Abstract
Background
Polymorphisms in SNCA, MAPT and LRRK2 genes have recently been confirmed as risk factors for Parkinson’s disease (PD), although with small individual attributable risk. Here we investigated the association of PD with interactions between variants of these genes.
Methods
As part of a previous study of PD susceptibility genes 119 SNCA, MAPT, and LRRK2 haplotype tagging single nucleotide polymorphisms (SNPs) and two variable number tandem repeats (VNTRs) were genotyped in 1,098 PD cases from the upper Midwest, USA and 1,098 matched controls. Twenty-six of these SNPs were selected for SNP-SNP (or SNP-VNTR or VNTR-VNTR) interaction analysis (256 interaction pairs). Case-control analyses were performed to study association of pairwise SNP interactions with PD susceptibility.
Results
Out of the 256 interaction pairs investigated, 10 had uncorrected p-values <0.05. These represented six SNCA-LRRK2 pairs, three SNCA-MAPT pairs, and one MAPT-LRRK2 pair. However, none of these pairwise interactions were significant after correction for multiple testing. Secondary analyses in strata defined by type of control (sibling or unrelated), sex, or age at onset of the case also did not reveal any significant interactions after accounting for multiple testing.
Conclusions
This study provides no statistically significant evidence of gene-gene interaction effects for the three confirmed genetic susceptibility loci for PD. However, this does not exclude the possibility that other genomic loci or environmental risk factors interact with these genes.
Keywords: Parkinson’s disease, gene-gene interaction, alpha-synuclein, microtubule associated protein tau, leucine rich repeat kinase 2
Introduction
Parkinson’s disease (PD) is of unknown etiology, but is generally assumed to have complex substrates, both genetic and environmental. Numerous factors associated with PD have been reported, but the attributable risk of each is small; no single genetic or environmental factor appears to make a substantial contribution to PD risk. Although individual factors operating independently account for little PD risk, the contribution from interactions between such factors could be more substantial.
The association of three genes with PD risk has been reproducibly documented in four recent and independent genome-wide association studies (GWAS) in large PD cohorts: alpha-synuclein (SNCA)[1–4], microtubule associated protein tau (MAPT)[1, 3, 4], and leucine rich repeat kinase 2 (LRRK2)[2, 3]. Although each of these three genes has small attributable PD risk, they all have substantial biologic plausibility from additional lines of evidence. Firstly, linkage studies revealed that pathogenic point mutations in SNCA[5] or LRRK2[6] result in familial Parkinson’s disease with phenotypic fidelity to sporadic PD; and SNCA triplications or duplications similarly cause familial PD[7]. Secondly, candidate gene studies revealed that polymorphisms of SNCA[8] and MAPT[9] each confer increased PD risk. Thirdly, immunohistochemical studies revealed that the neuropathologic hallmark of PD, the Lewy body, contains not only alpha-synuclein, but also MAPT[10] and LRRK2[11] proteins.
Interactions between the products of these three genes might conceivably amplify their individual pathogenic contributions to PD. Laboratory evidence suggests that such interactions may indeed occur. It has been shown that MAPT induces fibrillization of alpha-synuclein[12]; such fibrillization is proposed to be an initial step in the generation of PD-pathogenic alpha-synuclein aggregates. Alpha-synuclein not only interacts with tau protein in vitro[13], it also binds to tau and stimulates its phosphorylation; hyperphosphorylation destabilizes tau and impairs protein transport[14]. LRRK2, which is thought to be an upstream factor in the neurodegenerative pathway[15], induces SNCA expression[16]. Whereas tau is a microtubule-associated protein, LRRK2 is known to interact with microtubules[17]. Finally, in vivo studies suggest that tau and alpha-synuclein may interact to promote aggregation and accumulation of each protein[18].
Human post-mortem studies also suggest interactions among these three genes. Namely, there is a robust correlation between alpha-synuclein-labeled Lewy bodies and MAPT-labeled neurofibrillary tangles in sporadic PD[19] Moreover, whereas most cases of LRRK2 parkinsonism display alpha-synuclein-labeled Lewy bodies, occasionally LRRK2 cases are marked by MAPT-positive tangles[15].
