Abstract
Incidence rates of Parkinson’s disease (PD) are higher in men than women at all ages, and these differences may be due to the neuroprotective effects of estrogen on the nigrostriatal pathway. We investigated the association of common variants in four estrogen-related genes with PD. Tagging single nucleotide polymorphisms (SNPs) in the CYP19A1, ESR1, ESR2, and PRDM2 genes were selected from the International Haplotype Map and genotyped in 1,103 PD cases from the Upper Midwest, USA and in 1,103 individually matched controls (654 unaffected siblings, and 449 unrelated controls from the same region). Out of 137 informative SNPs, two PRDM2 SNPs were significantly associated with an increased risk of PD at the Bonferroni-corrected significance level of 0.0004 (rs2744690: OR = 1.54, 99.96% CI = 1.05 – 2.26, uncorrected P = 0.0001; rs2744687: OR = 1.53, 99.96% CI = 1.03 – 2.29, uncorrected P = 0.0002); the association was significant in the women only stratum but not in the men only stratum. An additional six SNPs in PRDM2, two in ESR1, one in ESR2, and one in CYP19A1 had significant P-values in the overall sample before Bonferroni correction. None of the SNPs were significantly associated with age at onset of PD after Bonferroni correction. Our results confirm the association of PRDM2 variants with PD susceptibility, especially in women.
INTRODUCTION
Parkinson’s disease (PD) is a chronic progressive neurodegenerative disorder that affects 2% of men and 1.3% of women during their life.1 Epidemiological studies report higher incidence rates of PD in men compared with women at all ages,2, 3 which may be due to the neuroprotective role of female sex hormones such as estrogen.
There is evidence for a neuroprotective effect of estrogen on the nigrostriatal pathway in mice.4–15 The neuroprotective properties of estrogen have also been demonstrated in primate models.16 Those laboratory findings are consistent with clinical and epidemiologic studies documenting possible neuroprotective roles of estrogen.17–19 We recently reported that both unilateral and bilateral oophorectomy performed prior to menopause may be associated with an increased risk of PD.20
Common variants of the estrogen receptor 1 gene (ESR1) are associated with earlier age at natural menopause and increased risk of surgical menopause.21 We previously failed to observe significant associations of two estrogen receptor gene variants with PD susceptibility in a smaller study.22 However, our genome-wide association study highlighted a significant association of the single-nucleotide polymorphism (SNP) rs2245218 in the estrogen-related PRDM2 gene with PD susceptibility.23 We expanded the scope of our earlier genetic studies to include multiple SNP variants in four estrogen-related genes in a much larger sample.
SUBJECTS AND METHODS
Cases and Controls
We conducted case-control (PD susceptibility) and case-only (age at onset) analyses. Cases were patients with PD referred sequentially to the Department of Neurology of the Mayo Clinic in Rochester, MN, from June 1, 1996 through June 30, 2007, who resided in the 5-state region including Minnesota, Wisconsin, Iowa, North Dakota, and South Dakota. The diagnosis of PD was made by a movement disorder specialist using previously reported criteria.2 Control subjects consisted primarily of unaffected siblings of PD cases who screened negative for PD or parkinsonism via telephone interview, or siblings who screened positive but were free of parkinsonism at clinical examination.24, 25 Cases were matched to a single participating sibling first by sex (when possible) and then by closest age. Cases without an available sibling were matched to unrelated controls living in the same 5-state region and of same sex and age (same year of birth ± 2 years). Controls of age 65 or older were randomly selected for contact from the Centers for Medicare and Medicaid Services (CMS) lists, while those younger than 65 years were selected for contact using random digit dialing, according to standard techniques.26, 27
Because unrelated controls screening positive for PD or parkinsonism could not be examined as part of the study, they were excluded from the list of potential controls. All examinations (cases and siblings screening positive) were performed in a standardized fashion by neurologists specialized in Movement Disorders, and employing a protocol for clinical assessment. The Institutional Review Board of the Mayo Clinic approved the study, and all subjects provided written informed consent.
Genotyping
For all subjects, genomic DNA was collected, extracted, and stored as previously described.28, 29 We studied the estrogen receptor 1 (ESR1) and estrogen receptor 2 (ESR2) genes because they encode the receptors to which estrogen binds. The genes that estrogens regulate via their cognate receptors are considered to include apoptotic/antiapoptotic genes, neurotrophins and growth factors, and genes that mediate structural alterations in neurons and synaptogenesis.30 We studied the cytochrome P450, family 19, subfamily A, polypeptide 1 (CYP19A1) gene because it encodes the rate-limiting step in estrogen metabolism (the enzyme aromatase). Finally, we studied the PR domain containing 2, with ZNF domain (PRDM2) gene in light of our prior genome-wide association study findings and because it encodes a protein that binds to the estrogen receptor and functions as a specific effector of estrogen action. SNPs were selected for these 4 estrogen-related genes using the International HapMap Project unrelated Centre d’Etude du Polymorphisme Humain collection (CEPH) samples.31
The LDSelect program was used to identify tag SNPs using a linkage disequilibrium (LD) r2 threshold of 0.8 and with minor allele frequencies of 5% or higher. Two tag SNPs were selected for each LD bin when the number of SNPs in the bin was 10 or more. We excluded SNPs with Illumina platform design scores < 0.4 and those within 60 bp of another SNP that had already been chosen.
We genotyped 141 SNPs using an Illumina GoldenGate custom SNP panel (Illumina Inc., San Diego, California, USA). This included 38 SNPs in CYP19A1, 58 SNPs in ESR1, 18 SNPs in ESR2, and 27 SNPS in PRDM2. Four SNPs failed genotyping (one in CYP19A1, one in ESR1, and two in PRDM2), whereas 137 SNPs were successfully genotyped and included in our analyses. The average call rate for these SNPs was 99.1%. Quality control was monitored by inclusion of DNA from a CEPH family trio in each plate (Father, mother, daughter from CEPH/UTAH pedigree 1347 from the Coreill Institute for Medical Research); concordance was 100%.
