Abstract
Microscale technologies offer the capability to generate in vitro artificial cellular microenvironments that recapitulate the spatial, biochemical, and biophysical characteristics of the native extracellular matrices and enable systematic, quantitative, and high-throughput studies of cell fate in their respective environments. We developed a microfluidic platform for the generation of two-dimensional arrays of micrometer-size cell-laden hydrogel modules (HMs) for cell encapsulation and culture. Fibroblast cells (NIH 3T3) and non-adherent T cells (EL4) encapsulated in HMs showed high viability and proliferation. The platform was used for real-time studies of the effect of spatial constraints and structural and mechanical properties of HMs on cell growth, both on the level of individual cells. Due to the large number of cell-laden HMs and stochastic cell distribution, cell studies were conducted in a time- and labor efficient manner. The platform has a broad range of applications in the exploration of the role of chemical and biophysical cues on individual cells, studies of in vitro cell migration, and the examination of cell-extracellular matrix and cell-cell interactions.
I. INTRODUCTION
Local extracellular matrix (ECM) is a key component of cellular microenvironments that serves as a scaffold supporting cells and provides regulatory cues to control cell behavior in spatiotemporal multicellular processes.1–3 Artificial ECMs mimicking some of the key biophysical and biochemical characteristics of their naturally derived counterparts have been extensively studied, with the ultimate goal of using them in tissue transplantation, regenerative medicine, and tissue engineering.4 Among these materials used as instructive artificial ECMs, polymer hydrogels are particularly promising due to their intrinsic porous structure and mechanical, biophysical, and chemical properties that can closely resemble those of natural ECMs.5–14
The exploratory work on instructive artificial ECMs has greatly benefited from microscale technologies, including photolithography,7,11,15 microprinting,16,17 and microfluidics,18 as these platforms paved the way for efficient, systematic, and quantitative studies of cell-ECM interactions and enabled high-throughput reproducible studies of cells at the level of a single cell or a small number of spatially confined cells. Photolithography was used for in situ generation of photopolymerized hydrogels with a spatial identity and desired topography; however, the utilization of ultraviolet radiation and the use of radicals may affect cell fate.19,20 Bioprinting of arrays of cells and biological molecules is a powerful method of cell seeding, yet, controlling cell viability and long-term functionality remains a challenge.21 Microfluidics (MFs) enabled the encapsulation of cells in uniform micrometer-sized hydrogel particles with composition and physical properties tuned in a high-throughput manner.22–25 This method offered the capability to generate libraries of cell-laden artificial instructive ECMs;26,27 however, subsequent analysis of cell fate relied on averaged characteristics over the entire population of encapsulated cells and did not examine the behavior of individual cells in their respective ECM, which is important in studies of rare diseases and gene mutations.28,29 An alternative MFs-based approach would be the development of two-dimensional (2D) arrays of cell-laden microscale hydrogel modules (HMs). The capability to enumerate (or index) individual HMs would enable in situ monitoring, manipulation, and analysis of cells in their respective microenvironments in a real-time manner. This platform resembles cell analysis in a microwell plate format; however, it utilizes a smaller amount of high-cost reagents, reduces evaporation of water, enables automated loading and analysis of samples, and provides an enhanced ability to study individual cells. Two-dimensional arrays of droplets have been produced by immobilizing pre-formed droplets in predesigned locations,30,31 by using a Slipchip method,32 and by utilizing surface patterning techniques.33,34 These methods enabled the generation of high-density indexed arrays of droplets and allowed direct studies of the properties of species compartmentalized within droplets, e.g., the neurotoxin-response of Caenorhabditis elegans,30 protein crystallization,35,36 and enzyme activity.37 The utilization of 2D arrays of cell-laden polymer hydrogels that can be used as instructive artificial ECMs was, however, hampered by the complexity of microfluidic devices, e.g., the use of digital valves.38
In the present work, we developed a MF platform for the generation of high-density 2D arrays of cell-laden polymer HMs. We used an elegant approach proposed by Chiu et al., who utilized self-digitalization of liquid streams into droplets using micrometer-size compartments.39 This cost-efficient and straightforward method has been used to study polymorph crystallization39 and DNA amplification;40 however, it was not extended toward studies of cell behavior in 2D arrays of hydrogels. In the present work, we used self-digitization of suspensions of cells in a gelling polymer solution to form high density 2D arrays of cell-laden HMs. Notably, HMs could be formed from a variety of physically or chemically gelling polymers, with the requirement of their gelation occurring after the formation of cell-laden droplets. We selected agarose as an exemplary physically gelling polymer for two reasons. Agarose forms gels by thermosetting, that is, upon cooling and it is non-cytotoxic and biocompatible.26,41 If needed, agarose can be readily functionalized with growth factors or peptide fragments to make it bioactive.42,43 The concentration of fluorescein isothiocyanate conjugated agarose (FITC-agarose) was selected at 2 wt. % for characterization of the shape and the size distribution of droplets and HMs, since the physical properties of the FITC-agarose solution at this concentration are comparable with the physical properties of unmodified agarose solution at higher concentration (eg. 3–5 wt. %). The concentration of agarose in HMs for culturing cells was selected from 3 to 5 wt. %, since the Young's modulus of agarose microgels at polymer concentration of 3–5 wt. % at 37 °C was 0.5–4.3 kPa,44 spanning the range of elastic moduli of normal to malignant breast tissues.45,46
The approach to 2D arrays of cell-laden HMs (i) enabled efficient generation of micrometer-size artificial ECMs from precursor droplets, which avoided problems caused by wetting and/or coalescence of droplets;47 (ii) produced libraries of cell-laden HMs with varying structural and mechanical properties; (iii) offered the capability to monitor individual cells in situ in their respective environments and in a real-time manner; and (iv) provided the ability to explore the effect of spatial constraints on cell fate. In the present work, we selected NIH 3T3 fibroblasts and EL4 T cells as two model cell lines representing adherent cells and non-adherent cells. We selected 3 days as cell culture period in HMs, as it is a standard period for proliferation and passaging of NIH 3T3 in cell culture flasks,48,49 and EL4 T cells with a 3 day culture period were comparable with the fibroblasts cultured under the same conditions. This MF platform has a broad range of applications, including studies of the effect of biochemical and biophysical cues on individual cell fate, studies of in vitro cell migration, and the examination of cell-ECM and cell-cell interactions.
II. EXPERIMENT
A. Fabrication of the microfluidic device
Fabrication of the MF device was carried out using a soft lithography method.39 The device was assembled from three layers of polydimethylsiloxane (PDMS, Sylgard184, Dow Corning) and a glass slide (Corning 75 × 50 mm Plain Microscope Slides, Product #2947-75X50) (see supplementary material, Fig. S1(a)).50 The assembled device was placed in an oven for 12 h at 115 °C to allow it to recover its hydrophobicity. The surface of microchannels was further modified by infusing a 10 wt. % solution of trichloro(1H,1H,2H,2H-perfluorooctyl) silane (97% pure, Sigma, USA, #448931-10G) in ethanol in the microchannel, heating the MF device at 45 °C for 3 min, and washing the silane solution away with 100% ethanol and deionized water.