While the main effects of SNCA, MAPT and LRRK2 as causal or risk factors for PD are well established, there has been only limited study of the joint effects or statistical interaction of the effects of these genes in PD. The causal variants in these genes are rare and have limited attributable risk and the common susceptibility variants in these genes have small effect sizes. We hypothesized, however, that the interactions of common variants in the SNCA, MAPT, and LRRK2 genes may have substantially larger effect sizes and therefore result in appreciable attributable risk in PD.
Specifically in this study we assessed whether SNCA, MAPT, and LRRK2 genes have not simply additive effects on the log odds of PD, but rather multiplicative effects on PD susceptibility. Thus, we comprehensively assessed the association of PD with pairwise SNP or VNTR interactions of these three genes in a large case-control study.
Subjects and Methods
Study subjects
The Institutional Review Board of the Mayo Clinic approved the study, and all subjects provided written informed consent. The enrollment of matched cases and controls has been previously described[20, 21]. PD cases were recruited from patients seen at the Department of Neurology of the Mayo Clinic in Rochester, MN between 1996 and 2007. All cases were residents of Minnesota or one of the surrounding four states (Wisconsin, Iowa, South Dakota, and North Dakota). The diagnosis of PD was made by movement disorders specialists using established criteria[22]. The age at onset of PD was defined as the time of the first observed cardinal motor sign (rest tremor, bradykinesia, rigidity, or postural instability), as reported by the patient or a family member at the time of clinical assessment for the study. Controls included unaffected siblings of cases or unrelated controls when there were no siblings available. Potential controls were screened for parkinsonism using a validated telephone instrument[23]. Only potential controls who screened negative for PD, or who were confirmed not to have PD via clinical assessment (despite having screened positive by telephone interview), were included in the study. Cases were matched to a single participating sibling first by sex (when possible) and then by closest age. Cases without an available sibling were matched to unrelated controls living in the same 5-state region and of same sex and age (same year of birth ± 2 years). Initially 1,103 cases and matched controls were enrolled in the study[20, 21]. Genomic DNA was collected, extracted, and stored as previously described[20]. Five cases were subsequently excluded because of indeterminate diagnoses. Thus 1,098 cases and matched controls were used in the analyses.
Genotyping
Single nucleotide polymorphisms (SNPs) in species-conserved regions of 13 PARK locus or related genes, including SNCA, MAPT and LRRK2, were detected via sequencing in 25 cases and 25 controls (see Chung et al.[21] for details). Additional tag SNPs were then selected for these genes from the HapMap database using the LDSelect program with a linkage disequilibrium (LD) r2 threshold of 0.8 and minor allele frequencies (MAF) > 0.05. Two tag SNPs were selected for each LD bin when the number of SNPs in the bin was 10 or more. In total, 19 SNPs in SNCA, 35 in MAPT, and 65 in LRRK2 were successfully genotyped using a bead array platform (Illumina GoldenGate). In addition two variable number tandem repeats (VNTRs) (SNCA REP1, MAPT H1/H2 haplotype) were genotyped using a sequencing platform (Applied Biosystems).
Selection of SNPs for gene-gene interaction analysis
Variants with MAF < 0.05 or showing departures from Hardy-Weinberg equilibrium (p < 0.001) were excluded from the analyses. Kooperberg and LeBlanc[24] demonstrated that testing for SNP-SNP interactions in a subset of SNPs selected using a screening step based on univariate SNP analysis (i.e. single SNP effects) tends to be more powerful than the alternative strategy of testing all possible pairs of SNPs. To determine the optimal p-value threshold for the screening step, we used the Splus library powerGWASinteraction (http://cran.r-project.org/web/packages/powerGWASinteraction/index.html), a program for estimating power under a range of scenarios corresponding to different screening-step p-value thresholds. Based on these calculations we selected p=0.2 in the single SNP analysis as the threshold for inclusion of SNPs in the interaction analysis. This strategy excludes SNPs with no evidence of single-SNP association with PD (based on the threshold p>0.2) while retaining SNPs with significant marginal effects. Using the threshold of p=0.2 attempts to retain SNPs that have weak marginal associations that are not detectable at traditionally used significance levels with the available sample size. Finally, to avoid redundancy of tests due to testing of SNPs in high LD, and to reduce the total number of tests performed, we further applied a tag-SNP selection strategy to the resulting SNP list. Tagging SNP selection was performed using the pairwise Tagger algorithm with r2=0.9 implemented in Haploview 4.2[25].