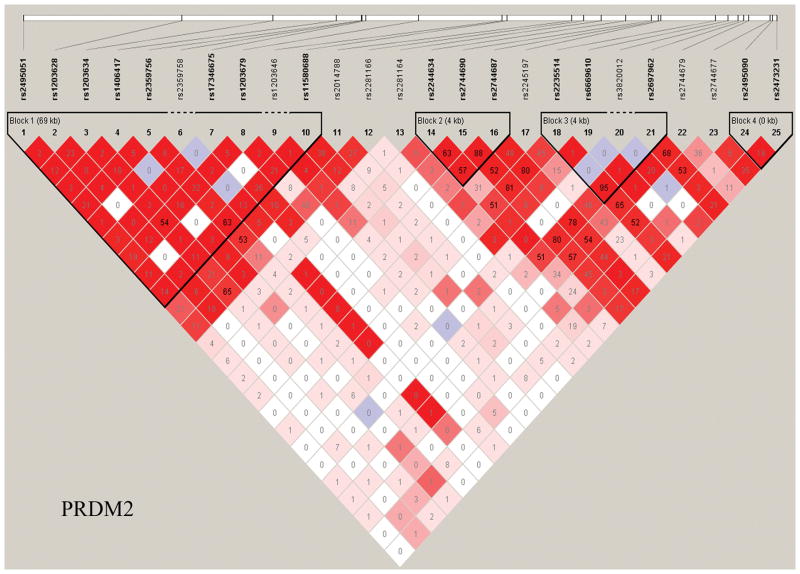
Haploview was used to generate LD maps for each gene using data from the controls.32 Only variants with minor allele frequencies > 0.01 and in Hardy-Weinberg equilibrium (P > 0.001) were included. Figure 1 shows the LD map for the PRDM2 gene. Supplementary Figure 1 provides the LD maps for all four genes.
Figure 1. Linkage disequilibrium (LD) maps.
Haplotype blocks of the PRDM2 gene. The LD values measured using r2 are given by numbers and the LD values measured by D′ are shown by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data).
Statistical Analyses
In the case-control analyses, we studied the association of each genetic variant with PD susceptibility in the overall sample, using conditional logistic regression with a log additive genotype coding scheme.33 We also performed analyses using dominant or recessive coding schemes. A log additive coding specifies that the log odds ratio for heterozygotes falls midway between the log odds ratios for the homozygotes, whereas dominant or recessive coding combines the heterozygotes with one or the other of the homozygote groups. The analyses were adjusted for age at study (continuous variable) and sex to remove possible residual confounding. For each genetic variant, we calculated an odds ratio (OR), a 95% confidence interval (CI), and a two-tailed P value. In addition, we performed similar analyses separately in strata defined by type of control (case-unaffected sibling pairs versus case-unrelated control pairs), sex, and age at onset of PD. Analyses in the sex-specific strata excluded 216 sib pairs of discordant sex, since not every case had an available sibling of the same sex.
In the case-only analyses, we studied the association of each genetic variant with age at onset of PD using Cox proportional hazard models and the same coding schemes described earlier.34 The analyses were adjusted for sex. For each genetic variant we calculated a hazard ratio (HR), a 95% CI, and a two-tailed P value. We performed similar analyses of age at onset of PD in men and women separately.
The P values from primary analyses were assessed for significance using a Bonferroni corrected significance level of 0.05/137=0.0004. However, because the Bonferroni correction may be conservative due to residual LD between the selected tag-SNPs, we also used permutation techniques to correct for multiple testing in the primary analyses.35 Case-control status was randomly permuted within matched pairs and each permuted data set was re-analyzed. This procedure was repeated 5000 times. Correction for multiple testing of each p-value in the original data analysis was achieved by counting the proportion of permutations in which at least one of the SNPs had a smaller p-value. The statistical packages SAS® (version 9.1; SAS Institute Inc., Cary, NC) and S-Plus® (version 8.0.1; MathSoft, Seattle, WA) were used for all analyses.
RESULTS
Sample
There were 1,103 cases and 1,103 controls included in the study (654 case-unaffected sibling pairs and 449 case-unrelated control pairs). There were 555 men-men pairs (290 case-sibling pairs and 265 case-unrelated control pairs), 332 women-women pairs (173 case-sibling pairs and 159 case-unrelated control pairs), and 216 men-women pairs (case-sibling pairs only). Patients with PD were more often men (64.1%) than women (35.9%). Approximately 17% of cases reported having a first degree relative with PD. The demographic characteristics of the sample are summarized in Table 1.
Table 1.