B. Formation of 2D array of droplets and hydrogel modules in the MF device
Self-digitization of the aqueous stream supplied to the MF device occurred in several steps. First, the surface of PDMS in the device was treated with fluorinated oil (HFE 7500, 3M, Canada) mixed with 0.1 wt. % block copolymer perfluorinated polyether-b–(polypropylene glycol-polyethylene glycol-polypropylene glycol)–b-perfluorinated polyether (PFPE-b-(PPG-PEG-PPG)-b-PFPE).51 In cell encapsulation experiments, a 1 wt. % solution of PFPE-b-(PPG-PEG-PPG)-b-PFPE in fluorinated oil was used. Then, the entire MF device was filled with either an agarose solution or a suspension of cells in an agarose solution. Next, the oil phase was introduced in the MF device to replace the aqueous phase in the supplying microchannel, but retaining aqueous droplets in the wells. Finally, the entire MF device was cooled down to 4 °C for 45 min to transform the precursor droplets into HMs, and the oil phase in the supplying channel was replaced with the cell culture medium.
C. Characterization of volumes of droplets and hydrogel modules
The average volume of precursor droplets in the microwells was calculated as the product of the average 2D projection area of the droplets and the average height of the wells (see supplementary material Fig. S150 for the dimensions of the channels and wells). The average volume of HMs in the wells was calculated as the product of the average 2D projection area of the HMs and the average height of the wells.
D. Cell culture
NIH 3T3 fibroblast cells were cultured in Dulbecco's Modified Eagle Medium with 4.5 g l−1 glucose, L-glutamine, and sodium pyruvate (DMEM, GIBCO, #11995-065) supplemented with 10% (v/v) fetal bovine serum (FBS, Invitrogen, #16000-036) and 1% (v/v) Penicillin/Streptomycin (Sigma-Aldrich, #P4333–100ML) in an incubator supplied with 5% CO2 at 37 °C. A 0.25 wt. % Trypsin-EDTA solution (GIBCO, #25200-056) was used to detach cells from the plastic flask. After cell detachment, fresh nutrition medium was added, and the cell suspension was centrifuged at 1000 rpm for 3 min. The supernatant was then disposed, and the cells were re-dispersed in fresh medium. For cell passaging, 5 vol. % of the cell suspension were transferred to the fresh medium every 3 days.
EL4 cells were cultured in Iscove's Modified Dulbecco's Medium (IMDM, GIBCO, #12440–053) with 10% (v/v) fetal bovine serum and 1% (v/v) Penicillin/Streptomycin (Sigma-Aldrich, #P4333-100ML) and incubated at 37 °C and at 5% CO2. Cells were washed with Hank's Balanced Salt Solution (HBSS, GIBCO, #14175-095), centrifuged and dispersed in the fresh medium at the same conditions, as described above. For cell passaging, 5 vol. % of the cell suspension were transferred to the fresh medium every 3 days.
E. Cell culture in hydrogel modules
Following the formation of NIH 3T3 fibroblast-laden HMs, the MF device was placed in an incubator containing 5% CO2 at 37 °C. A cell culture medium was continuously supplied to the MF device at the flow rate of 0.05, 0.08, or 0.11 ml/h via a perfluoroalkoxy alkane tubing (IDEX health and Science) using a syringe pump. EL4 cells encapsulated in HMs were cultured under the same conditions as the NIH 3T3 fibroblast cells, with the flow rate of the cell culture medium of 0.08 ml/h.
F. Live/dead assay and fluorescent staining of cells
Live cells were identified by staining them with Calcein AM (Invitrogen, Carlsbad, CA; green fluorescence), and dead cells were identified by staining them with Ethidium Homodimer I (Invitrogen, Carlsbad, CA; red fluorescence). The live and dead cells stained with fluorescence dyes were imaged by confocal fluorescence microscopy (CFM) (Nikon, Eclipse Ti-E, Japan). The viability of individual cells encapsulated in HMs on day 0 was characterized as the ratio of live cells to the total number of cells, that is, the sum of the numbers of live and dead cells. For cell colonies grown from individual cells on day 3, the viability was characterized as the ratio of live colonies to the total number of colonies. The non-proliferated cells on day 3 were also accounted as colonies in the determination of viability on day 3. The total number of cells was also confirmed by staining cell nuclei with Hoechst (Sigma-Aldrich Canada; blue fluorescence).
A solution of Calcein AM (4 μM), Ethidium Homodimer I (4 μM), or Hoechst 33342 (10 μg/ml) in HBSS was perfused through the MF device compartmentalizing cell-laden HM for 5 min, and the MF device was placed in the incubator at 37 °C for cell staining for 60 min.
G. Confocal fluorescence microscopy imaging
The 3D structure of the FITC-agarose HM was characterized using an inverted confocal microscope (Nikon, Eclipse Ti-E, Japan). The images of individual HMs were obtained by 3D reconstruction of the CFM images with a step of 10 μm. The images of the stained cells and cell colonies in the HMs were obtained by 3D reconstruction of the CFM images with a step of 3 and 5 μm, respectively.
H. Determination of the microstructure of the agarose gels
Agarose solutions were prepared by dissolving agarose powder (ultra-low gelling temperature, Lonza Rockland Inc., USA) in HBSS at 70 °C for 2 h. After 1 h incubation of the solution at 37 °C, gel slabs were prepared by cooling ∼0.15 ml of the solution at 4 °C for 45 min, following the conditions of the preparation of HMs. The gels were instantly frozen by liquid nitrogen, and the water was removed by freeze drying for 3 days under −40 °C at ∼200 Pa. Prior to imaging, the cross-section of the gel slabs was coated with Au nanoparticles using a SC7640 High Resolution Sputter Coater (Quorum Technologies) for 15 s at 2.0 kV. Scanning Electron Microscopy (SEM) imaging was carried out on the Quanta FEI Scanning Electron Microscope.
I. Statistical analysis
Statistical significance of experimental results was verified using the Student's t-test. The experimental results are expressed as the mean value plus/minus standard deviation (SD) for at least three independent experiments.
III. RESULTS
A. Preparation and characterization of precursor droplets and hydrogel modules
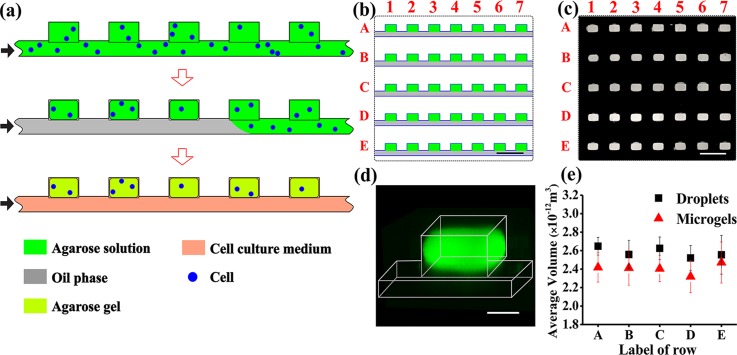
Five rows, each containing 100 wells with dimensions of 200 × 130 × 97 μm and a straight supplying microchannel (used for the delivery of cell suspension in an agarose solution), were placed in the MF device. The supplying microchannel was used for the delivery of cell suspension in an agarose solution to the wells and for the supply of the nutrition medium to cell-laden HMs. Fig. 1(a) shows a “top view” schematic of the preparation of the 2D array of HMs. After wetting the surface of the MF channel with fluorinated oil, the supplying channel and wells were filled with either an aqueous agarose solution at polymer concentration, Cagar, or a suspension of cells in the agarose solution (Fig. 1(a), top). Next, the aqueous phase in the supplying channel was replaced with the oil phase. This step led to the formation of an array of droplets of agarose solution or cell suspension in agarose solution, which were compartmentalized in the wells (Fig. 1(a), middle). The entire MF device was cooled down to 4 °C for 45 min to transform the droplets into HMs, and the oil phase in the supplying channel was replaced with the cell culture medium (Fig. 1(a), bottom). Fig. 1(b) shows a “top view” schematic of the fragment of the MF multi-well device. The wells and the rows were enumerated by numbers (1, 2, 3…, etc.) and letters (A, B, C, D, and E), respectively. The individual inlets and outlets enabled the delivery of distinct species in and out of each row of wells.