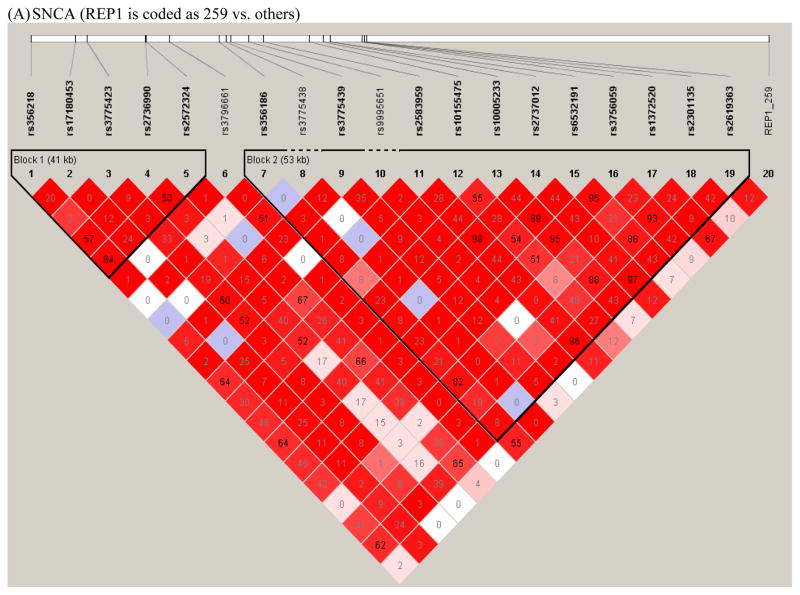
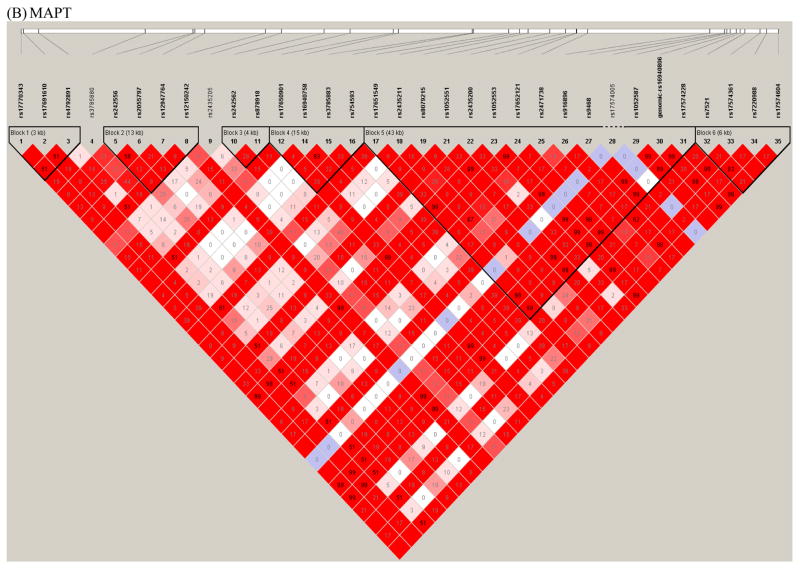
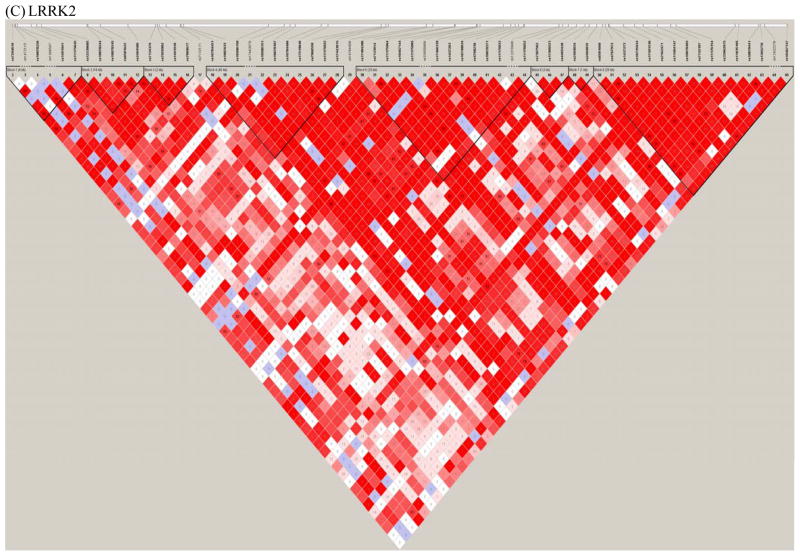
This procedure resulted in the selection of 10 SNPs in SNCA, 8 SNPs in MAPT, and 8 SNPs in LRRK2 that had p-values<0.2 in a trend test for association with PD (i.e. marginal test of association with PD under the assumption of log-additive allele effects). In addition, SNCA REP1 (coded based on the number of 259 bp alleles and the number of 263 bp alleles) and the MAPT VNTR that distinguishes the H1/H2 haplotype were included in the analyses, as these variables also showed marginal evidence of association at the p<0.2 level. The polymorphisms examined in the interaction analyses are listed in Table 1. LD plots for the three genes highlighting the SNPs and VNTRs used for the interaction analysis are shown in Figure 1 in the supplement.
Table 1.
Genetic variants used in gene-gene interaction analysis
| GENE | VARIANT | SNP location or known function | MINOR ALLELE FREQUENCY | P-VALUE3 |
|---|---|---|---|---|
| SNCA | rs1372520 | intron | 0.1848 | 0.0056 |
| rs2572324 | intron; associated with the extent of neurofibrillary pathology | 0.3226 | 0.0090 | |
| rs2583959 | intron | 0.3013 | 0.0506 | |
| rs2736990 | intron | 0.4902 | 0.0017 | |
| rs356186 | intron | 0.1686 | 0.0119 | |
| rs356218 | 3′ downstream | 0.3545 | 0.0419 | |