Demographic Characteristics of Parkinson’s Disease (PD) Cases, Unaffected Siblings, and Unrelated Controls
| General Characteristics | PD Case-Sibling Pairs
|
PD Case-Unrelated Control Pairs
|
All PD Case-Control Pairs
|
|||
|---|---|---|---|---|---|---|
| PD Cases | Sibling Controls | PD Cases | Unrelated Controls | PD Cases | All Controls | |
| Total sample, n | 654 | 654 | 449 | 449 | 1,103 | 1,103 |
| Men, n (%) | 417 (63.8) | 329 (50.3) | 290 (64.6) | 290 (64.6) | 707 (64.1) | 619 (56.1) |
| Women, n (%) | 237 (36.2) | 325 (49.7) | 159 (35.4) | 159 (35.4) | 396 (35.9) | 484 (43.9) |
| Age at onset of PD, median year (range) | 60.4 (28.2 – 86.9) | — | 64.7 (23.3 – 88.0) | — | 62.2 (23.3 – 88.0) | — |
| Age at study, median year (range)a | 66.3 (30.8 – 91.4) | 65.1 (32.0 – 90.4) | 70.3 (44.5 – 90.4) | 71.8 (44.9 – 92.8) | 68.0 (30.8 – 91.4) | 67.6 (32.0 – 92.8) |
| Duration of PD, median year (range) | 3.2 (0.1 – 23.6) | — | 3.9 (0 – 54.2) | — | 3.4 (0 – 54.2) | — |
| Region of origin of parentsb | ||||||
| Both parents of European origin, n (%) | 570 (87.2) | 557 (85.2) | 361 (80.4) | 391 (87.1) | 931 (84.4) | 948 (85.9) |
| Both parents Northern European, n (%) | 154 (27.0) | 148 (26.6) | 116 (32.1) | 126 (32.2) | 270 (29.0) | 274 (28.9) |
| Both parents Central European, n (%) | 233 (40.9) | 219 (39.3) | 119 (33.0) | 124 (31.7) | 352 (37.8) | 343 (36.2) |
| Both parents Southern European, n (%) | 3 (0.5) | 3 (0.5) | 3 (0.8) | 4 (1.0) | 6 (0.6) | 7 (0.7) |
| Both parents European, mixed region, n (%) | 180 (31.6) | 187 (33.6) | 123 (34.1) | 137 (35.0) | 303 (32.5) | 324 (34.2) |
| Only one parent of European origin, n (%)c | 53 (8.1) | 61 (9.3) | 60 (13.4) | 41 (9.1) | 113 (10.2) | 102 (9.2) |
| One parent declared “American”, n (%)d | 2 (0.3) | 1 (0.2) | 1 (0.2) | 4 (0.9) | 3 (0.3) | 5 (0.5) |
| Both parents declared “American”, n (%)d | 19 (2.9) | 20 (3.1) | 13 (2.9) | 7 (1.6) | 32 (2.9) | 27 (2.4) |
| Both parents Asian, n (%) | 3 (0.5) | 3 (0.5) | 5 (1.1) | 0 (0.0) | 8 (0.7) | 3 (0.3) |
| Both parents Mexican, n (%) | 1 (0.2) | 1 (0.2) | 1 (0.2) | 1 (0.2) | 2 (0.2) | 2 (0.2) |
| Unknown, n (%) | 6 (0.9) | 11 (1.7) | 8 (1.8) | 5 (1.1) | 14 (1.3) | 16 (1.5) |
Age at blood draw.
Self-reported by subjects. “Northern European” includes Scandinavian, Swedish, Norwegian, Finnish, Danish, Irish, or British origins. “Central European” includes French, Belgian, Dutch, Swiss, Luxemburgian, German, Austrian, Hungarian, Polish, Czechoslovakian, or Russian origins. “Southern European” includes Italian, Spanish, Portuguese, Greek, or Yugoslavian origins.
Includes subjects for whom origin of one parent is unknown.
These subjects were all Caucasians and not Native Americans.
Case-Control Analyses of Genetic Variants and PD Susceptibility
For the overall sample, 12 of the 137 SNPs were associated with PD susceptibility at an uncorrected significance level of 0.05, with a log-additive model for allele effects (Table 2). These included eight SNPs in PRDM2, two SNPs in ESR1, one SNP in CYP19A1, and one SNP in ESR2. The main effects of these variants were modest, with ORs ranging between 0.75 and 1.54. Two SNPs in the PRDM2 gene remained significantly associated with PD susceptibility when using the Bonferroni corrected significance level of 0.0004 (rs2744690: OR = 1.54, 95% CI = 1.24 – 1.90 (99.96% CI = 1.05 – 2.26), P = 0.0001; rs2744687: OR = 1.53, 95% CI = 1.23 – 1.91, P = 0.0002, log additive model). With a less conservative permutation-based correction for multiple testing, one additional PRDM2 SNP was significantly associated with PD susceptibility (rs2697962: OR = 1.45, 95% CI = 1.18 – 1.79 (99.96% CI = 1.03 – 2.29), p=0.0005, corrected P = 0.0436).
Table 2.
Single Nucleotide Polymorphisms (SNPs) in Estrogen-related Genes Significantly Associated with PD Susceptibility in the Overall Sample (n = 12)a
| Gene | SNP | Chromosome | Positionb | Type of Variant SNPc | Allele | Minor Allele Frequencies
|
Log Additive Model OR (95% CI)d | Log Additive Model P valuee | |
|---|---|---|---|---|---|---|---|---|---|
| Cases % | Controls % | ||||||||
| PRDM2 | rs2744690 | 1 | 14010388 | Intronic | A/C | 16.5 | 12.3 | 1.54 (1.24 – 1.90) | 0.0001 |
| PRDM2 | rs2744687 | 1 | 14011400 | Intronic | T/G | 15.1 | 11.4 | 1.53 (1.23 – 1.91) | 0.0002 |
| PRDM2 | rs2697962 | 1 | 14023579 | 3′ UTR | C/T | 16.7 | 13.0 | 1.45 (1.18 – 1.79) | 0.0005 |
| PRDM2 | rs2744679 | 1 | 14024257 | Near 3′ UTR | A/G | 20.4 | 16.6 | 1.39 (1.15 – 1.67) | 0.0006 |
| PRDM2 | rs2244634 | 1 | 14006741 | Intronic | T/G | 21.5 | 17.7 | 1.38 (1.14 – 1.66) | 0.0009 |
| PRDM2 | rs2235514 | 1 | 14019522 | Intronic | A/G | 16.7 | 13.1 | 1.41 (1.15 – 1.73) | 0.001 |
| PRDM2 | rs2245197 | 1 | 14011783 | Intronic | C/T | 23.6 | 19.9 | 1.34 (1.12 – 1.61) | 0.002 |
| ESR1 | rs3778099 | 6 | 152460268 | Intronic | C/T | 10.8 | 9.1 | 1.37 (1.07 – 1.76) | 0.01 |
| CYP19A1 | rs2470157 | 15 | 49377192 | Intronic | A/G | 9.0 | 11.0 | 0.75 (0.59 – 0.94) | 0.01 |
| ESR2 | rs12434245 | 14 | 63761606 | Intronic | C/T | 9.2 | 7.6 | 1.32 (1.03 – 1.69) | 0.03 |
| ESR1 | rs2813543 | 6 | 152466171 | Near 3′ UTR | A/G | 22.4 | 24.3 | 0.83 (0.70 – 0.99) | 0.03 |
| PRDM2 | rs2744677 | 1 | 14027310 | Near 3′ UTR | A/C | 22.5 | 20.0 | 1.19 (1.00 – 1.42) | 0.05 |
The SNPs are listed in order of decreasing statistical significance as indicated by the uncorrected P values.