FIG. 1.
Microfluidic generation of hydrogel modules (HMs). (a) Schematic of the MF generation of the 2D array of cell-laden agarose HMs from top view of the channel and wells. Step 1: cell suspension in an agarose solution is perfused through the microfluidic device. Step 2: the agarose solution in the supplying channel is switched to an oil phase. Droplets of agarose solution are confined in the wells, and cells are entrapped into the droplets. Step 3: the temperature is lowered to 4 °C to transform the droplets of agarose solution into gels (HMs). After HM formation, the oil phase in the supplying channel is replaced with a cell culture medium. The black arrows show the direction of infusion of liquid phases: top to bottom: agarose solution or cell suspension in the agarose solution, oil, and nutrition medium. (b) Schematic of the top view fragment of the 2D array containing five rows with seven microwells, each (the entire row contained 100 wells). (c) Fluorescence microscopy image of the top view fragment of the 2D array (shown in (b)), with wells filled with a 2 wt. % solution of agarose conjugated with fluorescein isothiocyanate (FITC). (d) Three-dimensional CFM image of an individual HM formed in the well from FITC-conjugated agarose. The image was obtained by 3D reconstruction of the images with a step of 10 μm. (e) Variation in the average volume of droplets of agarose solution at Cagar = 2 wt. % (▪) and resulting microgels (▲), shown for rows A–E. The dimensions of 90 droplets or microgels in each row were analyzed for the calculations of the average volume. The scale bar is 500 μm in (b) and (c) and 100 μm in (d). Error bars in (e) show standard deviation (SD).
We first examined the formation of HMs from the cell-free agarose solution. An aqueous 2 wt. % solution of agarose conjugated with FITC52 was introduced into the MF device at 37 °C and compartmentalized in the wells (Fig. 1(c), top view of the droplets), thereby forming a 2D array of droplets (serving as HM precursors). In the wells, the droplets had a cuboid shape with truncated corners, as determined by 3D reconstruction of CFM images (Fig. 1(d)). The droplets had an average volume of (2.6 ± 0.2) × 10−12 m3 (Fig. 1(e)). The coefficient of variation (CV), defined as a standard deviation in droplet volume divided by average droplet volume, did not exceed 6% for the entire population of droplets in five rows. The cuboid shape was preserved upon the transformation of the precursor droplets into agarose HMs. The average HM volume was (2.4 ± 0.2) × 10−12 m3 (Fig. 1(e)), that is, 7.7% smaller than the average volume of the precursor droplets. The CV of the entire population of HMs in five rows did not exceed 8%, indicating the uniformity of HM dimensions.
B. Encapsulation of cells in hydrogel modules
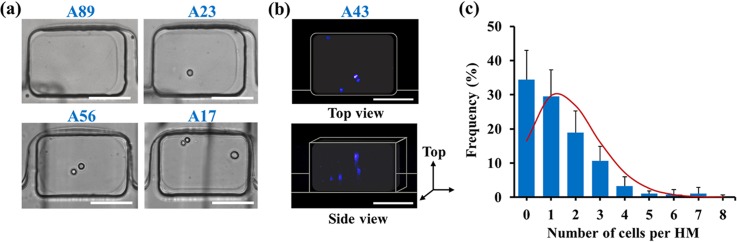
A suspension of NIH 3T3 fibroblasts in agarose solution at the concentration of cells of 7 × 105 cell ml−1 was introduced in the MF device at 37 °C. Following the formation of cell-laden droplets and their transformation into HMs, optical microscopy and CFM were used to examine the distribution of cells in HMs. A high transparency of agarose HMs enabled direct visualization of cells in each well. Fig. 2(a) shows optical microscopy images of the most representative populations of HMs containing 0, 1, 2, and 3 cells per HM. Representative CFM images show the lateral and vertical distributions of cells within the body of the same cell-laden HM (Fig. 2(b)).
FIG. 2.
Encapsulation of fibroblasts (NIH 3T3) in hydrogel modules. (a) Optical images (top view) of the cells compartmentalized in HMs prepared from cell suspension in 4 wt. % agarose solution. Cell concentration was 7 × 105 cells·ml−1. Scale bars are 100 μm. The vertical black lines show the features of the microchannel below the wells (see supplementary material).50 (b) Representative CFM images of cells, in which the nuclei were stained with Hoechst 33342. The top and the bottom images show the top and the side view, respectively, of the same cell-laden HM. Scale bars are 100 μm. (c) Experimentally determined distribution of cells per HM formed at Ccell = 7 × 105 cell·ml−1 and the volume of precursor droplets of (2.6 ± 0.2) × 10−12 m3. The solid red line shows the variation in the number of cells per HM based on the Poisson distribution. The experimental data were collected for 555 HMs from four independent experiments. Error bars present the standard deviation (SD).
Fig. 2(c) shows the distribution of the number of cells per HM, determined at the concentration of cells in the feeding suspension at Ccell = 7 × 105 cell·ml−1 and the average volume of precursor droplets of (2.6 + /−0.2) × 10−12 m3. A large fraction (58%) of HMs contained 1–3 cells, 34% of HMs were cell-free, and 6% of HMs contained more than 3 cells. The distribution of the number of cells per droplet (and per HM) was compared with Poisson distribution53
| (1) |
where P(x) is the fraction of HMs expected to contain x cells, and λ is the average number of cells per HM. The fraction of cell-free HMs was ∼18% higher than that determined from the Poisson distribution, while the fraction of HMs containing 2 cells was 8% lower. This effect had a hydrodynamic origin and stemmed from the competition of microscale laminar vortices and inertial cell focusing, as described elsewhere.54
A relatively high (34%) fraction of cell-free HMs was compensated by the large overall number of HMs on a chip: among 500 HMs, 300 HMs were laden with 1–3 cells. In our work, the value of Ccell was selected to be low to obtain a high fraction of HMs laden with 1–3 cells; however, in principle, based on Eq. (1), the number of cells encapsulated in HMs can be increased by using higher Ccell or by fabricating a MF device with larger dimensions of wells (thus increasing the size of HMs).