| rs3775423 | intron | 0.0903 | 0.0090 | |
| rs3775439 | intron | 0.1290 | 0.0716 | |
| rs9995651 | intron | 0.0519 | 0.1097 | |
| REP1-2591 | regulates SNCA gene expression | 0.2413 | 0.0345 | |
| REP1-2632 | regulates SNCA gene expression | 0.0749 | 0.0465 | |
| REP1 score | regulates SNCA gene expression | n.a. | 0.0118 | |
| MAPT | rs16940758 | intron; regulates MAPT gene expression | 0.1745 | 0.0844 |
| rs16940806 | 3′ UTR | 0.1800 | 0.0059 | |
| rs2435200 | Intron | 0.3968 | 0.0001 | |
| rs2435211 | intron; regulates MAPT gene expression | 0.3012 | 0.0856 | |
| rs4792891 | intron | 0.3016 | 0.0036 | |
| rs8079215 | intron | 0.2945 | 0.0660 | |
| rs878918 | intron | 0.3329 | 0.1437 | |
| MAPT H1/H2 | regulates MAPT gene expression | 0.1793 | 0.0042 | |
| LRRK2 | rs10784486 | intron | 0.3140 | 0.1691 |
| rs11175922 | intron | 0.4877 | 0.1736 | |
| rs1491939 | intron | 0.0601 | 0.1521 | |
| rs17466521 | intron | 0.2546 | 0.1961 | |
| rs17484286 | intron | 0.0927 | 0.0128 | |
| rs2404835 | intron | 0.3255 | 0.0872 | |
| rs6581668 | intron | 0.4727 | 0.1814 | |
| rs7307562 | intron | 0.4070 | 0.1669 |
REP1-259 denotes the SNCA VNTR REP1 coded as the number of 259 bp alleles
REP1-263 denotes the SNCA VNTR REP1 coded as the number of 263 bp alleles
P-value for trend test for association with PD. P-values are not corrected for multiple testing.