NCBI build 36 of the human genome.
SNP = single nucleotide polymorphism; UTR = untranslated region.
Log additive model; OR = odds ratio, CI = confidence interval
Log additive model; uncorrected P values. Only two PRDM2 gene variants remained significant after Bonferroni correction (rs2744690 and rs2744687) and three PRDM2 gene variants remained significant after permutation correction (rs2744690, rs2744687, and rs2697962).
For the 12 SNPs significantly associated with PD at the uncorrected level of p<0.05, ORs in the case-unaffected sibling pairs were similar in magnitude to ORs in the case-unrelated control pairs. An interaction analysis confirmed that the ORs did not differ significantly for the different types of controls. Similarly, the ORs obtained using men-men pairs and women-women pairs did not differ significantly, as demonstrated by an interaction analysis. However, we note that, although the differences were not significant, the ORs for the 12 SNPs in men-men pairs were considerably different from ORs in women-women pairs. In particular, SNPs in PRDM2 tended to show larger effects in the stratum of women, whereas SNPs in ESR1 and ESR2 tended to show larger effects in men. Supplementary Table 1 provides detailed results for the association with PD susceptibility of all genotyped SNPs in all four estrogen-related genes, including results for multiple coding schemes in the overall sample, and in strata defined by type of control, sex, and age at onset.
All eight PRDM2 SNPs associated with PD susceptibility map to blocks 2 and 3 of the LD map (Figure 1). The two PRDM2 SNPs (rs2744690 and rs2744687) that were significant after correction for multiple testing are in strong LD with each other (haplotype block 2). The additional PRDM2 SNP (rs2697962) that was significant after permutation testing is in block 3, and is also in LD with the two significant SNPs in block 2 (r2 > 0.78). All of these SNPs are also in high LD with the PRDM2 SNP rs2245218 that was significantly associated with PD in our previous genome-wide wide study (rs2744690: r2 = 0.636, D′ = 1.0; rs2744687: r2 = 0.578, D′ = 0.824; and rs2697962: r2 = 1.0, D′ = 1.0).23
In analyses stratified by sex, 11 SNPs were associated with PD susceptibility in men (eight in ESR1, two in ESR2, and one in CYP19A1) and 11 SNPs in women (eight in PRDM2, two in CYP19A1, and one in ESR2) at an uncorrected level of P < 0.05 (Table 3). Of these, two PRDM2 SNPs (rs2744690 and rs2744687) were significant after Bonferroni correction in women only. The SNPs significantly associated with PD susceptibility were different for men and women. In women, six of the seven SNPS in PRDM2 significantly associated with PD susceptibility in the overall sample remained significant and had stronger effects (ORs more remote from 1.0).
Table 3.
Single Nucleotide Polymorphisms (SNPs) in Estrogen-related Genes Significantly Associated with PD Susceptibility in Sex-Specific Analysesa
| Gene | SNP | Chromosome | Positionb | Type of Variant SNPc | Allele | Minor Allele Frequencies
|
Log Additive Model OR (95% CI)d | Log Additive Model P valuee | |
|---|---|---|---|---|---|---|---|---|---|
| Cases % | Controls % | ||||||||
| Men-Men Pairs (n = 555 pairs) | |||||||||
| ESR1 | rs10484922 | 6 | 152174010 | Intronic | C/T | 9.7 | 8.7 | 1.64 (1.17 – 2.31) | 0.004 |
| ESR2 | rs12434245 | 14 | 63761606 | Intronic | C/T | 9.2 | 7.6 | 1.58 (1.13 – 2.22) | 0.007 |
| ESR1 | rs7761133 | 6 | 152193556 | Intronic | C/T | 15.8 | 14.8 | 1.44 (1.09 – 1.90) | 0.01 |
| ESR1 | rs9322331 | 6 | 152204010 | Intronic | C/T | 32.8 | 34.2 | 0.78 (0.64 – 0.95) | 0.02 |
| ESR2 | rs7159462 | 14 | 63828629 | Intronic | C/T | 9.4 | 8.0 | 1.49 (1.07 – 2.07) | 0.02 |
| ESR1 | rs3778099 | 6 | 152460268 | Intronic | C/T | 10.8 | 9.2 | 1.48 (1.06 – 2.06) | 0.02 |
| ESR1 | rs7761846 | 6 | 152254201 | Intronic | C/T | 7.8 | 9.0 | 0.65 (0.44 – 0.96) | 0.03 |
| ESR1 | rs1514348 | 6 | 152224008 | Intronic | T/G | 40.4 | 41.8 | 0.81 (0.66 – 0.99) | 0.04 |
| ESR1 | rs985694 | 6 | 152328318 | Intronic | C/T | 15.7 | 15.0 | 1.32 (1.01 – 1.71) | 0.04 |
| CYP19A1 | rs3751591 | 15 | 49394002 | Intronic | A/G | 17.1 | 16.3 | 1.31 (1.01 – 1.70) | 0.04 |
| ESR1 | rs2813543 | 6 | 152466171 | Near 3′ UTR | A/G | 22.4 | 24.3 | 0.78 (0.61 – 0.99) | 0.04 |
| Women-Women Pairs (n = 332 pairs) | |||||||||
| PRDM2 | rs2744690 | 1 | 14010388 | Intronic | A/C | 16.5 | 12.3 | 1.98 (1.34 – 2.92) | 0.0006 |
| PRDM2 | rs2744687 | 1 | 14011400 | Intronic | T/G | 15.1 | 11.4 | 1.97 (1.31 – 2.96) | 0.001 |
| PRDM2 | rs2744679 | 1 | 14024257 | Near 3′ UTR | A/G | 20.4 | 16.6 | 1.75 (1.24 – 2.46) | 0.001 |
| PRDM2 | rs2697962 | 1 | 14023579 | 3′ UTR | C/T | 16.7 | 13.0 | 1.81 (1.25 – 2.62) | 0.002 |
| PRDM2 | rs2235514 | 1 | 14019522 | Intronic | A/G | 16.7 | 13.1 | 1.80 (1.25 – 2.59) | 0.002 |
| PRDM2 | rs2244634 | 1 | 14006741 | Intronic | T/G | 21.5 | 17.7 | 1.64 (1.16 – 2.32) | 0.005 |
| PRDM2 | rs6669610 | 1 | 14021343 | Intronic | C/T | 6.8 | 7.8 | 0.52 (0.32 – 0.84) | 0.008 |
| CYP19A1 | rs2470157 | 15 | 49377192 | Intronic | A/G | 9.0 | 11.0 | 0.58 (0.37 – 0.89) | 0.01 |
| PRDM2 | rs2473231 | 1 | 14028265 | — | C/G | 41.1 | 39.2 | 1.37 (1.06 – 1.77) | 0.02 |
| CYP19A1 | rs730154 | 15 | 49378496 | Intronic | C/T | 14.4 | 16.2 | 0.68 (0.48 – 0.97) | 0.03 |
| ESR2 | rs1255998 | 14 | 63763624 | 3′ UTR | C/G | 10.2 | 9.9 | 1.68 (1.01 – 2.78) | 0.04 |
The SNPs are listed in order of decreasing statistical significance as indicated by the uncorrected P values. None of the SNPs had significant interactions with sex after correction for multiple testing.
