Abstract
We designed a new microfluidic device that uses pillars on the same order as the diameter of a cell (20 μm) to isolate and enrich rare cell samples from background. These cell-scale microstructures improve viability, trapping efficiency, and throughput while reducing pearl chaining. The area where cells trap on each pillar is small, such that only one or two cells trap while fluid flow carries away excess cells. We employed contactless dielectrophoresis in which a thin PDMS membrane separates the cell suspension from the electrodes, improving cell viability for off-chip collection and analysis. We compared viability and trapping efficiency of a highly aggressive Mouse Ovarian Surface Epithelial (MOSE) cell line in this 20 μm pillar device to measurements in an earlier device with the same layout but pillars of 100 μm diameter. We found that MOSE cells in the new device with 20 μm pillars had higher viability at 350 VRMS, 30 kHz, and 1.2 ml/h (control 77%, untrapped 71%, trapped 81%) than in the previous generation device (untrapped 47%, trapped 42%). The new device can trap up to 6 times more cells under the same conditions. Our new device can sort cells with a high flow rate of 2.2 ml/h and throughput of a few million cells per hour while maintaining a viable population of cells for off-chip analysis. By using the device to separate subpopulations of tumor cells while maintaining their viability at large sample sizes, this technology can be used in developing personalized treatments that target the most aggressive cancerous cells.
I. INTRODUCTION
The isolation of specific cells of interest from a heterogeneous suspension has the potential to advance our understanding of a wide range of diseases, especially in diagnosis and treatment of cancer.1–3 Typically, samples from primary tumor sites consist of tumor-associated cells and cancer cells with differences in diverse properties such as gene expression, size, phenotype, and tumorigenic potential. A pure sample of specific subpopulations could be used to develop an optimal individualized treatment regimen for patients by targeting the most aggressive tumor cells and preventing tumor repopulation. Established methods to purify cell samples are flow cytometric and magnetic sorting. However, many cancers, especially those originating in the epithelial tissues, are known to modulate the expression of surface proteins, making acquisition difficult due to changing surface antigens.4,5 In many cases, this issue compounds application of flow cytometric and magnetic sorting by requiring multiple expensive antibodies to target cells with sufficient accuracy and certainty. Stress caused to the cells via the use of several compounds may alter the behavior and phenotype of the cell. Further, intra- and intertumoral variations of gene expression levels due to the development of phenotypically similar but genetically distinct pathways could lead to these methods failing in variable proportions of the population, whereas testing for phenotypic properties could lead to a more robust enrichment. An alternative are microdevices that do not rely on binding of probes to specific surface molecules but use the biophysical properties of cells for separation. For example, cells can be sorted based on size, shape, deformability, density, optical and electrical properties.6–10
Dielectrophoresis (DEP) is a useful separation method that offers label-free isolation of cells based on geometrical and electrical properties of cells in microfluidic devices. In contrast to some capture-based techniques, the cells remain viable and can be taken off-chip for further analysis. Of special interest are cells typically found in small concentrations such as white blood cells, circulating tumor cells and tumor-initiating cells.1,11–14 To separate similar cells, the device must be sensitive enough to distinguish small differences in electrical properties between cells. An example of such separation is the isolation of tumor-initiating cells (TICs), also known as cancer stem cells. They are a finite population of cancer cells having the unique ability to recapitulate the original tumors and are putatively responsible for the metastatic properties of tumors.15 TICs are hard to detect and separate even with the most sensitive techniques due to genetic similarity with other tumor cells. Furthermore, methods that rely on specific surface markers for detection are hampered by the lack of a defined genetic fingerprint of target cells even from the same tissue;16 thus, a pure population of specific cells still cannot be isolated.
High output cell viability is crucial if the device is to be used for isolating live cell populations for further in vivo or in vitro mechanistic studies. In the case of rare cancer cells, improving the viability of the sub-populations allows for efficient screening of chemotherapy drugs or alternative treatments, thus supporting treatment decisions and improving a patient's treatment outcome on an individualized basis. This is an advantage over other technologies such as affinity-based sorting that can successfully trap target cells based on their gene expression but that can permanently block signaling events or trigger cellular responses that can damage the cells or change the biochemical properties of the target cell population, impeding further off-chip analysis.1 DEP has the potential to isolate cell populations with minimal stress exerted to the cells. In order to capitalize on the potential for DEP application, electromechanical damage to the cells such as electroporation needs to be minimized using optimized device design.
Several different designs of DEP separation devices have been developed,17–20 with the inhomogeneity of the electric field generating the DEP force achieved either by the shapes of the electrodes, referred to as classical DEP,21,22 or by inclusion of insulating structures into the otherwise largely uniform field, referred to as insulator DEP (iDEP)23,24 and contactless DEP (cDEP).25,26 Dielectrophoretic separation is based on bioelectrical cell properties and is independent of the cells' genotype. Classical DEP uses metal electrodes to create a non-uniform electric field; at the edges of the electrodes, the electric field density can be locally high, damaging the cells. On the other hand, the amplitude of the DEP force decreases significantly when the cells move away from the electrodes. Viability of mammalian cells in negative DEP devices, where the DEP force is pushing the cells away from electrodes, can be as high as 97%;27 however, to the authors' knowledge, no viability study has been published on trapping-based high-throughput DEP systems. Development of 3D electrodes has allowed for extended range of the DEP force and higher throughput20,28 at the cost of more complex fabrication. Alternatively, the DEP force can be generated by placing insulating structures to distort an otherwise uniform electric field. In iDEP devices, the electric field is applied along the microfluidic channel and insulating structures distort the electric field, creating trapping regions for cells.23 These structures, typically pillars, are fabricated within the base substrate containing the channel and traverse the entire channel depth, making them amendable for mass fabrication. The channels can be large for high-throughput cell sorting; however, the metal electrodes are in contact with cell suspension, which could lead to deleterious electrochemical effects.
In cDEP devices, electrodes are separated from the main channel by a thin insulating membrane; this negates electrochemical damage such as electrolysis and minimizes electroosmosis within the sample. The method of cDEP utilizes insulating pillars to distort the electric field and a thin insulating membrane separating the electrode from the cell suspension to allow for maximum density of the field in the channel while maintaining the electric field at a low enough magnitude to minimize electrical damage to the cells.25,26,29 In classical DEP, a gap between electrodes is typically in the range of 0.1 mm, while for iDEP and cDEP they are a few millimeters apart, necessitating a high-voltage AC signal source.
Dielectrophoretic sorting devices are typically designed assuming that the density of the cell suspension is low enough that cell-to-cell interactions can be neglected. The effective polarizability of cells in a chain is different, hence the DEP force is also different.30,31 These cell-to-cell interactions lead to higher heterogeneity in the trapped population and diminish the specificity of sorting, a critical aspect in sub-population establishment and in separation of similar populations such as TICs from tumor cells. Diluting the cell suspension may eliminate cell-to-cell interaction for continuous flow through DEP devices; however, it does not eliminate the interaction in DEP devices where insulating pillars are used to create the non-uniform field and trap cells in the areas of highest electric field density (insulating and contactless DEP). The force between induced dipoles (cells) contributes to the DEP force on a single cell level and pearl chaining of cells is difficult to avoid. Sorting of cells in typical iDEP and cDEP devices with 100 μm pillars is sensitive to the density of cells (number of cells per ml) as cell-to-cell interactions change probability of trapping.