C. Viability and proliferation of fibroblast cells in hydrogel modules
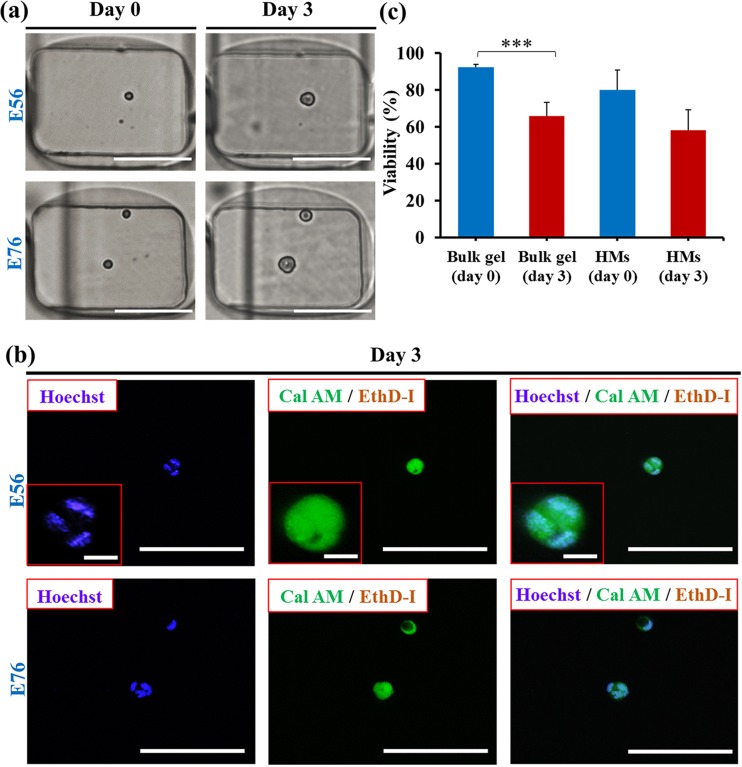
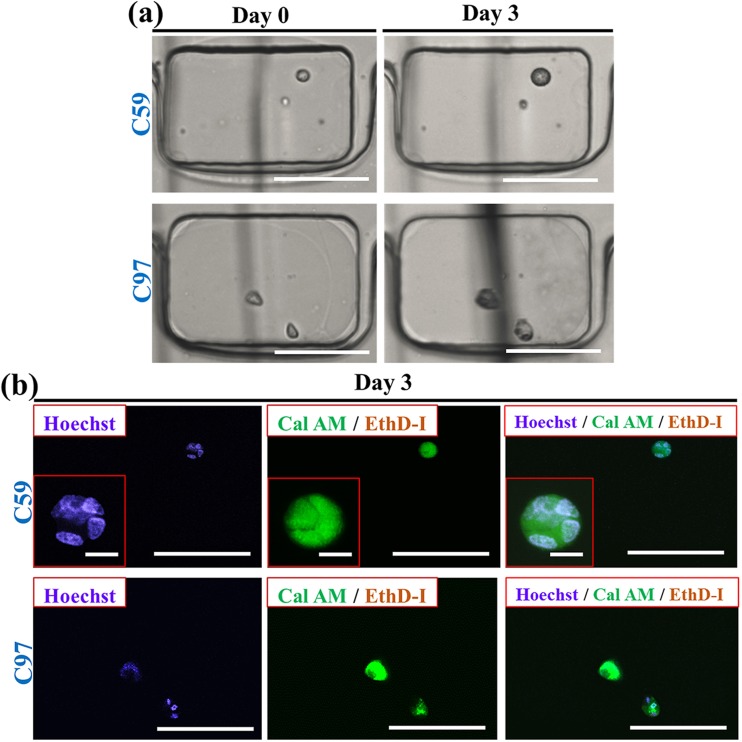
Encapsulation of cells in HMs provided the ability to monitor the behavior of individual cells in their respective microenvironments in a real-time manner. Fig. 3(a), left (top and bottom) shows representative HMs (Cagar = 4 wt. %), encapsulating one and two cells, respectively, imaged 60 min after cell encapsulation. Following 3 day culture, individual cells showed significant growth, as shown by the bright field images in Fig. 3(a), right. To explore cells growth, cell nuclei were stained by Hoechst (blue fluorescence) (Fig. 3(b)). The presence of multiple nuclei in each of the spots revealed that after 3 days, colonies contained multiple cells.
FIG. 3.
Viability and growth of fibroblasts (NIH 3T3) in hydrogel modules. (a) Optical microscopy images of NIH 3T3 fibroblast cells compartmentalized in HMs. Phase contrast images (left column) show one and two cells (top and bottom, respectively) per HM (Cagar = 4 wt. %). Phase contrast images (right column) show the same cells after culturing 3 days. Scale bars are 100 μm. The vertical black lines show the features of the microchannel below the wells (see supplementary material).50 (b) CFM images of the cells shown in (a) after 3 day culture. Top and bottom images show growth of fibroblast colonies starting from one and two cells (top and bottom) shown in (a), respectively. The CFM images show Hoechst 33342 staining, Calcein AM (Cal AM), and Ethidium Homodimer I (EthD-I) staining and merged image of cell staining by all three dyes, respectively. Nuclei staining by Hoechst 33342 shows the formation of cell colonies after 3 day culture. Scale bars are 100 μm. Scale bars in the insets are 10 μm. (c) Viability of cells encapsulated in macroscopic agarose gel and in 4 wt. % agarose HMs on day 0 and day 3. Cell-laden macroscopic gel was prepared under the same conditions as cell-laden HMs. The concentration of cells in the suspension was 7 × 105 cell ml−1. Error bars represent the standard deviation (SD). The bracket above the data bars in (c) indicates that the two data sets were compared using Student's t-test. The triple stars denote p < 0.001. For all other pairs of data sets in (c), p > 0.05, signifying no significant difference between any two of these data sets.
In addition, live stain by Calcein AM (green fluorescence) and dead stain by Ethidium Homodimer I (red fluorescence) revealed the viability of the cells in the colonies after 3 day culture. In the control experiment, cells were encapsulated in the macroscopic 4 wt. % agarose gels prepared using the same protocol as HMs (supplementary material, Fig. S250).
Fig. 3(b) shows that the viability of cells encapsulated in HMs on day 0 was ∼10% lower than in a macroscopic gel. After 3 day culture, cells encapsulated in the macroscopic agarose gel and in the agarose HMs both exhibited ∼20% decrease in viability, which implied that the fibroblasts immobilized in the agarose HMs represented similar levels of viability to those in macroscopic agarose gels.
D. Analysis of spatial constraints on growth of fibroblast cells
The capability to visualize and identify encapsulated cells in the course of cell culture enabled the characterization of the effect of spatial constraints, that is, the number of cells per HM, on cell growth. Importantly, the variation in HM volumes did not exceed 8%. By analyzing the 2D projected area of the cells and colonies with image J,55 we calculated the averaged diameter of the individual cell, , on day 0 and cell colony, , on day 3 and characterized the growth of individual cells in HMs as
| (2) |
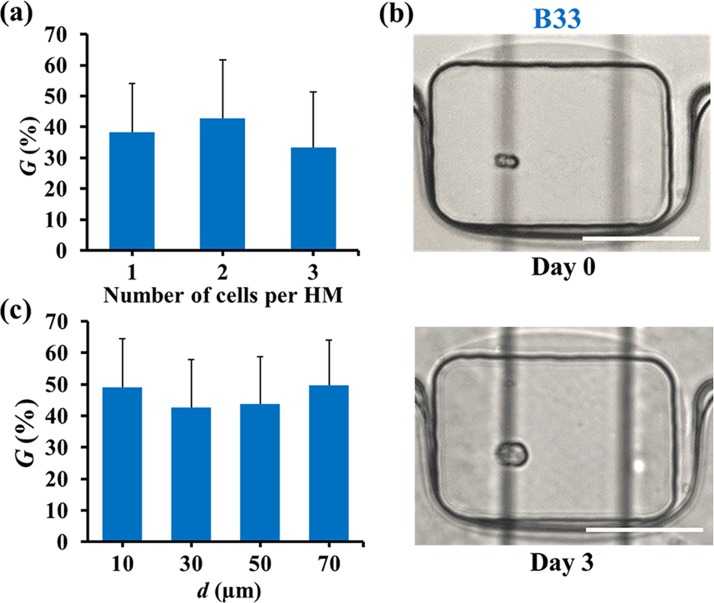
Fig. 4(a) shows the averaged values of G on day 3, for the HMs containing 1, 2, and 3 cells (the number of wells containing more than 3 cells was represented at significantly lower frequency, as shown in Fig. 2(c)). No significant dependence of cell growth on the number of cells per HM was observed for HMs compartmentalizing 1–3 cells, as shown in Fig. 4(a), with G ≈ 40%.