NCBI build 36 of the human genome.
UTR = untranslated region.
Log additive model; OR = odds ratio, CI = confidence interval.
Log additive model; uncorrected P values. No SNPs remained significant after Bonferroni correction or after permutation correction for multiple comparisons.
Case-Only Analyses of Genetic Variants and Age at Onset of PD
Nine SNPs (four in CYP19A1: rs8031463, rs16964258, rs730154, and rs10459592; four in ESR1: rs1709183, rs9322335, rs6912184, and rs2347923; one in ESR2: rs1256063; and none in PRDM2) were associated with age at onset of PD in the overall sample. However, none of these associations remained significant after Bonferroni correction. Fourteen SNPs had significant uncorrected P values in analyses restricted to men and nine SNPs in analyses restricted to women. Supplementary Table 1 provides the details for associations of all SNPs in the four genes with age at onset of PD, including results for multiple coding schemes in the overall sample, and for men and women separately.
DISCUSSION
This study confirmed the association of common variations in the PRDM2 gene with PD susceptibility, but showed limited evidence of association of common variations in other estrogen-related genes with PD. Three SNPs in PRDM2 (rs2744690, rs2744687, and rs2697962) that were significantly associated with PD susceptibility in the overall sample after permutation-based correction for multiple testing, were also significant (uncorrected P values) in the women only stratum but not in the men only stratum. The mechanism of this potentially sex-specific difference in genetic architecture relating to PD susceptibility remains uncertain. Nevertheless, the sex-specific nature of these associations is interesting and warrants further investigation of the functional role of these genetic variations in the susceptibility to PD in men and women.
The present findings are consistent with the findings from our previous genome-wide association study that included 443 PD cases and 443 matched controls genotyped for 198,345 informative genomic SNPs (tier 1 sample); and an additional 332 PD cases and 332 matched controls (tier 2 sample) genotyped for the SNPs that were significantly associated with PD in the tier 1 sample. That study yielded suggestive findings for the PRDM2 SNP rs2245218 (OR = 1.67, 95% CI = 1.29 – 2.14, P value = 4.61×10−5); however, the association was not significant after correction for multiple testing.23 In the current study, three SNPs in PRDM2 (rs2744690, rs2744687, and rs2697962) were associated with PD susceptibility in the overall sample (corrected P values), and in the stratum of women (uncorrected P values). These PRDM2 SNPs were in LD with the PRDM2 SNP rs2245218 highlighted by our previous genome-wide association study.23 The results presented here are not an independent replication because 374 case-control pairs and an additional 69 cases included in this study had also been included in the prior genome-wide study, and 11 of the 143 SNPs investigated here were included in the prior study. However, the current study had a considerably larger sample size and a greater coverage of genetic variation in the four estrogen-related genes. Furthermore, the significantly associated SNPs remained nominally significant even after removing the cases and controls that had been included in the prior GWAS study (p<0.005 for the two PRDM2 SNPs that were significant after Bonferroni-correction).
By contrast, four other genome-wide association studies highlighted no associations of estrogen-related genes with PD.36–39 The study by Fung and colleagues employed more SNP markers than our original genome-wide association study, but included only 276 PD cases and 276 unmatched controls. The study of Pankratz and colleagues employed more SNP markers than our original genome-wide association study and more subjects (857 PD cases and 867 controls); however, their sample included only familial PD cases. Neither of those two studies highlighted any of the estrogen-related genes independently or in pooled analyses. However, both of these studies had a smaller sample size than the current study. The studies of Simon-Sanchez and Satake included 7,208 PD cases and 27,184 controls collectively, and genotyped more than 500,000 SNPs. However our candidate gene study approach required a much smaller multiple-testing correction because we had a focused a priori hypothesis. Also, by selecting tagSNPs based on patterns of linkage disequilibrium, our candidate gene approach provided high coverage of the four estrogen-related genes. By contrast to this study, none of the five genome-wide studies selected genetic variants to comprehensively cover the four estrogen-related genes, nor did they report analyses stratified by sex.
We also studied the association of common variants in estrogen-related genes with age at onset of PD. We observed associations for nine SNPs at an uncorrected significance level of 0.05 (four in CYP19A1, four in ESR1, and one in ESR2). Thus far, there has been only one genome-wide association study of age at onset of PD that showed no significant genomic SNP associations after Bonferroni correction.40 Additional studies are needed to determine whether common variations in these estrogen-related genes are associated with age at onset of PD.
