Abstract
One of the objectives of genome science is the discovery and accurate annotation of all protein-coding genes. Proteogenomics has emerged as a methodology that provides orthogonal information to traditional forms of evidence used for genome annotation. By this method, peptides that are identified via tandem mass spectrometry are used to refine protein-coding gene models. Namely, these peptides are used to confirm the translation of predicted protein-coding genes, as evidence of novel genes or for correction of current gene models. Proteogenomics requires deep and broad sampling of the proteome in order to generate sufficient numbers of unique peptides. Therefore, we propose that proteogenomic projects are designed so that the generated peptides can also be used to create a comprehensive protein atlas that quantitatively catalogues protein abundance changes during development and in response to environmental stimulus.
Keywords: Proteogenomics, Proteomics, Atlas, Annotation
1. Introduction
The primary goal of genome annotation efforts is the discovery and accurate annotation of all protein-coding genes. A complete and accurately annotated proteome provides the building blocks for hypothesis-driven research seeking to enhance our understanding of biology. Genome annotation is a complex process involving multiple integrated tools, which have been described in detail [1–5] and are beyond the scope of this review. Briefly, traditional methods of genome annotation rely on combining various forms of evidence. This includes de novo gene prediction, which utilizes only patterns in the genomic sequence to infer gene structure. Additionally, transcript sequences from cDNA libraries can be leveraged to enhance gene prediction. Lastly, sequence conservation with related species can be incorporated into annotation pipelines. While DNA/RNA-based genome annotation approaches perform remarkably well, given the complexity of the challenge, they are currently unable to accurately predict all protein coding genes and their structure. Experimental evidence is required to determine if a transcript is translated and if the predicted protein sequence is correct.
The field of proteogenomics has emerged as a genome-wide method to improve genome annotations as well as to characterize the pattern of gene expression at the protein level. The concept of proteogenomics was introduced, by Jaffe and colleagues [6], as a method that utilizes peptides identified from their tandem mass spectra, for genome annotation (reviewed by [2,7–9]). Since its introduction, proteogenomics has successfully aided in the annotation of numerous prokaryotic and eukaryotic organisms. These studies have demonstrated that deep and broad sampling of the proteome is necessary, for proteogenomics, requiring the generation of hundreds of millions of mass spectra. Furthermore, protein accumulation depends upon development and environmental conditions so spectra must be generated from a diverse set of samples to enable deep coverage of the proteome. Such broad sampling enables the additional use of the identified peptides for creation of a protein atlas that catalogs where, when, and how much of a given protein is present.
2. Proteogenomic enabled annotation
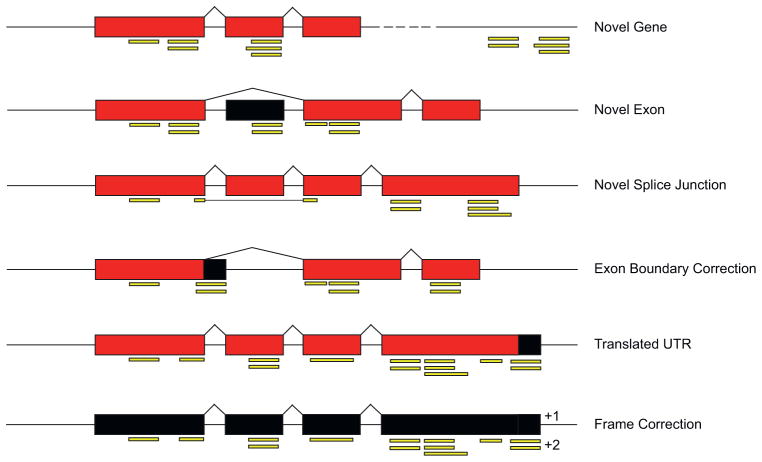
Proteogenomics provides a high-throughput method to incorporate protein level information into genome annotation. For this, tandem mass spectra are generated and then used to search genomic databases for peptide identification. The standard database utilized in proteogenomic pipelines is a six-frame translation of the genome [6]. Additionally, specialized types of databases such as an exon–splice graph, which is compact representation of predicted gene structures and splice junctions, have also been exploited [10]. The identified peptides fall into two categories. Namely, confirming peptides that match the current genome annotation and novel peptides, which do not (Fig. 1). It is important to emphasize that the confirming peptides represent critical events, as they directly confirm both the current structural annotation of a gene and demonstrate that the gene encodes a translated protein.