FIG. 4.
Growth of fibroblast colonies in hydrogel modules formed at Cagar = 4 wt. %. (a) Variation in the growth of colonies G (determined using Eq. (2)) for the different cell encapsulation rate. (b) Optical microscopy images of HMs containing cells in intimate contact on day 0 (top) and day 3 (bottom). Scale bars are 100 μm. The vertical black lines show the features of the microchannel below the wells (see supplementary material).50 (c) Variation in the value of G for cell at different original center-to-center distance d. In (a), the average diameter of cells and colonies in HMs containing one, two, and three cells per HM on day 0 was determined for 35, 30, and 24 cells, respectively. In (c), cell-to-cell distance was determined for 36 cells. The value of d was determined for 36 cells. The number of cells analyzed was 12, 6, 12, and 6 for d of 10, 30, 50, and 70 μm, respectively. The concentration of cells in the suspension was 7 × 105 cell ml−1. Error bars correspond to standard deviation (SD). Student's t-test: p > 0.05 for all pairs of data sets in (a) and (c), indicating no significant difference between any two data sets.
To investigate the role of spatial distribution of cells in HMs, we examined the dependence of cell colony growth on the original cell-to-cell distance, d, which was defined as the center-to-center distance (binned at ±10 μm) between the two adjacent cells located in the same (±10 μm) plane of HM. These studies were conducted for HMs encapsulating two cells only. No pre-selection has been performed from the subset of data in Fig. 4(a). The value of d changed from 10 to 70 μm, with d = 10 μm for two cells in intimate contact (Fig. 4(b)). Fig. 4(c) shows that no significant effect of the cell-to-cell distance on colony growth was observed after 3 day cell culture.
E. Effect of the mechanical properties and structure of HMs on colony growth
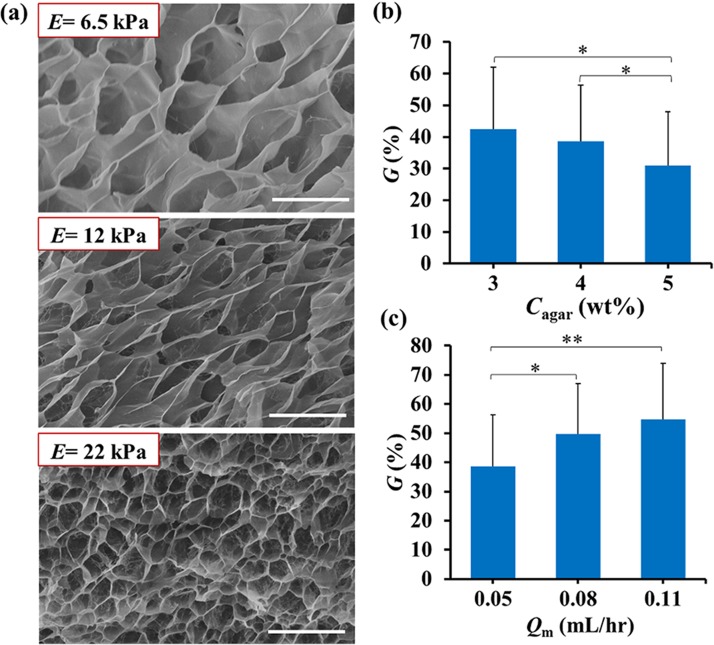
The 2D array of cell-laden HMs was used to study the effect of the structural and mechanical properties of the hydrogel on colony growth. The cell-laden HMs were prepared in three parallel rows of wells from precursor solutions with Cagar of 3, 4, and 5 wt. %. At 37 °C, the Young's modulus, E, of these microgels was 0.5, 1.5, and 4.3 kPa, respectively.44 Fig. 5(a) shows representative scanning electron microscopy images of these hydrogels. All the gels had a “honeycomb” microstructure, with a pore size reducing at higher polymer concentration.
FIG. 5.
Growth of fibroblasts in hydrogel modules with different Young's moduli and under different flow rate of the culture medium. (a) Scanning electron microscopy (SEM) images of HMs at Cagar (top to bottom) of 3.0, 4.0, and 5.0 wt. %. Scale bar is 10 μm. The insets show the Young's modulus of HMs at room temperature. (b) Colony growth in HMs with varying Cagar, on day 3. The flow rate of the cell culture medium Qm = 0.05 ml·h−1. (c) Effect of the flow rate of the cell culture medium, Qm, on colony growth in HMs on day 3 (Cagar = 4 wt. %). In (b), G of the cells in agarose HMs at 3, 4, and 5 wt. % was determined for 20, 37, and 30 cells, respectively. In (c), G of the cells in agarose HMs at 4 wt. % at Qm of 0.05, 0.08, and 0.11 ml/h was determined for 86, 18, and 17 cells, respectively. The brackets above the data bars in (b) and (c) indicate the two data sets compared in Student's t-test. The single and double stars denote the p values: * p < 0.05, ** p < 0.01. For all other pairs of data sets in (b) and (c), p > 0.05.
Fibroblast NIH 3T3 cells encapsulated into these HMs and cultured for 3 days exhibited ∼10% decrease in the growth of colonies, when Cagar increased from 3 to 5 wt. % (Fig. 5(b)). For cells compartmentalized in HMs, we also examined the effect of the flow rate, Qm, of the cell culture medium in the supplying microchannel on the colony growth. Fig. 5(c) shows that ∼10% increase in cell growth occurred, when Qm increased from 0.05 to 0.11 ml/h. When the flow rate of medium was ∼0.02 ml/h, the viability of cells after 3 day culture reduced to 0%. At Qm ∼0.15 ml/h, the gels partly disintegrated due to a strong shear force exerted on the HMs by the liquid medium.
F. Culture of T-cells
The 2D array of HMs was also utilized for the encapsulation and culture of non-adherent T cells (EL4 cells). Fig. 6(a) (left, top and bottom) shows bright field optical microscopy images of one and two EL4 cells per HM, respectively, imaged 60 min after cell encapsulation. On day 3, individual cells formed colonies (Fig. 6(a), right). The presence of multiple cell nuclei was confirmed by staining them with Hoechst (blue fluorescence), as shown in Fig. 6(b). The viability of EL4 cells was examined by staining cells with Calcein AM (live stain, green fluorescence) and Ethidium Homodimer I (dead stain, red fluorescence), as shown in Fig. 6(b). The viability of cells in HMs at Cagar = 4 wt. % and Qm = 0.08 ml/h was ∼87% after 3 day culture. In the control experiments conducted with cells encapsulated in macroscopic agarose gels, a small fraction of dead cells (∼10%) in colonies was also found in 4 wt. % macroscopic agarose gels after 3 day culture (supplementary material, Fig. S350).