Here, we describe a new approach to cDEP devices in which the insulating pillars needed to drive DEP are on the same scale as the diameter of a typical cell. This device improves the efficiency of trapping compared to previous devices due to the steeper drop in the electric field gradient, which is strong in an area about the size of one cell.32 The drag force around the pillar prevents accumulation of multiple cells on one pillar. The device specificity eliminates pearl chaining and clumping on the pillars, which leads to further improvements in the specificity of trapping by reducing the impact of cell-to-cell interactions on the DEP force. Therefore, we can also use a high density cell suspension. Due to the steep gradient, the gradient-to-electric field ratio is small such that the electric field in the device with small pillars is strong enough to trap cells but not damage them. The improved specificity and viability of the device output could support off-chip analysis of the enriched cell populations, such as specific genotype and drug response, and thereby aid individualized treatment decisions.
II. THEORY
DEP is the motion of polarized particles in a non-uniform electric field toward the high (positive DEP) or low (negative DEP) electric field gradients. The direction of the force depends on the particle's polarizability compared to the dielectric properties of the medium. The time-average DEP force on a spherical particle is described as
| (1) |
where is the permittivity of the suspending medium, r is the radius of the particle, and is the root mean square of the electric field.33 is the real part of the Clausius-Mossotti factor, , given by
| (2) |
where and are the complex electrical permittivities () of the particle and medium, respectively, is the relative electrical permittivity, is electrical conductivity, and is the angular frequency of the electric field. Equivalent complex permittivity of a cell can be calculated using a single shell model as
| (3) |
where d is the thickness of the cell membrane and and are the complex permittivities of the cytoplasm and membrane, respectively. The complex permittivity of the particle, , is a function of the frequency of the electric field, morphological, and electrical properties of the cell including the cytoskeleton, membrane morphology, and capacitance as well as cytoplasm conductivity. We compared the single-shell with the triple-shell model and found that internal structures such as the cell nucleus and endoplasmic reticulum have negligible effects on FDEP at frequencies below 100 kHz. Therefore, the single-shell model sufficiently describes a cell at the frequencies used in this study.
To induce a strong FDEP on a cell, it must be exposed to a strong electric field gradient. However, when the induced transmembrane voltage (ITV) is too high, pores form in the cell membrane, resulting in the process called electroporation.34 The induced transmembrane voltage for a spherical cell in suspension can be described with the following equation:35,36
| (4) |
where θ is a polar angle, measured from the center of the cell between the position on the membrane and the applied field direction and τ is the relaxation time of the cell membrane. To preserve the viability of cells, a device must distort the electric field to maximize and keep low enough to keep the cell membrane intact.
A cell follows the streamlines of the fluid flow until it approaches the solid structures (walls and pillars), collides with other cells or the FDEP changes its trajectory. Interactions of cells with these solid structures are challenging to describe, as cells have proteins in/on the membrane and cytoskeleton that determine the complex shape of the cell and its deformability. When the cell is freely flowing in the medium its motion is determined by the hydrodynamic frictional force,
| (5) |
where and are the velocities of the particle and medium, respectively, and is viscosity of the medium.
III. MATERIALS AND METHODS
A. Computational modeling
COMSOL Multiphysics V4.3 (Comsol, Inc., Burlington, MA) was used for the numerical computations. The physics modules used were COMSOL Electric Currents and Creeping Flow. All computations were made using a 3D geometry. As the viable cells in suspension are spherical, pillars are cylindrical in shape and the channel is 50 μm deep, therefore using a 2D approximation would not be valid. A section of the cDEP device that included all geometries of interest in a 5 × 5 pillar area was modeled and the variables around the central pillar were observed. Taking advantage of device symmetry in the z-axis, the modeled geometry was 25 μm high with a symmetry plane on the top to represent the full 50 μm deep channel. The diameter of the pillars varied from 20 μm to 100 μm, while the gap between pillars was held constant at 60 μm for all pillar sizes. When a cell was included in the model, it was positioned 1 μm away, centered in front of the pillar, as observed from the source of fluid flow. The voltage was applied perpendicularly to the fluid flow. The voltage-to-distance ratio between electrodes was kept constant at 300 V/cm for all pillar sizes with an applied frequency of 30 kHz. The planes on each side of the device were set to potential and ground, respectively, while all other boundaries were set to insulation. Inflow velocity of the fluid was set to 100 μm/s and outlet pressure to zero. The device was approximated as infinitely wide by applying symmetry to both sides of the channel. As mentioned previously, the boundary condition of the top plane was also symmetry, while the bottom plane and pillar surfaces were assigned a no-slip boundary condition. Values of the parameters used are given in Table I. Different geometric meshes in COMSOL were used for computations of electric field and fluid flow. For the electric field, the maximum element size was 3 μm. To model fluid flow, the maximum element size was 6 μm and the mesh elements close to any surface were further refined to accurately describe the fluid dynamics phenomena. To verify quality of computations, the meshes were refined and differences between computations of were less than 2% on 99.8% of the area shown in Fig. 3. Differences between computations of fluid velocity were less than 2% on 99.99% of the area. Variables were plotted on a virtual cut plane 12.5 μm from the floor of the device.
TABLE I.
Values of parameters used for computations.
| Description | Value | Source | |
|---|---|---|---|
| r | Cell radius | 8.5 × 10−6 m | Measured |
| d | Cell membrane thickness | 4 × 10−9 m | Ref. 37 |
| σe | Conductivity of medium | 0.01 S/m | Measured |
| εe | Permittivity of medium | 6.9 × 10−10 As/V m | Ref. 38 |
| σm | Conductivity of cell membrane | 3 × 10−7 S/m | Ref. 39 |
| εm | Permittivity of cell membrane | 6.2 × 10−11 As/V m | Ref. 40 |
| σi | Conductivity of cytoplasm | 0.5 S/m | Ref. 41 |
| εi | Permittivity of cytoplasm | 5.3 × 10−10 As/V m | Ref. 40 |
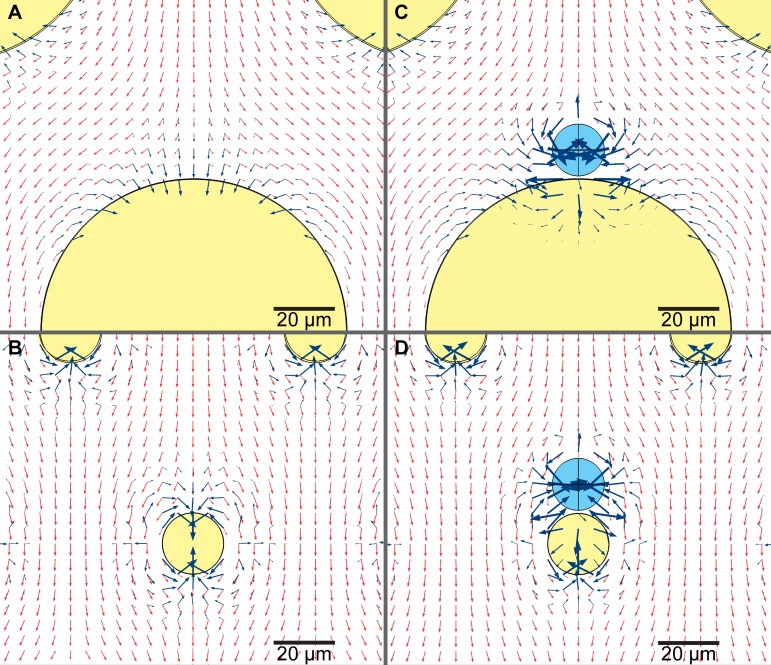
FIG. 3.
