi: 10.1523/JNEUROSCI.1184-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd SA, Wensley B, Faherty CJ, Smeyne RJ. Regional differences in cortical dendrite morphology following in utero exposure to cocaine. Developmental Brain Research. 2003;147:59–66. doi: 10.1016/j.devbrainres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Martone ME, Zhang S, Gupta A, Qian X, He H, Price DL, et al. The cell-centered database: a database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics. 2003;1:379–395. doi: 10.1385/NI:1:4:379. [DOI] [PubMed] [Google Scholar]
- Marx M, Feldmeyer D. Morphology and Physiology of Excitatory Neurons in Layer 6b of the Somatosensory Rat Barrel Cortex. Cerebral Cortex. 2012;23(12):2803–17. doi: 10.1093/cercor/bhs254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megias M, Emri Z, Freund TF, Gulyas AI. Total number and distribution of inhibitory and excitatory synapses on hippocampal CA1 pyramidal cells. Neuroscience. 2001;102:527–540. doi: 10.1016/s0306-4522(00)00496-6. [DOI] [PubMed] [Google Scholar]
- Meijering E. Neuron tracing in perspective. Cytometry A. 2010;77:693–704. doi: 10.1002/cyto.a.20895. [DOI] [PubMed] [Google Scholar]
- Multani P, Myers RH, Blume HW, Schomer DL, Sotrel A. Neocortical dendritic pathology in human partial epilepsy: a quantitative Golgi study. Epilepsia. 1994;35:728–736. doi: 10.1111/j.1528-1157.1994.tb02503.x. [DOI] [PubMed] [Google Scholar]
- Niblock MM, Brunso-Bechtold JK, Riddle DR. Insulin-like growth factor I stimulates dendritic growth in primary somatosensory cortex. Journal of Neuroscience. 2000;20:4165–4176. doi: 10.1523/JNEUROSCI.20-11-04165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordman JC, Kabbani N. An interaction between α7 nicotinic receptors and a G-protein pathway complex regulates neurite growth in neural cells. Journal of Cell Science. 2012;125:5502–5513. doi: 10.1242/jcs.110379. [DOI] [PubMed] [Google Scholar]
- Nunez-Abades PA, He F, Barrionuevo G, Cameron WE. Morphology of developing rat genioglossal motoneurons studied in vitro: changes in length, branching pattern, and spatial distribution of dendrites. Journal of Comparative Neurology. 1994;339:401–420. doi: 10.1002/cne.903390308. [DOI] [PubMed] [Google Scholar]
- Oberlaender M, Boudewijns ZSRM, Kleele T, Mansvelder HD, Sakmann B, de Kock CPJ. Three-dimensional axon morphologies of individual layer 5 neurons indicate cell type-specific intracortical pathways for whisker motion and touch. Proceedings of the National Academy of Sciences. 2011;108:4188–4193. doi: 10.1073/pnas.1100647108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver DL, Kuwada S, Yin TC, Haberly LB, Henkel CK. Dendritic and axonal morphology of HRP-injected neurons in the inferior colliculus of the cat. Journal of Comparative Neurology. 1991;303:75–100. doi: 10.1002/cne.903030108. [DOI] [PubMed] [Google Scholar]
- Palmer AR, Wallace MN, Arnott RH, Shackleton TM. Morphology of physiologically characterised ventral cochlear nucleus stellate cells. Experimental Brain Research. 2003;153:418– 426. doi: 10.1007/s00221-003-1602-6. [DOI] [PubMed] [Google Scholar]
- Parekh R, Ascoli GA. Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron. 2013;77:1017–1038. doi: 10.1016/j.neuron.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent M, Parent A. Single-axon tracing and three-dimensional reconstruction of centre median-parafascicular thalamic neurons in primates. Journal of Comparative Neurology. 2005;481:127–144. doi: 10.1002/cne.20348. [DOI] [PubMed] [Google Scholar]