Fig. 1.
Examples of gene model revision. Currently annotated exons are shown in red. Gene model revision suggested by novel peptides is depicted in black. Proteogenomically identified peptides are shown in yellow.
The novel peptides themselves can be further divided into two types of events. One category includes intergenic peptides, which map outside of known genes, and thus reveal the presence of novel genes. A second category is intragenic peptides that fall within a known locus, but do not match the currently annotated gene model. Intragenic peptides include those demonstrating the translation of 5′ or 3′ untranslated regions (UTR), alternative start/stop sites, proteins out of frame, incorrect exon boundaries, novel exons or novel splice sites. While one may assume that the identification these types of novel intergenic and intragenic peptides by proteogenomics to be rare, they are actually commonly found, even in well annotated model organisms (i.e. organisms that have been subjected to multiple rounds of genome annotation) (Table 1). This demonstrates that proteogenomics is a necessary addition to any comprehensive genome annotation effort.
Table 1.
Proteogenomic publications in plants. (If Novel Genes and Model Revision were not clearly identified all values went into the Model Revision Column.)
| Organism | Peptides | Proteins | Novel peptides | Novel genes | Model revision | Citation |
|---|---|---|---|---|---|---|
| Arabidopsis thaliana | 86,456 | 13,029 | 261 | 22 | 35 | [28] |
| Arabidopsis thaliana | 144,079 | 12,769 | 18,024 | 778 | 695 | [13] |
| Populus deltoides | 4943 | 56 | [34] | |||
| Chlamydomonas reinhardtii | 9336 | 932 | 3 | 65 | [35] | |
| Oryza sativa | 15,121 | 5034 | 166 | 40 | [36] | |
| Medicago truncatula | 78,647 | 9843 | 1568 | 32 | 293 | [37] |
| Zea mays | 225,166 | 14,615 | 24,782 | 165 | 1904 | [38] |
| Triticum aestivum | 203 | 17 | 5 | 8 | [39] |
3. Proteome sampling for proteogenomics
Deep and broad sampling of the proteome is necessary for comprehensive proteogenomic efforts. There are numerous strategies that have been developed for proteogenomic experiments to aid in maximizing the number of unique peptides identified by mass spectrometry [7,9,11]. Briefly, fractionation methods such as one-dimensional and two-dimensional gel electrophoresis, as well as gel-free chromatography based separations of proteins and peptides, aid in deep proteome sampling. Specialized sample preparations can also be used to sample subsets of the proteome such as phosphoproteins, basic proteins, small proteins, and N-terminal peptides [7,8,12–14]. Additionally, use of multiple proteases (examples include trypsin, chymotrypsin, Glu-C, and Lsy-C) helps to increase the percentage of sequence covered for a given protein. Another consideration is that the proteome composition depends on both developmental and environmental factors. Thus, analyzing a diverse array of samples is critical for achieving comprehensive proteome coverage [12,13].
4. Proteome atlas
The extensive sampling required for a comprehensive proteogenomic project enables the dual use of the generated peptides for creation of a proteome atlas, which catalogues protein abundance throughout developmental time and/or in response to environmental stimulus. This type of catalogue is relatively common at the mRNA level, where extensive transcriptional atlases have been created for a range of plant species including Arabidopsis thaliana [15,16], barley [17], Oryza sativa [18,19], Medicago truncatula [20], Glycine max [21], Solanum tuberosum [22], Zea mays [23,24], Rosa chinensis [25], Vitis vinifera [26], and Lotus japonicus [27]. However, to our knowledge, there are only a handful of proteome atlas publications in plants, which we define as covering at least several thousand proteins from three or more cell-types and/or plant anatomical structures (Table 2) [28–31]. Well there are only a handful of proteome atlas publications there are several web-based resources including pep2pro [32] and MASCP Gator [33] that aggregate proteome datasets into a single information portal. Finally, an ideal comprehensive protein atlas would provide proteome-wide coverage and include multiple developmental stages, for each organ, as well as a range of environmental perturbations. While this is a daunting task, the ability to leverage the generated peptides for both proteogenomics, as well as building a protein atlas provides a considerable resource for the scientific community.
Table 2.