Gradient of electric field squared (dark blue arrows) and velocity of fluid flow uf (light red arrows) in a device with ((a) and (c)) 100 μm pillars and ((b) and (d)) 20 μm pillars, when no cells are present ((a) and (b)) and when one cell is already trapped ((c) and (d)). Insulating pillars are shaded with yellow and cells with light blue. Log values of both vectors are plotted however they are scaled arbitrarily (not to compare size of and uf) while the same scale is used in all panels. If a cell is already trapped in front of a pillar the next cells will be exposed to different forces. In front of a large 100 μm pillar there is enough space that cells can form a chain, while in front of a small 20 μm pillar the balance of DEP and drag force is different and cells are unlikely to form a chain.
B. Device fabrication
The mask design was drawn in AutoCAD (Autodesk, Inc., San Rafael, CA) and standard photolithography was used to pattern the mold. Deep reactive-ion etching (DRIE) was used to fabricate the main channel as described in detail in Shafiee et al.25 AZ 9260 (AZ Electronic Materials, Somerville, NJ) photoresist was spun onto a silicon wafer and soft-baked. The wafer was then exposed to UV light through a printed glass mask. The photoresist was then removed using developer AZ400K followed by another hard baking. DRIE was used to etch the silicon master stamp to 50 μm height. Surface roughness on the side walls was removed by 5 min wet etching with tetramethylammonium hydroxide. SU-8 was used to fabricate a master stamp for the electrode layer, as the aspect ratio is low and the features are relatively large. Following the manufacturers protocol, SU-8 2035 (MicroChem, Westborough, MA) was spun onto a silicon wafer 100 μm thick, soft-baked, patterned, and hard-baked. The photoresist was then removed using SU-8 developer and cleaned with isopropyl alcohol.
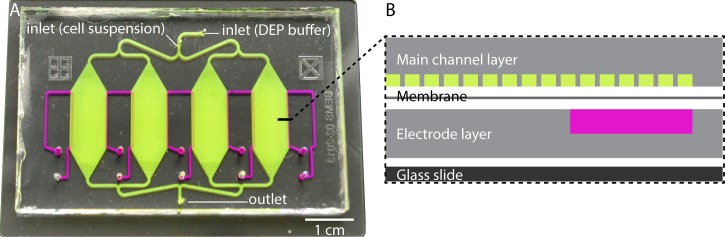
The liquid PDMS was made by mixing PDMS monomers and a curing agent in a 10:1 ratio (Sylgard 184, Dow Corning, Midland, MI). To fabricate the electrode and main channel layers, degassed PDMS liquid was poured onto the silicon master, cured for 45 minutes at 100 °C, and then peeled off from the silicon mold. The membrane was fabricated using a 5:1 ratio of PDMS by spinning PDMS on a blank silicon wafer at 500 rpm for 5 s followed by 4000 rpm for 45 s, resulting in a 13 μm thick membrane. The PDMS electrode layer was bonded to the membrane by exposing the surface to air plasma for 40 s (Harrick Plasma, Ithaca, NY) and then bonded to the main channel layer, sandwiching the membrane (Fig. 1). Both layers have patterns with alignment marks and were visually aligned before bonding. Fluidic connections to the channels were punched (Harris Uni-Core, Redding, CA), and the PDMS sandwich was bonded to the glass slide via exposure to plasma.
FIG. 1.
20 μm pillar contactless dielectrophoresis device. (a) Top view photo and (b) exploded schematic of a cross section across a row of pillars with white space added to visually separate layers. The main channel is colored green and contains inlets for both cell suspension and DEP buffer. Electrode channels are colored purple, they are filled with 10 × PBS and connected to the high voltage generator. An electric field is applied across each chamber, perpendicular to the direction of fluid flow.
Two device designs were used; the new design with 20 μm pillars (68 664 pillars in 4 chambers) was compared to a device with 100 μm pillars (16 412 pillars). Both had the same electrode layer, membrane, and outer dimensions of the 4 parallel chambers. The edge-to-edge distance between the pillars was also identical but as the diameter of the pillars was different, the number of pillars in the device was also different.
C. Cell preparation
The mouse ovarian surface epithelial (MOSE) cell model has been developed from C57BL/6 mice and represents the progressive stages of ovarian cancer as described.42,43 The MOSE-LFFLv used in this study is a highly aggressive cell type generated by intraperitoneal injection of tumorigenic MOSE cells (MOSE-L) into syngeneic mice and recovery of cells in the ascites; injection of MOSE-LFFLv results in rapidly developing tumors,44,45 therefore suggesting TIC character.46 For the purpose of this study, cells were cultured in DMEM (Sigma Aldrich, St. Louis, MO) supplemented with 4% of FBS (Atlanta Biologicals, Flowery Branch, GA) and 100 μg/ml each of penicillin and streptomycin (Life Technologies, Carlsbad, CA) at 37 °C in a humidified incubator with 5% CO2. Cells were trypsinized, labeled with Calcein AM (Life Technologies), centrifuged and washed in low-conductivity medium (DEP buffer). The buffer consisted of 8.5% sucrose [w/v], 0.3% glucose [w/v], 0.725% RPMI [v/v] (Life Technologies),47 modified with 0.1% of BSA [w/v] and 0.1% of Kolliphor P188 [w/v] (both Sigma Aldrich) as described48,49 and 0.1 mM EDTA (Boston Bio Products, Ashland, MA). Cells were centrifuged and resuspended in DEP buffer without BSA, Kolliphor and EDTA to a concentration of 2.5 × 106 cells/ml. The final conductivity of the cell suspension was between 110 and 125 μS/cm, measured by a conductivity meter (B-173, Horiba, Kyoto, Japan).
D. Experimental set-up
cDEP microfluidic devices as shown in Fig. 1 were kept under vacuum before experiments, then primed with DEP buffer. Electrode channels were filled with 10 × PBS. The voltage was delivered to the electrodes by combination of a function generator (U2761A, Agilent, now Keysight Technologies, Santa Rosa, CA) and high-voltage amplifier Trek 2205 (Trek, Medina, NY). Oscilloscope U2701A (Keysight Technologies) was used to monitor the voltage delivered. An inverted microscope (DMI6000B, Leica Microsystems, Bannockburn, IL) was used to observe flow of cells through the channels. Teflon tubing gauge 30 (Cole Palmer, Vernon Hills, IL) was used to connect one inlet of the cDEP device to a 1 ml syringe filled with DEP buffer and a second inlet to a syringe containing the cell suspension. A remote syringe pump 11 Elite Nanomite (Harvard Apparatus, Holliston, MA) was used to sequentially pump cells and DEP buffer through the device. The syringe was tilted 45°–60° from the horizontal plane in order to allow for uniform cell density while pumping cells into the device.