While our most significant findings (significant after Bonferroni correction) were for two PRDM2 SNPs (rs2744690 and rs2744687), it remains unknown whether these intronic SNPs have a functional effect or whether they are markers of other functional variants. An additional SNP (rs2697962) that was significant with permutation testing maps to the 3′ untranslated region of the PRDM2 gene. It is not known whether that SNP modifies gene expression. Further studies are needed to replicate our association findings for these PRDM2 gene SNPs with PD susceptibility and to fine map and define functional variants within the gene.
This study has several strengths. First, our large sample size (1,103 PD cases and 1,103 controls) provided sufficient statistical power to detect a wide range of ORs (and HRs) for a range of minor allele frequencies (log additive model). Second, we studied four estrogen-related genes that have plausibility as candidate genes for PD. Third, we studied multiple LD tagging SNPs in each gene. Fourth, we observed a strong concordance of ORs of each variant associated with PD susceptibility between case-unaffected sibling and case-unrelated control pairs (internal replication). Fifth, we studied the association of genetic variants with age at onset of PD as well as with susceptibility.
Our study also has some limitations. First, our sample was not population-based. However, population-based case-control studies are often not large enough to detect the small effects of common genetic variants. We tried to limit sampling bias by recruiting cases prospectively from a defined geographic region (the upper Midwest, USA). We previously showed that for approximately half of our patients with PD (residing within 120 miles of the Mayo Clinic in Rochester, MN), the demographic characteristics are similar to those of an incidence cohort of PD patients from Olmsted County, MN. By contrast, the other half of our patients with PD (residing within a broader five-state region) were younger, possibly increasing the genetic load.2, 41, 42 Although all PD patients were recruited from a single tertiary specialty clinic, the risk of referral bias is expected to be minimal unless clinical characteristics of referred PD patients have a different genetic basis from non-referred patients. Notably, frequency of family history was similar in our study to that observed in other population-based studies, which suggests the role of genetic factors may not differ greatly between this population and other populations with PD. However, the genetic associations and corresponding effect sizes observed in this study are based on cases seen in a tertiary clinic, and may not generalize to all patient populations
Second, our controls were primarily unaffected siblings to limit possible population stratification bias and to maximize participation rates. Unaffected sibling controls may be overmatched for genetic and environmental factors, leading to false negative findings (reduced statistical power). For this reason, we performed sensitivity analyses, which showed similar ORs in separate analyses for case-unaffected sibling pairs or case-unrelated control pairs.
Third, while our overall sample size was large, our sample size within strata defined by type of control or by sex were more modest. Fourth, we did not re-sequence the four genes in all subjects to detect rare point mutations or copy number variations that were associated with PD. Such studies are expected to become feasible in the coming years when the costs for next-generation sequencing technologies will decline.43, 44 Fifth, we performed multiple statistical tests, increasing the likelihood of chance findings. Therefore, we employed Bonferroni correction for our primary analysis to identify genetic associations that exceeded chance expectations.
Sixth, we did not study gene-gene or gene-environment interactions (beyond the scope of this initial exploratory study). Finally, we did not replicate our significant findings in independent samples (also beyond the scope of this initial exploratory study). Large-scale replications of genetic association studies of PD are feasible within existing large consortia.45, 46
Supplementary Material
Haplotype blocks of the four estrogen-related genes that we studied (CYP19A1, ESR1, ESR2, and PRDM2 genes). The LD values measured using r2 are given by numbers and the LD values measured by D′ are shown by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data).
Case-control analyses for susceptibility and cases-only analyses for age at onset of PD for all of the 137 informative SNPs in the four estrogen-related genes in the overall sample and in strata, and using multiple SNP codings corresponding to different allele effect models (log additive, dominant, and recessive).
Acknowledgments
The major aspects of the research program were funded by the National Institutes of Health (7 R01 ES010751-10). We thank the many members of Mayo’s Molecular Epidemiology of Parkinson’s Disease research team for their efforts, and especially our Mayo Clinic patients and their families for their participation.
Footnotes
Author Roles: Sun Ju Chung– Dr. Chung was involved in Research Project (Execution), Statistical Analysis (Review and Critique), Manuscript (Writing of the first draft); Sebastian M. Armasu–Mr. Armasu was involved in Statistical Analysis (Design, Execution, Review and Critique), Manuscript (Review and Critique); Joanna M. Biernacka–Dr. Biernacka was involved in Statistical Analysis (Design, Review and Critique), Manuscript (Review and Critique); Timothy G. Lesnick–Mr. Lesnick was involved in Research Project (Conception, Organization, and Execution), Statistical Analyses (Design, Execution, and Review and Critique), Manuscript (Writing of the first draft, Review and Critique); David N. Rider–Mr. Rider was involved in Statistical Analysis (Execution, Review and Critique), Manuscript (Review and Critique); Julie M. Cunningham–Dr. Cunningham was involved in Manuscript (Review and Critique); Demetrius M. Maraganore–Dr. Maraganore was involved in Research Project (Conception, Organization, Execution), Statistical Analysis (Design, Execution, Review and Critique), Manuscript (Writing of the first draft, Review and Critique).