FIG. 6.
Viability and growth of T cells encapsulated in hydrogel modules. (a) Optical microscopy images of EL4 cells compartmentalized in HMs. Phase contrast images (left column, top and bottom) show HMs laden with one and two cells, respectively. Cagar = 4 wt. %. Phase contrast images (right column, top and bottom) show the same HMs after 3 day cell culture. Scale bars are 100 μm. The vertical black lines show the features of the microchannel below the wells (see supplementary material).50 (b) CFM images of EL4 cells shown in (a) after 3 day culture. Top and bottom images show growth of T cell colonies starting from one and two cells (top and bottom) shown in (a), respectively. The CFM images show (left to right) cell colonies stained with Hoechst 33342, Calcein AM (Cal AM), and Ethidium Homodimer I (EthD-I) and a merged image of cells stained by all three dyes. Nuclei staining by Hoechst 33342 shows the formation of cell colonies after 3 day culture. The concentration of cells in the suspension was 7 × 105 cell ml−1. Scale bars are 100 μm. Scale bars in the insets are 10 μm.
IV. DISCUSSION
The MF platform for the generation of 2D arrays of microscale artificial ECMs enables screening of their chemical and biophysical properties, as well as real-time studies of spatial constraints on cell behavior in their respective microenvironments. The distinct features of the platform include a uniform size of HMs (CV < 8%), the use of a small amount of reagents, suppressed evaporation of water, and an automated loading and analysis of samples. In the present work, the array of 500 cell-laden enumerated HMs with controllable mechanical properties and structures was produced within 90 min.
The applicability of the MF platform was demonstrated by analyzing cell viability and growth in HMs formed in comprehensively studied, well-characterized agarose hydrogels. Both adherent and non-adherent cells—NIH 3T3 fibroblast cells and EL4 T cells—showed high viability and growth. A large number of call-laden HMs with a stochastic distribution of encapsulated cells enabled time- and labour-efficient studies of the effect of inter-cell distance and the encapsulation rate on colony growth. The platform provided the capability to study cell fate on the level of individual cells, which is important in studies of rare cells and cell-cell interactions.26,56
Fibroblasts encapsulated in HMs exhibited high viability and proliferation rate after 3 day culture; however, they did not spread in the inherently inert agarose HMs, consistent with earlier work.42 Proliferation of cells could be prohibited by the lack of bioactive ligands in the supporting matrix.57 Presumably, due to the lack of cell binding sites, we also did not observe the effect of inter-cell distance on fibroblast colony growth. The platform was particularly advantageous for studies of the effect of hydrogel properties on cell fate. A large number of cell-laden HMs with identical gel compositions and properties in the same row provided statistically significant cell characterization, while HMs in parallel rows varied in agarose concentration, and thus in Young's modulus and structure of the hydrogel. Cell culture in HMs with a higher Cagar showed a decreased proliferation, which was consistent with earlier findings on the inhibited cell proliferation in matrices with a high stiffness.58,59 In addition, a larger pore size in the gels formed at a lower Cagar favored cell proliferation, thus enabling more cells to achieve 3D confluency.60–62 A poor viability of cells cultured at medium flow rate ≤ 0.02 ml/h was ascribed to the poor delivery of nutrients to cell-laden HMs and an insufficient removal of waste from the cells. The MF platform was also utilized for the fabrication of 2D arrays of agarose HMs encapsulating non-adherent T cells. After 3 day culture, T cells exhibited high viability and proliferation rate. The platform offered the capability for in situ monitoring of T cell migration in artificial ECMs, important for T cell activation, differentiation, survival, and interaction with target cells.63–66
A MF approach to the generation of 2D arrays of cell-laden HMs paves the way for efficient studies of cell fate in instructive artificial ECMs. Real-time imaging of cells and cell colonies encapsulated in HMs provided sufficient resolution for the determination of cell viability and spatial factors on cell growth. Imaging of cell-laden HMs by confocal fluorescence microscopy provided sufficient signal intensity to resolve individual cells and quantify the viability of individual cells and cell colonies. Moreover, real-time imaging of cells and colonies by optical microscopy enabled direct visualization and subsequent quantification of spatial factors, e.g., cell-to-cell distance, as well as the dimensions of individual cells and colonies, which would be otherwise challenging when overall fluorescence intensity of a large number of cells is utilized.
The platform offers the capability to acquire information on the role of spatial effects on individual cell behavior, e.g., the effect of the distance between the neighboring cells in HMs. While in macroscopic cell-laden gels, the results of such studies are generally averaged, by encapsulating a small number of cells into HMs and by varying the cell-to-cell distance within a limited range the effect of cell-cell interactions can be more accurately investigated. Furthermore, 2D HM arrays offer at least two advantages in the growth of cancer spheroids. Due to the uniform HM size distribution, the diffusion path length for the delivery of soluble molecular cues, e.g., drugs and nutrients to the cells, will be identical, which would provide quantitative, statistically significant data. Second, in-flow delivery of drugs to cancer spheroids allows mimicking drug delivery in vivo.
While agarose HMs were used for the proof of principle work, studies of other hydrogels of synthetic and biological nature are a clear next step for in-depth cell analysis. Another important direction of research on the level of individual cells includes studies of in vitro cell migration and cell-cell interactions in the respective HMs. The integration of 2D arrays of cell-laden HMs with microplate readers would make the MF platform particularly beneficial in fundamental cell biology research, drug discovery, and bioassay validation.
V. CONCLUSION
In conclusion, we have demonstrated MF generation of 2D arrays of uniform, micrometer-size hydrogel modules for cell encapsulation and culture. Advantages of the platform include the uniform dimensions of the modules, the use of small reagent volumes, suppressed evaporation of water, and an automated loading and analysis of samples. Due to the large number of cell-laden modules and stochastic distribution of cells in the modules, cell studies can be conducted in a time- and labor efficient manner. Both NIH 3T3 fibroblast cells and EL4 T cells showed good viability and proliferation in hydrogel modules after 3 day culture. The platform was used—on the level of individual cells—for studies of the effect of spatial constraints on cell growth. The 2D array of hydrogel modules was also used to examine the role of structural and mechanical properties of the hydrogel on cell growth.
The growth of cell colonies is an important factor determining cell fate. It plays a vital role in, e.g., stem cell proliferation and differentiation,67 tumorigenicity,55 intercellular signaling,68 and mediation of cell morphology.69 The application of the platform in cell culture, starting from a single cell or a very small number of cells, will provide valuable information on colony growth in real time.
Other types of biopolymers, e.g., collagen and gelatin and synthetic polymers, as well as polymer mixtures can be used for the microfluidic generation of 2D arrays of HMs. The mechanism of gelation can include thermosetting, as in the current work, or photo- and thermocrosslinking of polymers or monomers. The requirement is that polymer gelation occurs after the formation of precursor droplets. The platform can also be used for the exploration of the effect of chemical and biophysical cues on individual cells, studies of in vitro cell migration, and the examination of cell-ECM and cell-cell interactions by co-encapsulating different cells in the hydrogel modules.
In the present work, we used a five-row microwell MF device and tested five different conditions of cell culture in HMs. In principle, the described platform can utilize up to 20 000 HMs per chip with dimensions of 5 cm × 8 cm. In this situation, tens to hundreds of independent conditions of cell culture can be investigated on a single chip.