Sinusoidal voltage with a frequency of 30 kHz and amplitude of 250–350 VRMS was applied, and 50 μl of cell suspensions were pumped through the cDEP device at 20–36 μl/min. Part of the population was selectively trapped and held in the device while untrapped cells continued to flow through and were collected in the first Eppendorf tube. Next, the syringe containing cell suspension on the pump was switched to pure DEP buffer and 50 μl of buffer was pumped through the device at the same flow rate and voltage to wash the untrapped cells from the device. Finally, the voltage was turned off and trapped cells were released, washed out with 50 μl of DEP buffer and collected in the second tube. A sample of cells in each bin was mixed with equal volume of DMEM and with Propidium Iodide at a concentration of 20 μg/ml within 5–10 min after releasing from the device. These samples were then pipetted into a hemocytometer and analyzed for viability. The duration of one cell separation batch process was 4.2 and 7.5 min at 36 and 20 μl/min, respectively, with an additional minute for manually changing syringes on the pump and collection tubes.
IV. RESULTS AND DISCUSSION
Insulator DEP and contactless DEP both use insulating pillars to distort the electric field in order to trap cells of interest.23,25 Devices have been used to successfully sort cells with different electrical/geometrical properties, such as live and dead cells, but sorting cells with small variations in their electrical properties is challenging. Typically pillars that distort the electric field have a much larger diameter (100 μm) than cells (less than 20 μm) and the dielectrophoretic forces accumulate several cells around pillars.23,50 To address this issue, we have developed a new approach—pillars with diameter the size of a cell, which generate small trapping regions and minimize cell-to-cell interactions.
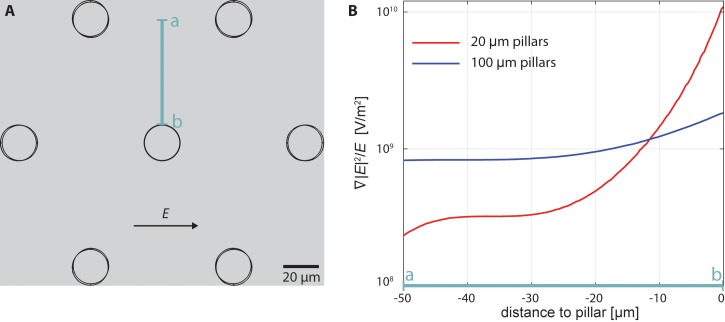
We modeled devices with either 20 or 100 μm pillars and applied the same voltage-to-distance ratio and the same velocity of the fluid flow. While the voltage to distance ratio is the same for both devices, the gradient of the electric field is steeper in the cell-scale-pillar device and this steep gradient is limited to close proximity of the pillar (Fig. 2). The steep gradient of the electric field is vital to keep a cell trapped near the post while a few tens of microns away the gradient is not high enough to accumulate more cells on each pillar. The gradient in front of the 100 μm pillars has small variation, which is beneficial in trapping cells, but it reduces specificity of trapping.
FIG. 2.
Cross-section of the ratio of the gradient electric field squared to electric field in front of a pillar. (a) Diagram of the pillar layout in the device. (b) The ratio in front of 20 μm and 100 μm pillars. At the same voltage to distance ratio, the gradient is much steeper in the 20 μm pillar device than in 100 μm pillar device, reducing accumulation of excessive cells at each pillar. Note that both designs have a 60 μm gap between pillars.
Furthermore, the direction of the gradient is pointing to the front center of the pillar (see Fig. 3), keeping the cell in a stable position just in front of the pillar. Arrow plots in the figure show directions of the gradient of electric field, , which is proportional to FDEP (blue arrows) and the velocity of fluid flow, uf, which is proportional to Fdrag (red arrows). Fluid flows tangentially to the surface and tends to move the cell further along the channel. In front of the 100 μm pillar, the gradient has a large normal component to the surface of the pillar attracting the cell, while the tangential component is in many situations too small to keep the cell trapped in the center. In the 20 μm pillar device, the tangential component is big enough to keep the cell trapped. Due to these factors, the electric field must be higher in the 100 μm pillar device than in the 20 μm one to trap a single cell.
We also modeled a scenario in which a cell is already trapped at the pillar and another cell approaches it and we compared the differences in DEP force in both devices. An approaching cell is exposed to different gradient of electric field when a cell is already trapped at the pillar. Especially in the 100 μm pillar device, the magnitude of the electric field gradient (and therefore the DEP force) increases tenfold (Figs. 3(c) and 3(d)). In the 100 μm pillar device, a cell flowing toward the pillar is added to the cells already trapped there and the cells form a chain. In the 20 μm pillar device, cells will be also attracted, though the drag force is pushing the cell forward, towards the side of the pillar. The DEP force is not strong enough to hold the cell on the side of the pillar, and therefore two possible scenarios typically occur: (a) the cell will not be trapped there, but will trap on next empty pillar or (b) the first cell will move so that both cells can be trapped centrally aligned one next to each other. If a third cell approaches the pillar with two cells already trapped, then in the 100 μm pillar device it will just trap next to the other cells. In the 20 μm pillar device that cell will be still attracted to the side of the other two cells, but it will not be trapped because the drag force is too strong. The advantage of the 20 μm pillar device is the balance of DEP and drag forces because there is simply not enough space to trap more than two cells next to each other in front of the pillar when using moderate intensity electric fields. The channel is 50 μm high and cells have a diameter of 12 μm, so they could potentially stack in the vertical (z-axis) direction. However, the cells sediment by the time they reach the trapping region. At 36 μl/min they travel from inlet to the trapping region in 4.4 s on average and at a sedimentation rate of 7.8 μm/s.51 Their vertical position is between 6 μm (average radius) and 10 μm from the floor of the device. Two particles with the same sign of the Clausius-Mossotti factor are attracted to each other when the pair is aligned along the electric field and they repel when the pair is aligned perpendicularly to the field.52,53 When two cells are close together in our device they tend to align with the electric field across the channel and form a pair in front of a pillar at the same vertical position. Theoretically, if the fluid flow brings two cells one on top of the other, DEP force pushes them apart and they do not form a vertical chain. We observed that this mechanism greatly reduces the probability of cell stacking. The computational tools used to model the DEP and drag forces are excellent for optimizing the design of the microfluidic device. However, the cytoskeleton and membrane proteins greatly determine cellular interactions, and some cells may even adhere to the pillars without an electric field present. Typically, these properties of cells are not well known and they can change with time, so including them in the model to predict cell sorting would be computationally demanding and it still would not predict some cellular interactions. By using models to optimize the design of the chip and validating these models with experimental data, it is possible to develop a device that is able to account for complex cellular interactions and biological features.