Financial disclosures: Sun Ju Chung–Employment: Assistant Professor, Department of Neurology, Asan Medical Center, University of Ulsan College of Medicine, Seoul, South Korea, Disclosures: Dr. Chung has received funding for travel from Glaxo-SmithKline Korea; Sebastian M. Armasu–Employment: Statistician I, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA, Disclosures: Mr. Armasu has received funding from the National Institutes of Health (7 R01 ES010751-10 [Co-Investigator], 1 P50 CA136393-01A1 [Co-Investigator], 5 U01 HG004735-02 [Co-Investigator]); Joanna M. Biernacka–Employment: Assistant Professor of Biostatistics and Psychiatry, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA, Disclosures: Dr. Biernacka has received funding from the National Institutes of Health (1 P20 AA017830-01 [Co-Investigator], 7 R01 ES010751-10 [Co-Investigator], 1 R03 AA019570-01 [PI], 1 R01 MH079261-01A2 [Co-Investigator]); Timothy G. Lesnick–Employment: Statistician III, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA, Disclosures: Mr. Lesnick may accrue revenue from pending patent applications related to the Prediction of Parkinson disease and receives research support from the National Institutes of Health (7 R01 ES010751-10 [Co-Investigator]); David N. Rider–Employment: Senior Analyst/Programmer, Division of Biomedical Statistics and Informatics, Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA, Disclosures: No disclosures to report; Julie M. Cunningham–Employment: Assistant Professor of Laboratory Medicine and Pathology, Mayo Genomics Research Center, Mayo Clinic, Rochester, MN, USA, Disclosures: No disclosures to report; Demetrius M. Maraganore–Employment: Ruth Cain Ruggles Chairman of the Department of Neurology, and Medical Director of the Neurological Institute, at NorthShore University HealthSystem, Evanston, IL, USA, Disclosures: Dr. Maraganore may accrue revenue from pending patent applications related to the prediction of Parkinson disease and the treatment of neurodegenerative disease; has received license fee payments and royalty payments from Alnylam Pharmaceuticals (method to treat Parkinson’s disease); and receives research support from the National Institutes of Health (7 R01 ES010751-10 [PI]).
References
- 1.Elbaz A, Bower JH, Maraganore DM, et al. Risk tables for parkinsonism and Parkinson’s disease. J Clin Epidemiol. 2002;55(1):25–31. doi: 10.1016/s0895-4356(01)00425-5. [DOI] [PubMed] [Google Scholar]
- 2.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Incidence and distribution of parkinsonism in Olmsted County, Minnesota, 1976–1990. Neurology. 1999;52(6):1214–1220. doi: 10.1212/wnl.52.6.1214. [DOI] [PubMed] [Google Scholar]
- 3.Baldereschi M, Di Carlo A, Rocca WA, et al. Parkinson’s disease and parkinsonism in a longitudinal study: two-fold higher incidence in men. ILSA Working Group. Italian Longitudinal Study on Aging. Neurology. 2000;55(9):1358–1363. doi: 10.1212/wnl.55.9.1358. [DOI] [PubMed] [Google Scholar]
- 4.Chan RS, Huey ED, Maecker HL, et al. Endocrine modulators of necrotic neuron death. Brain Pathol. 1996;6(4):481–491. doi: 10.1111/j.1750-3639.1996.tb00877.x. [DOI] [PubMed] [Google Scholar]
- 5.Regan RF, Guo Y. Estrogens attenuate neuronal injury due to hemoglobin, chemical hypoxia, and excitatory amino acids in murine cortical cultures. Brain Res. 1997;764(1–2):133–140. doi: 10.1016/s0006-8993(97)00437-x. [DOI] [PubMed] [Google Scholar]
- 6.Singer CA, Rogers KL, Strickland TM, Dorsa DM. Estrogen protects primary cortical neurons from glutamate toxicity. Neurosci Lett. 1996;212(1):13–16. doi: 10.1016/0304-3940(96)12760-9. [DOI] [PubMed] [Google Scholar]
- 7.Sribnick EA, Ray SK, Nowak MW, et al. 17beta-estradiol attenuates glutamate-induced apoptosis and preserves electrophysiologic function in primary cortical neurons. J Neurosci Res. 2004;76(5):688–696. doi: 10.1002/jnr.20124. [DOI] [PubMed] [Google Scholar]
- 8.Miller DB, Ali SF, O’Callaghan JP, Laws SC. The impact of gender and estrogen on striatal dopaminergic neurotoxicity. Ann N Y Acad Sci. 1998;844:153–165. [PubMed] [Google Scholar]
- 9.Bains M, Cousins JC, Roberts JL. Neuroprotection by estrogen against MPP+-induced dopamine neuron death is mediated by ERalpha in primary cultures of mouse mesencephalon. Exp Neurol. 2007;204(2):767–776. doi: 10.1016/j.expneurol.2007.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dluzen D. Estrogen decreases corpus striatal neurotoxicity in response to 6-hydroxydopamine. Brain Res. 1997;767(2):340–344. doi: 10.1016/s0006-8993(97)00630-6. [DOI] [PubMed] [Google Scholar]
- 11.Datla KP, Murray HE, Pillai AV, et al. Differences in dopaminergic neuroprotective effects of estrogen during estrous cycle. Neuroreport. 2003;14(1):47–50. doi: 10.1097/00001756-200301200-00009. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Dluzen DE. Tamoxifen abolishes estrogen’s neuroprotective effect upon methamphetamine neurotoxicity of the nigrostriatal dopaminergic system. Neuroscience. 2001;103(2):385–394. doi: 10.1016/s0306-4522(01)00014-8. [DOI] [PubMed] [Google Scholar]
- 13.Gajjar TM, Anderson LI, Dluzen DE. Acute effects of estrogen upon methamphetamine induced neurotoxicity of the nigrostriatal dopaminergic system. J Neural Transm. 2003;110(11):1215–1224. doi: 10.1007/s00702-003-0045-3. [DOI] [PubMed] [Google Scholar]
- 14.Dluzen DE, McDermott JL, Liu B. Estrogen as a neuroprotectant against MPTP-Induced neurotoxicity in C57/B1 mice. Neurotoxicol Teratol. 1996;18(5):603–606. doi: 10.1016/0892-0362(96)00086-4. [DOI] [PubMed] [Google Scholar]
- 15.Dluzen DE, McDermott JL, Liu B. Estrogen alters MPTP-induced neurotoxicity in female mice: effects on striatal dopamine concentrations and release. J Neurochem. 1996;66(2):658–666. doi: 10.1046/j.1471-4159.1996.66020658.x. [DOI] [PubMed] [Google Scholar]