ACKNOWLEDGMENTS
The authors thank NSERC Canada and CHIR Canada for financial support of this work. Y.W. acknowledges Microfluidic Applications and Training in Cardiovascular Health (MATCH) Scholarship. Y.L. acknowledges Banting Postdoctoral Fellowship. The ability to use the instrumentation in the Centre for Microfluidic Systems in Chemistry and Biology (University of Toronto) is highly appreciated. The authors declare no competing financial interests.
References
- 1. Vogel V. and Sheetz M., Nat. Rev. Mol. Cell Biol. 7, 265 (2006). 10.1038/nrm1890 [DOI] [PubMed] [Google Scholar]
- 2. Watt F. M. and Huck W. T. S., Nat. Rev. Mol. Cell Biol. 14, 467 (2013). 10.1038/nrm3620 [DOI] [PubMed] [Google Scholar]
- 3. Moore K. A. and Lemischka I. R., Science 311, 1880 (2006). 10.1126/science.1110542 [DOI] [PubMed] [Google Scholar]
- 4. Lutolf M. P. and Hubbell J. A., Nat. Biotechnol. 23, 47 (2005). 10.1038/nbt1055 [DOI] [PubMed] [Google Scholar]
- 5. Lutolf M. P., Gilbert P. M., and Blau H. M., Nature 462, 433 (2009). 10.1038/nature08602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Discher D. E., Mooney D. J., and Zandstra P. W., Science 324, 1673 (2009). 10.1126/science.1171643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee S. H., Moon J. J., and West J. L., Biomaterials 29, 2962 (2008). 10.1016/j.biomaterials.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nakayama K. H., Hou L., and Huang N. F., Adv. Healthcare Mater. 3, 628 (2014). 10.1002/adhm.201300620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J., Abdeen A. A., Zhang D., and Kilian K. A., Biomaterials 34, 8140 (2013). 10.1016/j.biomaterials.2013.07.074 [DOI] [PubMed] [Google Scholar]
- 10. Kloxin A. M., Kasko A. M., Salinas C. N., and Anseth K. S., Science 324, 59 (2009). 10.1126/science.1169494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hahn M. S., Miller J. S., and West J. L., Adv. Mater. 18, 2679 (2006). 10.1002/adma.200600647 [DOI] [Google Scholar]
- 12. Drury J. L. and Mooney D. J., Biomaterials 24, 4337 (2003). 10.1016/S0142-9612(03)00340-5 [DOI] [PubMed] [Google Scholar]
- 13. Oelker A. M., Morey S. M., Griffith L. G., and Hammond P. T., Soft Matter 8, 10887 (2012). 10.1039/c2sm26487k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gillette B. M., Jensen J. A., Tang B., Yang G. J., Bazargan-Lari A., Zhong M., and Sia S. K., Nat. Mater. 7, 636 (2008). 10.1038/nmat2203 [DOI] [PubMed] [Google Scholar]
- 15. Peppas N. A., Hilt J. Z., Khademhosseini A., and Langer R., Adv. Mater. 18, 1345 (2006). 10.1002/adma.200501612 [DOI] [Google Scholar]
- 16. Phillippi J. A., Miller E., Weiss L., Huard J., Waggoner A., and Campbell P., Stem Cells 26, 127 (2008). 10.1634/stemcells.2007-0520 [DOI] [PubMed] [Google Scholar]
- 17. Miller E. D., Fisher G. W., Weiss L. E., Walker L. M., and Campbell P. G., Biomaterials 27, 2213 (2006). 10.1016/j.biomaterials.2005.10.021 [DOI] [PubMed] [Google Scholar]
- 18. Golden A. P. and Tien J., Lab Chip 7, 720 (2007). 10.1039/b618409j [DOI] [PubMed] [Google Scholar]
- 19. Panda P., Ali S., Lo E., Chung B. G., Hatton T. A., Khademhosseini A., and Doyle P. S., Lab Chip 8, 1056 (2008). 10.1039/b804234a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nuttelman C. R., Anseth K. S., and Bryant S. J., J. Biomater. Sci., Polym. Ed. 11, 439 (2000). 10.1163/156856200743805 [DOI] [PubMed] [Google Scholar]
- 21. Tasoglu S. and Demirci U., Trends Biotechnol. 31, 10 (2013). 10.1016/j.tibtech.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chung B. G., Lee K.-H., Khademhosseini A., and Lee S.-H., Lab Chip 12, 45 (2012). 10.1039/C1LC20859D [DOI] [PubMed] [Google Scholar]
- 23. Li W., Lee S., Ma M., Kim S. M., Guye P., Pancoast J. R., Anderson D. G., Weiss R., Lee R. T., and Hammond P. T., Sci. Rep. 3, 2863 (2013). 10.1038/srep02863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rossow T., Heyman J. A., Ehrlicher A. J., Langhoff A., Weitz D. A., Haag R., and Seiffert S., J. Am. Chem. Soc. 134, 4983 (2012). 10.1021/ja300460p [DOI] [PubMed] [Google Scholar]
- 25. Tan W. H. and Takeuchi S., Adv. Mater. 19, 2696 (2007). 10.1002/adma.200700433 [DOI] [Google Scholar]
- 26. Tumarkin E., Tzadu L., Csaszar E., Seo M., Zhang H., Lee A., Peerani R., Purpura K., Zandstra P. W., and Kumacheva E., Integr. Biol. (Camb.) 3, 653 (2011). 10.1039/c1ib00002k [DOI] [PubMed] [Google Scholar]
- 27. Kumachev A., Greener J., Tumarkin E., Eiser E., Zandstra P. W., and Kumacheva E., Biomaterials 32, 1477 (2011). 10.1016/j.biomaterials.2010.10.033 [DOI] [PubMed] [Google Scholar]
- 28. Taly V., Pekin D., El Abed A., and Laurent-Puig P., Trends Mol. Med. 18, 405 (2012). 10.1016/j.molmed.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 29. Bardiya N., Choi J.-W., and Chang S.-I., BioChip J. 8, 15 (2014). 10.1007/s13206-014-8103-5 [DOI] [Google Scholar]
- 30. Shi W., Qin J., Ye N., and Lin B., Lab Chip 8, 1432 (2008). 10.1039/b808753a [DOI] [PubMed] [Google Scholar]
- 31. Shim J., Olguin L. F., Whyte G., Scott D., Babtie A., Abell C., Huck W. T. S., and Hollfelder F., J. Am. Chem. Soc. 131, 15251 (2009). 10.1021/ja904823z [DOI] [PubMed] [Google Scholar]
- 32. Shen F., Du W., Kreutz J. E., Fok A., and Ismagilov R. F., Lab Chip 10, 2666 (2010). 10.1039/c004521g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jackman R. J., Duffy D. C., Ostuni E., Willmore N. D., and Whitesides G. M., Anal. Chem. 70, 2280 (1998). 10.1021/ac971295a [DOI] [PubMed] [Google Scholar]
- 34. Boedicker J. Q., Vincent M. E., and Ismagilov R. F., Angew. Chem., Int. Ed. Engl. 48, 5908 (2009). 10.1002/anie.200901550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Du W., Li L., Nichols K. P., and Ismagilov R. F., Lab Chip 9, 2286 (2009). 10.1039/b908978k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li L., Du W., and Ismagilov R., J. Am. Chem. Soc. 132, 106 (2010). 10.1021/ja908555n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmitz C. H. J., Rowat A. C., Köster S., and Weitz D. A., Lab Chip 9, 44 (2009). 10.1039/B809670H [DOI] [PubMed] [Google Scholar]