- Perez-Cruz C, Nolte MW, van Gaalen MM, Rustay NR, Termont A, Tanghe A, et al. Reduced spine density in specific regions of CA1 pyramidal neurons in two transgenic mouse models of Alzheimer's disease. Journal of Neuroscience. 2011;31:3926–3934. doi: 10.1523/JNEUROSCI.6142-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Barch DM, Mitchell JP, Wager TD, Wagner AD, Devlin JT, et al. Toward open sharing of task-based fMRI data: the OpenfMRI project. Front Neuroinform. 2013;7:12. doi: 10.3389/fninf.2013.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyapali GK, Sik A, Penttonen M, Buzsaki G, Turner DA. Dendritic properties of hippocampal CA1 pyramidal neurons in the rat: intracellular staining in vivo and in vitro. Journal of Comparative Neurology. 1998;391:335–352. doi: 10.1002/(sici)1096-9861(19980216)391:3<335::aid-cne4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Pyapali GK, Turner DA. Denervation-induced dendritic alterations in CA1 pyramidal cells following kainic acid hippocampal lesions in rats. Brain Research. 1994;652:279–290. doi: 10.1016/0006-8993(94)90238-0. [DOI] [PubMed] [Google Scholar]
- Pyapali GK, Turner DA. Increased dendritic extent in hippocampal CA1 neurons from aged F344 rats. Neurobiology of Aging. 1996;17:601–611. doi: 10.1016/0197-4580(96)00034-6. [DOI] [PubMed] [Google Scholar]
- Rihn LL, Claiborne BJ. Dendritic growth and regression in rat dentate granule cells during late postnatal development. Developmental Brain Research. 1990;54:115–124. doi: 10.1016/0165-3806(90)90071-6. [DOI] [PubMed] [Google Scholar]
- Ropireddy D, Ascoli GA. Potential Synaptic Connectivity of Different Neurons onto Pyramidal Cells in a 3D Reconstruction of the Rat Hippocampus. Frontiers in Neuroinformatics. 2011;5:1–13. doi: 10.3389/fninf.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropireddy D, Bachus SE, Ascoli GA. Non-homogeneous stereological properties of the rat hippocampus from high-resolution 3D serial reconstruction of thin histological sections. Neuroscience. 2012;205:91–111. doi: 10.1016/j.neuroscience.2011.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropireddy D, Scorcioni R, Lasher B, Buzsáki G, Ascoli GA. Axonal morphometry of hippocampal pyramidal neurons semi-automatically reconstructed after in vivo labeling in different CA3 locations. Brain Structure and Function. 2011;216:1–15. doi: 10.1007/s00429-010-0291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak J, Kuβ A, Lamecker H, Zachow S, Hege H-C, Lienhard M, et al. The Digital Bee Brain: Integrating and Managing Neurons in a Common 3D Reference System. Frontiers in Systems Neuroscience. 2010;4 doi: 10.3389/fnsys.2010.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago LF, Rocha EG, Santos CLA, Pereira AJ, Franca JG, Picanco-Diniz CW. S1 to S2 hind- and forelimb projections in the agouti somatosensory cortex: axon fragments morphological analysis. Journal of Chemical Neuroanatomy. 2010;40:339–345. doi: 10.1016/j.jchemneu.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Schubert D, Kotter R, Luhmann HJ, Staiger JF. Morphology, electrophysiology and functional input connectivity of pyramidal neurons characterizes a genuine layer Va in the primary somatosensory cortex. Cerebral Cortex. 2006;16:223–236. doi: 10.1093/cercor/bhi100. [DOI] [PubMed] [Google Scholar]