- 16.Leranth C, Roth RH, Elsworth JD, et al. Estrogen is essential for maintaining nigrostriatal dopamine neurons in primates: implications for Parkinson’s disease and memory. J Neurosci. 2000;20(23):8604–8609. doi: 10.1523/JNEUROSCI.20-23-08604.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benedetti MD, Maraganore DM, Bower JH, et al. Hysterectomy, menopause, and estrogen use preceding Parkinson’s disease: an exploratory case-control study. Mov Disord. 2001;16(5):830–837. doi: 10.1002/mds.1170. [DOI] [PubMed] [Google Scholar]
- 18.Saunders-Pullman R, Gordon-Elliott J, Parides M, et al. The effect of estrogen replacement on early Parkinson’s disease. Neurology. 1999;52(7):1417. doi: 10.1212/wnl.52.7.1417. [DOI] [PubMed] [Google Scholar]
- 19.Currie LJ, Harrison MB, Trugman JM, et al. Postmenopausal estrogen use affects risk for Parkinson disease. Arch Neurol. 2004;61(6):886–888. doi: 10.1001/archneur.61.6.886. [DOI] [PubMed] [Google Scholar]
- 20.Rocca WA, Bower JH, Maraganore DM, et al. Increased risk of parkinsonism in women who underwent oophorectomy before menopause. Neurology. 2008;70(3):200–209. doi: 10.1212/01.wnl.0000280573.30975.6a. [DOI] [PubMed] [Google Scholar]
- 21.Weel AE, Uitterlinden AG, Westendorp IC, et al. Estrogen receptor polymorphism predicts the onset of natural and surgical menopause. J Clin Endocrinol Metab. 1999;84(9):3146–3150. doi: 10.1210/jcem.84.9.5981. [DOI] [PubMed] [Google Scholar]
- 22.Maraganore DM, Farrer MJ, McDonnell SK, et al. Case-control study of estrogen receptor gene polymorphisms in Parkinson’s disease. Mov Disord. 2002;17(3):509–512. doi: 10.1002/mds.1253. [DOI] [PubMed] [Google Scholar]
- 23.Maraganore DM, de Andrade M, Lesnick TG, et al. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maraganore DM. Blood is thicker than water: the strengths of family-based case-control studies. Neurology. 2005;64(3):408–409. doi: 10.1212/01.WNL.0000152585.76852.9C. [DOI] [PubMed] [Google Scholar]
- 25.Rocca WA, Maraganore DM, McDonnell SK, Schaid DJ. Validation of a telephone questionnaire for Parkinson’s disease. J Clin Epidemiol. 1998;51(6):517–523. doi: 10.1016/s0895-4356(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 26.Hartge P, Brinton LA, Rosenthal JF, et al. Random digit dialing in selecting a population-based control group. Am J Epidemiol. 1984;120(6):825–833. doi: 10.1093/oxfordjournals.aje.a113955. [DOI] [PubMed] [Google Scholar]
- 27.Potthoff RF. Telephone sampling in epidemiologic research: to reap the benefits, avoid the pitfalls. Am J Epidemiol. 1994;139(10):967–978. doi: 10.1093/oxfordjournals.aje.a116946. [DOI] [PubMed] [Google Scholar]
- 28.Brighina L, Frigerio R, Schneider NK, et al. {alpha}-synuclein, pesticides, and Parkinson disease: a case-control study. Neurology. 2008;70(16_Part_2):1461–1469. doi: 10.1212/01.wnl.0000304049.31377.f2. [DOI] [PubMed] [Google Scholar]
- 29.Brighina L, Schneider NK, Lesnick TG, et al. alpha-Synuclein, alcohol use disorders, and Parkinson disease: A case-control study. Parkinsonism Relat Disord. 2009 doi: 10.1016/j.parkreldis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wise PM, Dubal DB, Wilson ME, et al. Estrogens: trophic and protective factors in the adult brain. Front Neuroendocrinol. 2001;22(1):33–66. doi: 10.1006/frne.2000.0207. [DOI] [PubMed] [Google Scholar]
- 31.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 33.Breslow NE, Day NE. Statistical methods in cancer research. Volume I - The analysis of case-control studies. IARC Sci Publ. 1980;(32):5–338. [PubMed] [Google Scholar]
- 34.Cox DR. Regression models of life-tables (with discussion) J R Stat Soc [Ser B] 1972;34:187–220. [Google Scholar]
- 35.Welch WJ, Gutierrez LG. Robust permutation tests for matched-pairs designs. J Am Stat Assoc. 1988;83:450–455. [Google Scholar]
- 36.Fung HC, Scholz S, Matarin M, et al. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- 37.Pankratz N, Wilk JB, Latourelle JC, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 40.Latourelle JC, Pankratz N, Dumitriu A, et al. Genomewide association study for onset age in Parkinson disease. BMC Med Genet. 2009;10:98. doi: 10.1186/1471-2350-10-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Influence of strict, intermediate, and broad diagnostic criteria on the age- and sex-specific incidence of Parkinson’s disease. Mov Disord. 2000;15(5):819–825. doi: 10.1002/1531-8257(200009)15:5<819::aid-mds1009>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 42.Rocca WA, Peterson BJ, McDonnell SK, et al. The Mayo Clinic Family Study of Parkinson’s Disease: study design, instruments, and sample characteristics. Neuroepidemiology. 2005;24(3):151–167. doi: 10.1159/000083612. [DOI] [PubMed] [Google Scholar]
- 43.Kahvejian A, Quackenbush J, Thompson JF. What would you do if you could sequence everything? Nat. Biotechnol. 2008;26:1125–1133. doi: 10.1038/nbt1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mardis ER. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24(3):133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 45.Maraganore DM, de Andrade M, Elbaz A, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296(6):661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 46.Evangelou E, Maraganore DM, Annesi G, et al. Non-replication of association for six polymorphisms from meta-analysis of genome-wide association studies of Parkinson’s disease: Large-scale collaborative study. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Haplotype blocks of the four estrogen-related genes that we studied (CYP19A1, ESR1, ESR2, and PRDM2 genes). The LD values measured using r2 are given by numbers and the LD values measured by D′ are shown by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data).
Case-control analyses for susceptibility and cases-only analyses for age at onset of PD for all of the 137 informative SNPs in the four estrogen-related genes in the overall sample and in strata, and using multiple SNP codings corresponding to different allele effect models (log additive, dominant, and recessive).