- 38. Lii J., Hsu W.-J., Parsa H., Das A., Rouse R., and Sia S. K., Anal. Chem. 80, 3640 (2008). 10.1021/ac8000034 [DOI] [PubMed] [Google Scholar]
- 39. Cohen D. E., Schneider T., Wang M., and Chiu D. T., Anal. Chem. 82, 5707 (2010). 10.1021/ac100713u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gansen A., Herrick A. M., Dimov I. K., Lee L. P., and Chiu D. T., Lab Chip 12, 2247 (2012). 10.1039/c2lc21247a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen S. S., Fitzgerald W., Zimmerberg J., Kleinman H. K., and Margolis L., Stem Cells 25, 553 (2007). 10.1634/stemcells.2006-0419 [DOI] [PubMed] [Google Scholar]
- 42. Rahman N., Purpura K. A., Wylie R. G., Zandstra P. W., and Shoichet M. S., Biomaterials 31, 8262 (2010). 10.1016/j.biomaterials.2010.07.040 [DOI] [PubMed] [Google Scholar]
- 43. Luo Y. and Shoichet M. S., Nat. Mater. 3, 249 (2004). 10.1038/nmat1092 [DOI] [PubMed] [Google Scholar]
- 44. Kumachev A., Tumarkin E., Walker G. C., and Kumacheva E., Soft Matter 9, 2959 (2013). 10.1039/c3sm27400d [DOI] [Google Scholar]
- 45. Butcher D. T., Alliston T., and Weaver V. M., Nat. Rev. Cancer 9, 108 (2009). 10.1038/nrc2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Levental K. R., Yu H., Kass L., Lakins J. N., Erler J. T., Fong S. F. T., Csiszar K., Giaccia A., Yamauchi M., Gasser D. L., and Weaver V. M., Cell 139, 891 (2009). 10.1016/j.cell.2009.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roberts C. C., Rao R. R., Loewenberg M., Brooks C. F., Galambos P., Grillet A. M., and Nemer M. B., Lab Chip 12, 1540 (2012). 10.1039/c2lc21197a [DOI] [PubMed] [Google Scholar]
- 48. Shirbin S. J., Ladewig K., Fu Q., Klimak M., Zhang X., Duan W., and Qiao G. G., Biomacromolecules 16, 2463 (2015). 10.1021/acs.biomac.5b00692 [DOI] [PubMed] [Google Scholar]
- 49. Liu W., Li L., Wang X., Ren L., Wang X., Wang J., Tu Q., Huang X., and Wang J., Lab Chip 10, 1717 (2010). 10.1039/c001049a [DOI] [PubMed] [Google Scholar]
- 50.See supplementary material at http://dx.doi.org/10.1063/1.4940430E-BIOMGB-10-009601 for experimental design of the microfluidic platform, viability of fibroblasts in macroscopic agarose gels, and viability of EL4 T cells in macroscopic agarose gels.
- 51. Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angilè F. E., Schmitz C. H. J., Köster S., Duan H., Humphry K. J., Scanga R. A., Johnson J. S., Pisignano D., and Weitz D. A., Lab Chip 8, 1632 (2008). 10.1039/b806706f [DOI] [PubMed] [Google Scholar]
- 52. Horger K. S., Estes D. J., Capone R., and Mayer M., J. Am. Chem. Soc. 131, 1810 (2009). 10.1021/ja805625u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Thompson A. M., Paguirigan A. L., Kreutz J. E., Radich J. P., and Chiu D. T., Lab Chip 14, 3135 (2014). 10.1039/C4LC00175C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hur S. C., Mach A. J., and Di Carlo D., Biomicrofluidics 5, 22206 (2011). 10.1063/1.3576780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liu J., Tan Y., Zhang H., Zhang Y., Xu P., Chen J., Poh Y.-C., Tang K., Wang N., and Huang B., Nat. Mater. 11, 734 (2012). 10.1038/nmat3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhu Z., Zhang W., Leng X., Zhang M., Guan Z., Lu J., and Yang C. J., Lab Chip 12, 3907 (2012). 10.1039/c2lc40461c [DOI] [PubMed] [Google Scholar]
- 57. Lin C. and Anseth K. S., Proc. Natl. Acad. Sci. U. S. A. 108, 6380 (2011). 10.1073/pnas.1014026108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bott K., Upton Z., Schrobback K., Ehrbar M., Hubbell J. A., Lutolf M. P., and Rizzi S. C., Biomaterials 31, 8454 (2010). 10.1016/j.biomaterials.2010.07.046 [DOI] [PubMed] [Google Scholar]
- 59. Ulrich T. A., Jain A., Tanner K., MacKay J. L., and Kumar S., Biomaterials 31, 1875 (2010). 10.1016/j.biomaterials.2009.10.047 [DOI] [PubMed] [Google Scholar]
- 60. Mandal B. B. and Kundu S. C., Biomaterials 30, 2956 (2009). 10.1016/j.biomaterials.2009.02.006 [DOI] [PubMed] [Google Scholar]
- 61. Mygind T., Stiehler M., Baatrup A., Li H., Zou X., Flyvbjerg A., Kassem M., and Bünger C., Biomaterials 28, 1036 (2007). 10.1016/j.biomaterials.2006.10.003 [DOI] [PubMed] [Google Scholar]
- 62. Takahashi Y., Yamamoto M., and Tabata Y., Biomaterials 26, 3587 (2005). 10.1016/j.biomaterials.2004.09.046 [DOI] [PubMed] [Google Scholar]
- 63. Overstreet M. G., Gaylo A., Angermann B. R., Hughson A., Hyun Y.-M., Lambert K., Acharya M., Billroth-Maclurg A. C., Rosenberg A. F., Topham D. J., Yagita H., Kim M., Lacy-Hulbert A., Meier-Schellersheim M., and Fowell D. J., Nat. Immunol. 14, 949 (2013). 10.1038/ni.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Friedl P. and Weigelin B., Nat. Immunol. 9, 960 (2008). 10.1038/ni.f.212 [DOI] [PubMed] [Google Scholar]
- 65. Weninger W., Biro M., and Jain R., Nat. Rev. Immunol. 14, 232 (2014). 10.1038/nri3641 [DOI] [PubMed] [Google Scholar]
- 66. Dustin M. L. and de Fougerolles A. R., Curr. Opin. Immunol. 13, 286 (2001). 10.1016/S0952-7915(00)00217-X [DOI] [PubMed] [Google Scholar]
- 67. Peerani R., Rao B. M., Bauwens C., Yin T., Wood G. A., Nagy A., Kumacheva E., and Zandstra P. W., EMBO J. 26, 4744 (2007). 10.1038/sj.emboj.7601896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lin R.-Z., Moreno-Luna R., Li D., Jaminet S.-C., Greene A. K., and Melero-Martin J. M., Proc. Natl. Acad. Sci. U. S. A. 111, 10137 (2014). 10.1073/pnas.1405388111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Maldonado M., Wong L. Y., Echeverria C., Ico G., Low K., Fujimoto T., Johnson J. K., and Nam J., Biomaterials 50, 10 (2015). 10.1016/j.biomaterials.2015.01.037 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1063/1.4940430E-BIOMGB-10-009601 for experimental design of the microfluidic platform, viability of fibroblasts in macroscopic agarose gels, and viability of EL4 T cells in macroscopic agarose gels.