- Shepherd GMG, Svoboda K. Laminar and columnar organization of ascending excitatory projections to layer 2/3 pyramidal neurons in rat barrel cortex. Journal of Neuroscience. 2005;25:5670–5679. doi: 10.1523/JNEUROSCI.1173-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotrel A, Williams RS, Kaufmann WE, Myers RH. Evidence for neuronal degeneration and dendritic plasticity in cortical pyramidal neurons of Huntington's disease: a quantitative Golgi study. Neurology. 1993;43:2088–2096. doi: 10.1212/wnl.43.10.2088. [DOI] [PubMed] [Google Scholar]
- Staiger JF, Flagmeyer I, Schubert D, Zilles K, Kotter R, Luhmann HJ. Functional diversity of layer IV spiny neurons in rat somatosensory cortex: quantitative morphology of electrophysiologically characterized and biocytin labeled cells. Cerebral Cortex. 2004;14:690–701. doi: 10.1093/cercor/bhh029. [DOI] [PubMed] [Google Scholar]
- Stephens B, Mueller AJ, Shering AF, Hood SH, Taggart P, Arbuthnott GW, et al. Evidence of a breakdown of corticostriatal connections in Parkinson's disease. Neuroscience. 2005;132:741–754. doi: 10.1016/j.neuroscience.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Stuart G, Spruston N. Determinants of voltage attenuation in neocortical pyramidal neuron dendrites. Journal of Neuroscience. 1998;18:3501–3510. doi: 10.1523/JNEUROSCI.18-10-03501.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter CM, Stevens CF. A general principle of neuronal arbor branch diversity. Current Biology. 2011;21(24):2105–8. doi: 10.1016/j.cub.2011.11.013. [DOI] [PubMed] [Google Scholar]
- Toris CB, Eiesland JL, Miller RF. Morphology of ganglion cells in the neotenous tiger salamander retina. Journal of Comparative Neurology. 1995;352:535–559. doi: 10.1002/cne.903520405. [DOI] [PubMed] [Google Scholar]
- Turner DA, Li XG, Pyapali GK, Ylinen A, Buzsaki G. Morphometric and electrical properties of reconstructed hippocampal CA3 neurons recorded in vivo. Journal of Comparative Neurology. 1995;356:580–594. doi: 10.1002/cne.903560408. [DOI] [PubMed] [Google Scholar]
- van Ooyen A. Using theoretical models to analyse neural development. Nature Reviews Neuroscience. 2011;12:311–326. doi: 10.1038/nrn3031. [DOI] [PubMed] [Google Scholar]
- van Pelt J, Verwer RW. The exact probabilities of branching patterns under terminal and segmental growth hypotheses. Bulletin of Mathematical Biology. 1983;45:269–285. doi: 10.1007/BF02462361. [DOI] [PubMed] [Google Scholar]
- Vetter P, Roth A, Hausser M. Propagation of action potentials in dendrites depends on dendritic morphology. Journal of Neurophysiology. 2001;85:926–937. doi: 10.1152/jn.2001.85.2.926. [DOI] [PubMed] [Google Scholar]
- Wan X, Pavlidis P. Sharing and Reusing Gene Expression Profiling Data in Neuroscience. Neuroinformatics. 2007;5:161–175. doi: 10.1007/s12021-007-0012-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gupta A, Toledo-Rodriguez M, Wu CZ, Markram H. Anatomical, physiological, molecular and circuit properties of nest basket cells in the developing somatosensory cortex. Cerebral Cortex. 2002;12:395–410. doi: 10.1093/cercor/12.4.395. [DOI] [PubMed] [Google Scholar]
- Watson KK, Jones TK, Allman JM. Dendritic architecture of the von Economo neurons. Neuroscience. 2006;141:1107–1112. doi: 10.1016/j.neuroscience.2006.04.084. [DOI] [PubMed] [Google Scholar]
- Wei H, Jundi el B, Homberg U, Stengl M. Implementation of pigment-dispersing factor-immunoreactive neurons in a standardized atlas of the brain of the cockroach Leucophaea maderae. Journal of Comparative Neurology. 2010;518:4113–4133. doi: 10.1002/cne.22471. [DOI] [PubMed] [Google Scholar]
- Zubler F, Douglas R. A framework for modeling the growth and development of neurons and networks. Frontiers in Computational Neuroscience. 2009;3:25. doi: 10.3389/neuro.10.025.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]