Plant protein atlas publications. To be considered a protein atlas publication we required the quantification of protein abundance for at least several thousand proteins across three or more cell-types and/or plant anatomical structures.
| Organism | Proteome coverage | Samples | Phosphorylation | Citation |
|---|---|---|---|---|
| Arabidopsis thaliana | 13,029 | Multiple developmental stages from roots, leaves, flowers and seeds | No | [28] |
| Arabidopsis thaliana | 1995 | Six root cell types | No | [29] |
| Zea mays | 14,165 | Aleurone/pericarp as well as multiple developmental stages of endosperm and embryo | Yes | [31] |
| Populus tremula × alba | 7538 | Leaf, root and stem | No | [30] |
5. Perspective
Since its inception a decade ago proteogenomics has matured into a robust methodology, thanks in large part to rapid advances in mass spectrometry based proteomics. It is now possible to deeply sample the proteome identifying millions of mass spectra and hundreds of thousands of unique peptides. These unique peptides provide rich fodder not only for genome annotation but also for building protein atlases. Thus, in an ideal scenario all genome annotation pipelines would include proteogenomics and the proteogenomic component would be designed to enable the creation of a quantitative protein atlas.
Acknowledgments
This work was supported by National Science Foundation Grant 0924023 (to S.P.B.) and a National Institutes of Health National Research Service Award Postdoctoral Fellowship F32GM096707 (to J.W.W.).
References
- 1.Curwen V, Eyras E, Andrews TD, Clarke L, Mongin E, Searle SMJ, Clamp M. The Ensembl automatic gene annotation system. Genome Res. 2004;14:942–950. doi: 10.1101/gr.1858004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansong C, Purvine SO, Adkins JN, Lipton MS, Smith RD. Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief Funct Genomics Proteomics. 2008;7:50–62. doi: 10.1093/bfgp/eln010. [DOI] [PubMed] [Google Scholar]
- 3.Brent MR. Steady progress and recent breakthroughs in the accuracy of automated genome annotation. Nat Rev Genet. 2008;9:62–73. doi: 10.1038/nrg2220. [DOI] [PubMed] [Google Scholar]
- 4.Liang C, Mao L, Ware D, Stein L. Evidence-based gene predictions in plant genomes. Genome Res. 2009;19:1912–1923. doi: 10.1101/gr.088997.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yandell M, Ence D. A beginner’s guide to eukaryotic genome annotation. Nat Rev Genet. 2012;13:329–342. doi: 10.1038/nrg3174. [DOI] [PubMed] [Google Scholar]
- 6.Jaffe JD, Berg HC, Church GM. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- 7.Armengaud J. A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr Opin Microbiol. 2009;12:292–300. doi: 10.1016/j.mib.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 8.Castellana N, Bafna V. Proteogenomics to discover the full coding content of genomes: a computational perspective. J Proteomics. 2010;73:2124–2135. doi: 10.1016/j.jprot.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renuse S, Chaerkady R, Pandey A. Proteogenomics. Proteomics. 2011;11:620–630. doi: 10.1002/pmic.201000615. [DOI] [PubMed] [Google Scholar]
- 10.Tanner S, Shen Z, Ng J, Florea L, Guigo R, Briggs SP, Bafna V. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krug K, Nahnsen S, Macek B. Mass spectrometry at the interface of proteomics and genomics. Mol Biosyst. 2011;7:284–291. doi: 10.1039/c0mb00168f. [DOI] [PubMed] [Google Scholar]
- 12.Brunner E, et al. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol. 2007;25:576–583. doi: 10.1038/nbt1300. [DOI] [PubMed] [Google Scholar]
- 13.Castellana NE, Payne SH, Shen Z, Stanke M, Bafna V, Briggs SP. Discovery and revision of Arabidopsis genes by proteogenomics. Proc Natl Acad Sci U S A. 2008;105:21034–21038. doi: 10.1073/pnas.0811066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallien S, Perrodou E, Carapito C, Deshayes C, Reyrat JM, Van Dorsselaer A, Poch O, Schaeffer C, Lecompte O. Orthoproteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res. 2009;19:128–135. doi: 10.1101/gr.081901.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGA-TORArabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Druka A, et al. An atlas of gene expression from seed to seed through barley development. Funct Integr Genomics. 2006;6:202–211. doi: 10.1007/s10142-006-0025-4. [DOI] [PubMed] [Google Scholar]
- 18.Jiao Y, et al. A transcriptome atlas of rice cell types uncovers cellular, functional and developmental hierarchies. Nat Genet. 2009;41:258–263. doi: 10.1038/ng.282. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, Zhang Q. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010;61:752–766. doi: 10.1111/j.1365-313X.2009.04100.x. [DOI] [PubMed] [Google Scholar]