The membrane that separates the cell suspension from the electrodes is specific to cDEP. It prevents the deleterious effects of electrolysis. However, the electric field has to penetrate the thin membrane capacitively, causing these devices to have a low frequency limit, which mainly depends on material, thickness of the membrane and its surface.54 In single-layer devices with electric fields perpendicular to the direction of the fluid flow, the surface area of the membrane is limited by the height of the main channel. A solution to increase the membrane surface area is to use a multilayer approach and place the membrane below the main channel. The surface area of the membrane can be large enough to allow for the desired current to flow through main channel. Lower voltage on the electrodes can be used to achieve the same electric field and the low frequency limit can be sufficiently small. A multilayer design also has mechanical benefits. Single-layer cDEP devices have a thin vertical membrane, which needs to be bonded to a glass slide. Since the membrane is typically 50–200 μm tall, only 20 μm thick and has one side unattached, it may bend in the bonding process, resulting in a leakage. The membrane in the multilayer device has similar thickness as in the single layer devices. However, it spreads across the channel and is connected to all sides, so that a potential imperfection in bonding will not cause leakage. Multilayer devices have the area at the side of the main chamber on top of the electrodes where electric field is not sufficient for trapping of cells. If the device is 7.6 mm wide, two 0.5 mm wide bands over the electrode channels are relatively small. Therefore, a multilayer design is more suitable for wide cDEP devices and a single-layer design for narrow ones.
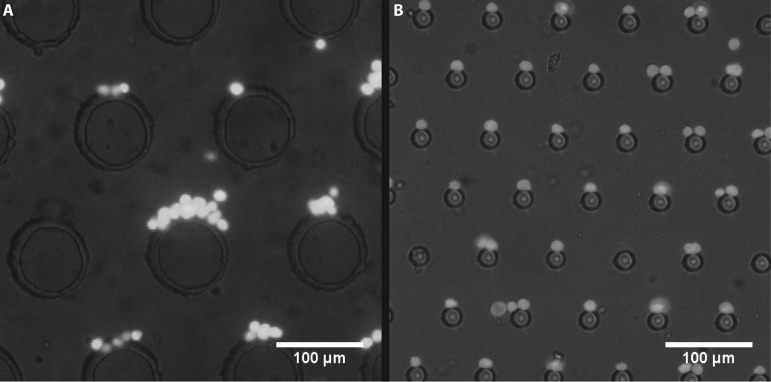
Based on computations and observations of real cells trapped on pillars, we designed a cDEP device with 20 μm diameter pillars—close to the diameter of typical mammalian cells. This allows cell trapping to be much closer to binary: cells can trap in the center of the pillar and stay there (up to 2 cells per pillar), or they do not trap and continue flowing through the device. The resulting device has about 17 000 insulating pillars in one channel with a 60 μm gap between them. The main channel is 12.3 mm long, 7.6 mm wide, and 50 μm high. We have developed a high-throughput batch-sorting cDEP device with 4 parallel channels and about 68 000 pillars, capable of trapping individual cells or pairs of cells on a pillar while maintaining high throughput (Fig. 1). For pillar size comparison, devices with 100 μm and 20 μm pillars were used. They had the same layout of the main channels and a common electrode layer design was used. Fig. 4 shows typical trapping in both devices at 30 kHz, 300 VRMS and 20 μl/min. Typically, either many cells or none trap at a 100 μm pillar, but there is rarely single cell trapping. A 20 μm pillar typically traps one or two cells and they are more uniformly distributed along the main chamber. Conditions for trapping cells in chains are different than trapping single cells. If two populations of cells only differ slightly in their electrical properties, then pearl chaining reduces the purity of sorted cells by introducing inhomogeneity into the trapping ranges. The design with 20 μm pillars reduces pearl chaining, because trapping regions are small and drag force of the fluid carries away the cells that could otherwise form a chain. By limiting trapping to one or two cells, the device is capable of distinguishing electrical properties of cells in order to trap, limiting the heterogeneity and potentially improving purity of sorted cells. In the computational model, we used cellular parameters measured from different mammalian cells reported in the literature (see Table I), as MOSE cells have not been fully characterized. The model predicted that cells would trap at the average inflow velocity of 100 μm/s, which corresponds to the flow rate of 6.8 μl/min in the actual device. The model allowed us to design a device and set the initial values of parameters for cell trapping, however, we were able to increase the flow rate and trap cells at flow rates from 20 to 36 μl/min.
FIG. 4.
Photo of trapped cells in cDEP devices. Direction of fluid flow is from top to bottom, while electric field is applied horizontally. (a) High density of cell suspension in a device with large (100 μm) pillars results in pearl-chaining and unspecific trapping. (b) In a device with small 20 μm pillars only one or two cells can trap on each pillar. When saturated, excessive cells will not accumulate, but continue to next available location.
Cells can be easily trapped at 100–1000 kHz; however, all types of MOSE cells have similar DEP response at those frequencies. The cells behave in a more distinct manner when the electric field is close to the crossover frequency, the frequency where the DEP force changes direction. The crossover frequency of MOSE-LFFLv cells is 11.9 kHz.55 At 30 kHz the theoretical model with single shell sphere (Eqs. (1)–(3)) shows that the value of the Clausius-Mossotti factor is 0.5. The model also predicts that 30 kHz may be optimal frequency for sorting of different types of MOSE cells, as differences in the Clausius-Mossotti factor are at a relative maximum just a few kHz above the crossover frequency, and the magnitude of the factor is still large enough to trap cells.