- 20.Benedito VA, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 21.Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. An integrated transcriptome atlas of the crop model Glycine max and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- 22.Massa AN, Childs KL, Lin H, Bryan GJ, Giuliano G, Buell CR. The transcriptome of the reference potato genome Solanum tuberosum Group Phureja clone DM1-3 516R44. PLoS ONE. 2011;6:e26801. doi: 10.1371/journal.pone.0026801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. Genome-wide atlas of transcription during maize development. Plant J. 2011;66:553–563. doi: 10.1111/j.1365-313X.2011.04527.x. [DOI] [PubMed] [Google Scholar]
- 24.Sekhon RS, Briskine R, Hirsch CN, Myers CL, Springer NM, Buell CR, de Leon N, Kaeppler SM. Maize gene atlas developed by RNA sequencing and comparative evaluation of transcriptomes based on RNA sequencing and microarrays. PLOS ONE. 2013;8:e61005. doi: 10.1371/journal.pone.0061005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois A, et al. Transcriptome database resource and gene expression atlas for the rose. BMC Genomics. 2012;13:638. doi: 10.1186/1471-2164-13-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasoli M, et al. The grapevine expression atlas reveals a deep transcriptome shift driving the entire plant into a maturation program. Plant Cell. 2012;24:3489–3505. doi: 10.1105/tpc.112.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verdier J, Torres-Jerez I, Wang M, Andriankaja A, Allen SN, He J, Tang Y, Murray JD, Udvardi MK. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 2013;74:351–362. doi: 10.1111/tpj.12119. [DOI] [PubMed] [Google Scholar]
- 28.Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 29.Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, Benfey PN. The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci U S A. 2012;109:6811–6818. doi: 10.1073/pnas.1202546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abraham P, Giannone RJ, Adams RM, Kalluri U, Tuskan GA, Hettich RL. Putting the pieces together: high-performance LC–MS/MS provides network-, pathway-, and protein-level perspectives in Populus. Mol Cell Proteomics. 2013;12:106–119. doi: 10.1074/mcp.M112.022996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walley JW, Shen Z, Sartor R, Wu KJ, Osborn J, Smith LG, Briggs SP. Reconstruction of protein networks from an atlas of maize seed proteotypes. Proc Natl Acad Sci U S A. 2013;110:E4808–E4817. doi: 10.1073/pnas.1319113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baerenfaller K, Hirsch-Hoffmann M, Svozil J, Hull R, Russenberger D, Bischof S, Lu Q, Gruissem W, Baginsky S. pep2pro: a new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr Biol. 2011;3(3):225–237. doi: 10.1039/c0ib00078g. [DOI] [PubMed] [Google Scholar]
- 33.Joshi HJ, et al. MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 2011;155:259–270. doi: 10.1104/pp.110.168195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, et al. Discovery and annotation of small proteins using genomics proteomics, and computational approaches. Genome Res. 2011;21:634–641. doi: 10.1101/gr.109280.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Specht M, Stanke M, Terashima M, Naumann-Busch B, Janßen I, Höhner R, Hom EFY, Liang C, Hippler M. Concerted action of the new Genomic Peptide Finder and AUGUSTUS allows for automated proteogenomic annotation of the Chlamydomonas reinhardtii genome. Proteomics. 2011;11:1814–1823. doi: 10.1002/pmic.201000621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helmy M, Tomita M, Ishihama Y. OryzaPG-DB: rice proteome database based on shotgun proteogenomics. BMC Plant Biol. 2011;11:63. doi: 10.1186/1471-2229-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkening JD, Bailey DJ, Rose CM, Grimsrud PA, Howes-Podoll M, Venkateshwaran M, Westphall MS, Ané JM, Coon JJ, Sussman MR. A proteogenomic survey of the Medicago truncatula genome. Mol Cell Proteomics. 2012;11:933–944. doi: 10.1074/mcp.M112.019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castellana NE, Shen Z, He Y, Walley JW, Cassidy CJ, Briggs SP, Bafna V. An automated proteogenomic method uses mass spectrometry to reveal novel genes in Zea mays. Mol Cell Proteomics. 2014;13:157–167. doi: 10.1074/mcp.M113.031260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer KFX, et al. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]