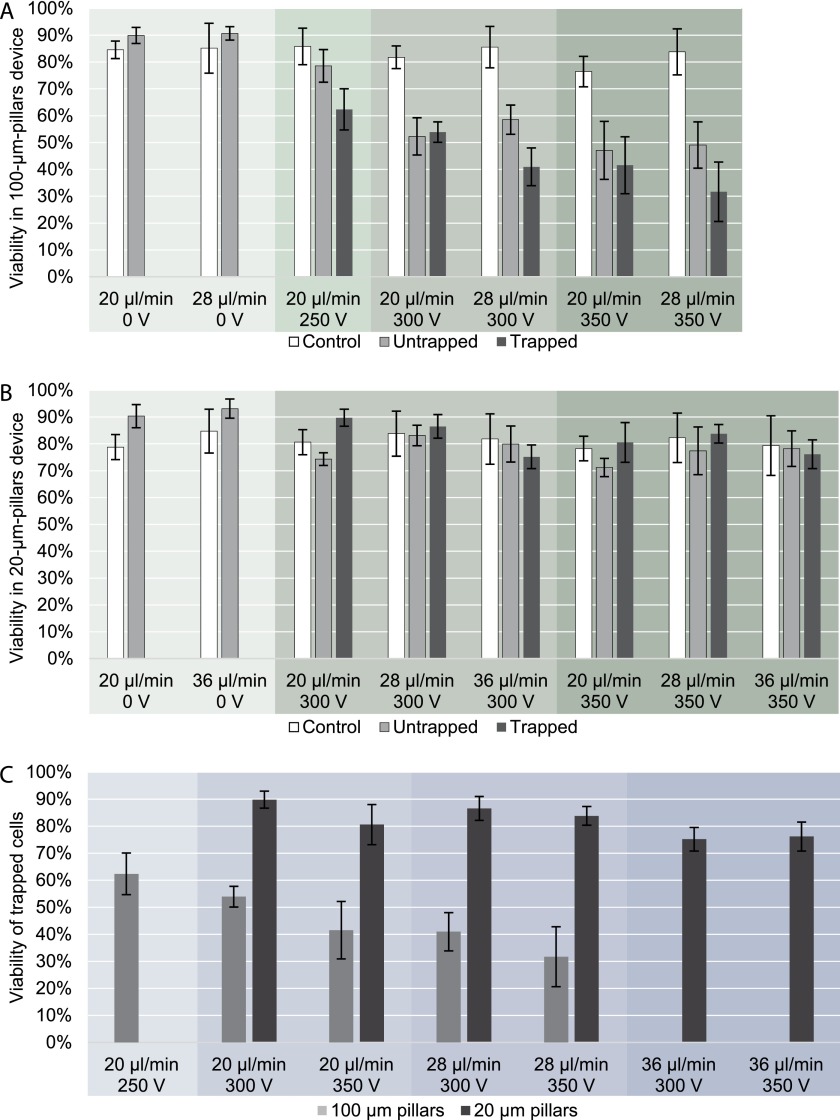
Easy release of trapped cells and preserved viability of cells is an important advantage of DEP cell sorting since the release of cells from affinity-based sorting devices can be challenging.1 To keep the cells alive, the suspension buffer must have physiological osmotic pressure and pH, shear stress must be minimized and electric field must be low enough not to electroporate cells. When no field is applied, the viability of the cells flowing through the device remains equal or higher than the control cells, which means that damage due to shear stress in the tubing, inlet and outlet channels is low. Dead cells can irreversibly adhere to the device walls before they reach the trapping region regardless of electric field. This can increase the apparent viability of untrapped and trapped cells in the output populations compared to the control cells. Viability studies of the target population of MOSE-L-FFLv cells show that small 20 μm pillars generate a sufficient gradient of electric field to trap cells, while the field itself is low enough not to damage them. Fig. 5(a) shows that viability decreases to 32% in the device with 100 μm pillars, while in the device with 20 μm pillars viability changes only slightly from the initial 78–85% to 71–90% (Fig. 5(b)). The viability of trapped cells also decreases with increased voltage and flow rate in both devices (Fig. 5(c)). A strong electric field can damage the cells; therefore it is important to keep it below the electroporation threshold. Viability of trapped cells in the 100 μm pillar device decreases with increasing voltage and/or flow rate; however, in much larger populations of untrapped cells viability is independent of flow rate. It is possible that the overall stress to cells slowly damages the cells so that their membrane is still a sufficient barrier to allow for strong polarization and trapping, but after few minutes it is damaged enough to allow propidium iodide to enter a cell. In the 20 μm pillar device such a trend is insignificant as the viability is practically the same between the control and the cells flown through the device at all flow rates used.
FIG. 5.
Viability of MOSE-L-FFLv cells in devices with 100 and 20 μm pillars at flow rate of 20, 28, and 36 μl/min, voltage of 0, 250, 300, and 350 VRMS and frequency of 30 kHz. The experiment was repeated 3 times for each set of parameters. (a) 100 μm pillar device, (b) 20 μm pillar device, (c) Viability of trapped cells, grouped by flow rate. 20 μm pillar device has little effect on viability at all voltages and flow rates, while the 100 μm pillar device significantly reduces cell viability. In both devices cell viability decreases with increasing voltage and flow rate.
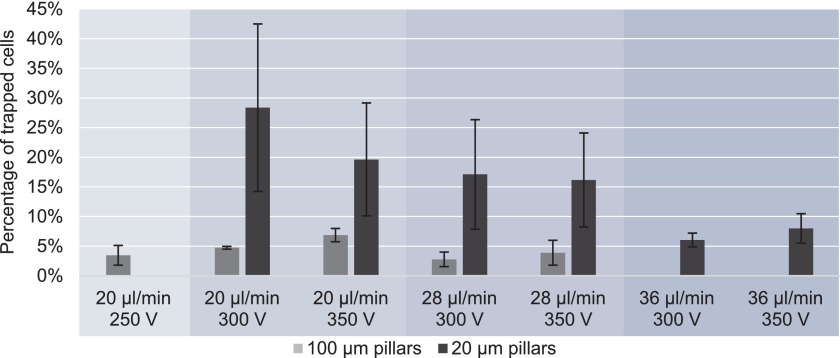
Fig. 6 presents the percentage of trapped cells at the same conditions for both device types that have the same channel dimensions and different number and diameter of the pillars. Up to 6 times more cells can be trapped in the device with 20 μm pillars in comparison to the older design with 100 μm pillars. In the device with 100 μm pillars at 36 μl/min, less than 2% of cells were trapped and an accurate analysis was not possible. In the device with 100 μm pillars, the percentage of trapped cells decreased with increasing flow rate and voltage, but it did not exceed 7%. More efficient trapping can be achieved at lower flow rate, however, that may cause excessive chaining of cells and is not suitable for potential high throughput cell separation. In the 20 μm pillar device, the trapping efficiency decreases at higher flow rates, but no difference was observed between 300 V and 350 V, allowing the desired percentage of trapped cells to be varied by adjusting the flow rate of cell suspension. Maintaining higher viability in the cell sub-populations flowing through the chip allows for improved analysis off-chip. Improving separability by working close to crossover frequency and improving device specificity by decreasing the heterogeneity of cell populations that can be trapped could lead to improved purities in cell sub-populations that can then be taken off-chip. Future efforts are needed to assess and quantify the enhanced selectivity of our devices over the more conventional platform.
FIG. 6.
Percentage of trapped MOSE-L-FFLv cells in devices with 100 and 20 μm pillars. At the same voltage, flow rate and density of cell suspension the device with small pillars can trap up to 6 times more cells than the device with large pillars.
A device with 100 μm pillars could potentially be used for selective cell sorting by significantly reducing density of cells in suspension (∼105 cells/ml) and thus minimizing cell-to-cell interactions. However, the throughput of such an approach is low. In the device with larger pillars, we also observed that trapping with low cell density is inconsistent, as the geometry of the pillars allows for lateral sliding around the pillar, which leads to variation in the effective DEP force that the cell is exposed to. Decreasing the flow rate can compensate for this, but also encourages cells with a wider range of properties to trap, decreasing the degree of potential separability. A better solution is a design in which each pillar has a trapping area that is small enough to trap individual cells while a sufficiently fast flow of medium carries away other cells. This design encourages 1 or 2 cell trapping, eliminating the inhomogeneities associated with clumps of cells or pearl-chaining as is seen in devices with larger pillars. The location of trapping is also strongly determined by the fluid flow and the drag force. If we consider just two main forces, a cell is trapped when the dielectrophoretic force can withstand the drag force. Observing in the direction of the flow, the lowest drag force on a cell is in the center in front of the pillar, while it is higher around the sides of the pillar. In moderate intensity electric fields, cells will not trap on the sides or in the back of a pillar, but only on its front. However, in devices with large pillars, due the low curvature and high radius of the pillar compared to the cell curvature and radius, cells tend to stick to the pillar in the center. Instability leads to them sliding one way or another, at which point they slide to the opposite back center of the cell and fall off. When multiple cells are added, this instability leads to pearl chaining around the pillar and eventually the cells form a clump.
Devices with pillars in the range of 100 μm were used in several studies demonstrating successful cell separation; however, the trapping area around each pillar is large enough that several cells per pillar were trapped if the cell density was sufficiently high. Under these conditions, trapping depends not only on the trapping voltage of specific cells, but also on the cell density (cells per ml) and on individual interactions between cells already trapped and cells flowing through the device, amplifying random variation from one cell interacting with one fabricated pillar to one cell interacting with a pillar and cell complex of variable electric properties. Theoretically, decreasing the diameter of the pillars reduces the area with large electric field gradients and therefore decreases the capacity of trapping, which is a bad choice for a continuous high throughput device. However, in batch sorting, cells have to distribute among all pillars as equally as possible to use the full capacity of the device, which is hard to implement in a large device with 100 μm pillars. The area of sufficiently high electric field gradient to trap cells in front of each pillar is large and cells tend to trap in the first rows of the main channel between the electrodes. In a device with 20 μm pillars, the trapping area is relatively small and cells have enough space to pass the first rows of pillars already saturated with cells. In separating tumor initiating cells from other cancer cells, differences between their electrical properties may be small compared to differences in electrical properties between two different cell types, meaning that a sensitive device should be able to trap a narrow range of cell properties and enrich target population of cells.
V. CONCLUDING REMARKS
We designed a new high-throughput cDEP device with cell-size pillars that alleviates the issues of cell-to-cell interactions of typical iDEP and cDEP devices. We observed reduced pearl chaining with the small pillar device. Theoretical modeling shows that cell isolation and enrichment in this new device should be more specific than in the previous designs. Future efforts are needed to assess and quantify the enhanced selectivity of our devices over the more conventional platform. The small trapping areas allow for cells to evenly distribute along the channel, increasing the throughput. We have shown that the new device can trap more cells at higher flow rate than the established devices with 100 μm pillars, while maintaining high viability. A multilayer design is used to fabricate robust devices that enable more of the electric field to penetrate through the insulating membrane, even at lower frequencies. Allowing the use of lower frequency electric fields further improves separability between similar cell types. The fabricated device is capable of sorting over a million cells per hour. By improving viability and throughput in the new device and minimizing non-specific accumulation of cells, a larger subpopulation of cells can be obtained for off-chip analysis. This population could then be used to tailor cancer treatment to target TIC cells.
ACKNOWLEDGMENTS
This work was supported by NIH 5R21 CA173092-01 and CIT MF13-037-LS. We would also like to acknowledge Virginia Biosciences Health Research Corporation (VBHRC) and NSF IGERT DGE-0966125 MultiSTEPS. R. V. Davalos has patents in the field of dielectrophoresis.
References
- 1. Yu L., Ng S. R., Xu Y., Dong H., Wang Y. J., and Li C. M., Lab Chip 13, 3163 (2013). 10.1039/c3lc00052d [DOI] [PubMed] [Google Scholar]
- 2. Gascoyne P. R. C., Anal. Chem. 81, 8878 (2009). 10.1021/ac901470z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nagrath S., Sequist L. V., Maheswaran S., Bell D. W., Irimia D., Ulkus L., Smith M. R., Kwak E. L., Digumarthy S., Muzikansky A., Ryan P., Balis U. J., Tompkins R. G., Haber D. A., and Toner M., Nature 450, 1235 (2007). 10.1038/nature06385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Denison T. A. and Bae Y. H., J. Controlled Release 164, 187 (2012). 10.1016/j.jconrel.2012.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alix-Panabières C. and Pantel K., Nat. Rev. Cancer 14, 623 (2014). 10.1038/nrc3820 [DOI] [PubMed] [Google Scholar]
- 6. Bhagat A., Bow H., Hou H., Tan S., Han J., and Lim C., Med. Biol. Eng. Comput. 48, 999 (2010). 10.1007/s11517-010-0611-4 [DOI] [PubMed] [Google Scholar]
- 7. Gossett D. R., Weaver W. M., Mach A. J., Hur S. C., Tse H. T. K., Lee W., Amini H., and Di Carlo D., Anal. Bioanal. Chem. 397, 3249 (2010). 10.1007/s00216-010-3721-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beech J. P., Holm S. H., Adolfsson K., and Tegenfeldt J. O., Lab Chip 12, 1048 (2012). 10.1039/c2lc21083e [DOI] [PubMed] [Google Scholar]
- 9. Jubery T. Z., Srivastava S. K., and Dutta P., Electrophoresis 35, 691 (2014). 10.1002/elps.201300424 [DOI] [PubMed] [Google Scholar]
- 10. Čemažar J., Miklavčič D., and Kotnik T., Inf. Midem-J. Microelectron. Electron. Compon. Mater. 43, 143 (2013); available at http://www.midem-drustvo.si/Journal%20papers/MIDEM_43(2013)3p143.pdf. [Google Scholar]
- 11. Vykoukal J., Vykoukal D. M., Freyberg S., Alt E. U., and Gascoyne P. R. C., Lab Chip 8, 1386 (2008). 10.1039/b717043b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pratt E. D., Huang C., Hawkins B. G., Gleghorn J. P., and Kirby B. J., Chem. Eng. Sci. 66, 1508 (2011). 10.1016/j.ces.2010.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hyun K.-A. and Jung H.-I., Electrophoresis 34, 1028 (2013). 10.1002/elps.201200417 [DOI] [PubMed] [Google Scholar]
- 14. Smith J. P., Huang C., and Kirby B. J., Biomicrofluidics 9, 014116 (2015). 10.1063/1.4908049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tirino V., Desiderio V., Paino F., Rosa A. D., Papaccio F., Noce M. L., Laino L., Francesco F. D., and Papaccio G., FASEB J. 27, 13 (2013). 10.1096/fj.12-218222 [DOI] [PubMed] [Google Scholar]
- 16. Doherty R. E., Haywood-Small S. L., Sisley K., and Cross N. A., Biochem. Biophys. Res. Commun. 414, 801 (2011). 10.1016/j.bbrc.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 17. Pethig R., Biomicrofluidics 4, 022811 (2010). 10.1063/1.3456626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Çetin B. and Li D., Electrophoresis 32, 2410 (2011). 10.1002/elps.201100167 [DOI] [PubMed] [Google Scholar]
- 19. Gagnon Z. R., Electrophoresis 32, 2466 (2011). 10.1002/elps.201100060 [DOI] [PubMed] [Google Scholar]
- 20. Martinez-Duarte R., Electrophoresis 33, 3110 (2012). 10.1002/elps.201200242 [DOI] [PubMed] [Google Scholar]
- 21. Li H. B. and Bashir R., Sens. Actuators B-Chem. 86, 215 (2002). 10.1016/S0925-4005(02)00172-7 [DOI] [Google Scholar]
- 22. Khoshmanesh K., Nahavandi S., Baratchi S., Mitchell A., and Kalantar-zadeh K., Biosens. Bioelectron. 26, 1800 (2011). 10.1016/j.bios.2010.09.022 [DOI] [PubMed] [Google Scholar]
- 23. Lapizco-Encinas B. H., Simmons B. A., Cummings E. B., and Fintschenko Y., Anal. Chem. 76, 1571 (2004). 10.1021/ac034804j [DOI] [PubMed] [Google Scholar]
- 24. Kang Y., Cetin B., Wu Z., and Li D., Electrochimica Acta 54, 1715 (2009). 10.1016/j.electacta.2008.09.062 [DOI] [Google Scholar]
- 25. Shafiee H., Caldwell J., Sano M., and Davalos R., Biomed. Microdevices 11, 997 (2009). 10.1007/s10544-009-9317-5 [DOI] [PubMed] [Google Scholar]
- 26. Salmanzadeh A., Romero L., Shafiee H., Gallo-Villanueva R. C., Stremler M. A., Cramer S. D., and Davalos R. V., Lab Chip 12, 182 (2012). 10.1039/C1LC20701F [DOI] [PubMed] [Google Scholar]
- 27. Gupta V., Jafferji I., Garza M., Melnikova V. O., Hasegawa D. K., Pethig R., and Davis D. W., Biomicrofluidics 6, 024133 (2012). 10.1063/1.4731647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Iliescu C., Yu L. M., Xu G. L., and Tay F. E. H., J. Microelectromech. Syst. 15, 1506 (2006). 10.1109/JMEMS.2006.883567 [DOI] [Google Scholar]
- 29. Sano M. B., Caldwell J. L., and Davalos R. V., Biosens. Bioelectron. 30, 13 (2011). 10.1016/j.bios.2011.07.048 [DOI] [PubMed] [Google Scholar]
- 30. Sancho M., Martínez G., Muñoz S., Sebastián J. L., and Pethig R., Biomicrofluidics 4, 022802 (2010). 10.1063/1.3454129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moncada-Hernandez H., Nagler E., and Minerick A. R., Electrophoresis 35, 1803 (2014). 10.1002/elps.201300636 [DOI] [PubMed] [Google Scholar]
- 32. Cemazar J., Anders L. M., Cramer S. D., and Davalos R. V., in Proc. 36th Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., Chicago (2014). [Google Scholar]
- 33. Pohl H. A. and Crane J. S., Biophys. J. 11, 711 (1971). 10.1016/S0006-3495(71)86249-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kotnik T., Kramar P., Pucihar G., Miklavčič D., and Tarek M., IEEE Electr. Insul. Mag. 28, 14 (2012). 10.1109/MEI.2012.6268438 [DOI] [Google Scholar]
- 35. Schwan H. P., in Biological Effects on Dosim. Nonionizing Radiation, edited by Grandolfo M., Michaelson S. M., and Rindi A. ( Springer US, 1983), pp. 213–231. [Google Scholar]
- 36. Marszalek P., Liu D. S., and Tsong T. Y., Biophys. J. 58, 1053 (1990). 10.1016/S0006-3495(90)82447-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alberts B., Johnson A., Lewis J., Raff M., Roberts K., and Walter P., Molecular Biology of the Cell, 4th ed. ( Garland Science, New York, 2002). [Google Scholar]
- 38. Buchner R., Hefter G. T., and May P. M., J. Phys. Chem. A 103, 1 (1998). 10.1021/jp982977k [DOI] [Google Scholar]
- 39. Gascoyne P. R. C., Pethig R., Burt J. P. H., and Becker F. F., Biochim. Biophys. Acta BBA - Biomembr. 1149, 119 (1993). 10.1016/0005-2736(93)90032-U [DOI] [PubMed] [Google Scholar]
- 40. Hu Q., Joshi R. P., and Beskok A., J. Appl. Phys. 106, 024701 (2009). 10.1063/1.3173344 [DOI] [Google Scholar]
- 41. Holzel R. and Lamprecht I., Biochim. Biophys. Acta 1104, 195 (1992). 10.1016/0005-2736(92)90150-K [DOI] [PubMed] [Google Scholar]
- 42. Roberts P. C., Mottillo E. P., Baxa A. C., Heng H. H. Q., Doyon-Reale N., Gregoire L., Lancaster W. D., Rabah R., and Schmelz E. M., Neoplasia 7, 944 (2005). 10.1593/neo.05358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Creekmore A. L., Silkworth W. T., Cimini D., Jensen R. V., Roberts P. C., and Schmelz E. M., PLoS ONE 6, e17676 (2011). 10.1371/journal.pone.0017676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cohen C. A., Shea A. A., Heffron C. L., Schmelz E. M., and Roberts P. C., PLoS ONE 8, e66477 (2013). 10.1371/journal.pone.0066477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cohen C. A., Shea A. A., Heffron C. L., Schmelz E. M., and Roberts P. C., Cancer Prev. Res. Phila. Pa 6, 1182 (2013) 10.1158/1940-6207.CAPR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderson A. S., Roberts P. C., Frisard M. I., Hulver M. W., and Schmelz E. M., Exp. Cell Res. 328, 44 (2014). 10.1016/j.yexcr.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Flanagan L. A., Lu J., Wang L., Marchenko S. A., Jeon N. L., Lee A. P., and Monuki E. S., Stem Cells 26, 656 (2008). 10.1634/stemcells.2007-0810 [DOI] [PubMed] [Google Scholar]
- 48. Francis G. L., Cytotechnology 62, 1 (2010). 10.1007/s10616-010-9263-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang Z., Al-Rubeai M., and Thomas C. R., Enzyme Microb. Technol. 14, 980 (1992). 10.1016/0141-0229(92)90081-X [DOI] [PubMed] [Google Scholar]
- 50. Srivastava S. K., Gencoglu A., and Minerick A. R., Anal. Bioanal. Chem. 399, 301 (2010). 10.1007/s00216-010-4222-6 [DOI] [PubMed] [Google Scholar]
- 51. Wang Z. and Belovich J. M., Biotechnol. Prog. 26, 1361 (2010). 10.1002/btpr.432 [DOI] [PubMed] [Google Scholar]
- 52. Morgan H. and Green N. G., AC Electrokinetics : Colloids and Nanoparticles ( Research Studies, Baldock, Hertfordshire, 2003). [Google Scholar]
- 53. Jones T. B., Electromechanics of Particles, Digitally printed 1st pbk. version ( Cambridge University Press, Cambridge, New York, 1995). [Google Scholar]
- 54. Sano M. B., Salmanzadeh A., and Davalos R. V., Electrophoresis 33, 1938 (2012). 10.1002/elps.201100677 [DOI] [PubMed] [Google Scholar]
- 55. Salmanzadeh A., Sano M. B., Gallo-Villanueva R. C., Roberts P. C., Schmelz E. M., and Davalos R. V., Biomicrofluidics 7, 11809 (2013) 10.1063/1.4788921. [DOI] [PMC free article] [PubMed] [Google Scholar]