Statistical Analyses
Pairwise interactions between genetic variants in SNCA, MAPT, and LRRK2 were assessed using conditional logistic regression analyses. To identify interactions between rather than within genes, only pairs of SNPs in two different genes were considered in the primary analysis. Thus a total of 256 pairwise interactions were tested. For SNPs, a log-additive genotype coding scheme was used, while for SNCA REP1 genotypes were converted to scores ranging from 0 to 4 as previously described[20]. Namely, the score for the REP1 genotype was calculated as the sum of two allele scores, with each 259 bp allele contributing 0 points, each 261 bp allele contributing 1 point, and each copy of a 263 bp allele contributing 2 points. In addition, the REP1 genotype coded both as the number of 259 alleles and the number of 263 alleles was used in the interaction analyses. All analyses were adjusted for age at study and sex. For each genetic variant we calculated the odds ratio (OR), 95% confidence interval (CI), and p value for the univariate effect in the conditional logistic regression model. In addition, the coefficient for the multiplicative interaction term and the associated p value were calculated.
We performed similar analyses of gene-gene interactions in subgroups, restricting either to case-unaffected sibling or case-unrelated control pairs, to men–men or women–women pairs, or to younger or older pairs as defined by median age at onset in the cases.
Finally, to understand the joint effects of variants within each of the three investigated genes, we tested for SNP-SNP or SNP-VNTR interactions within SNCA (63 pairs), within MAPT (28 pairs), and within LRRK2 (28 pairs) using the same conditional logistic regression approach.
The statistical packages SAS (version 9.1; SAS Institute Inc., Cary, NC) and S-Plus (version 8.0.1; MathSoft, Seattle, WA) were used for all analyses. In addition to the uncorrected p values, a Bonferroni correction was applied to correct p values for the number of tests performed.
Results
The demographic characteristics of the sample of 1,098 cases and 1,098 controls included in the study (653 case-unaffected sibling pairs and 445 case-unrelated control pairs) are summarized in Table 2.
Table 2.
Demographic Characteristics of Parkinson’s Disease (PD) Cases, Siblings, and Unrelated Controls
| General characteristics | PD Case – Sibling Pairs | PD Case-Unrelated Control Pairs | All PD Case- Control Pairs | |||
|---|---|---|---|---|---|---|
|
| ||||||
| PD Cases | Sibling Controls | PD Cases | Unrelated Controls | PD Cases | All Controls | |
| Total sample, n | 653 | 653 | 445 | 445 | 1,098 | 1,098 |
| Men, n (%) | 417 (63.8) | 329 (50.4) | 288 (64.7) | 288 (64.7) | 705 (64.2) | 617 (56.2) |
| Women, n (%) | 236 (36.1) | 324 (49.6) | 157 (35.3) | 157 (35.3) | 393 (35.8) | 481 (43.8) |
| Age at onset of PD, median (range) | 60.4 (28.2–86.9) | -- | 64.6 (23.3–88.0) | -- | 62.2 (23.3–88.0) | -- |
| Age at study, median (range)1 | 66.3 (30.8–91.4) | 65.1 (32.0–90.4) | 70.3 (44.5–90.4) | 71.8 (44.9–92.8) | 68.0 (30.8–91.4) | 67.6 (32.0–92.8) |
| Ethnicity: | ||||||
| Both parents of European origin, n(%) | 570 (87.3) | 557 (85.3) | 357 (80.2) | 388 (87.2) | 927 (84.4) | 945 (86.1) |
| One parents of European origin, n(%) | 52 (8.0) | 60 (9.2) | 60 (13.5) | 41 (9.2) | 112 (10.2) | 101 (9.2) |
| Family History of PD | ||||||
| First degree family history of PD, n (%) | 73 (16.4) | -- | 106 (16.2) | -- | 179 (16.3) | -- |
| At least third degree family history of PD, n (%) | 112 (27.4) | -- | 193 (29.6) | -- | 315 (28.7) | -- |
Age at blood draw
After correction for multiple testing, there were no significant pairwise interactions between the selected SNCA, MAPT and LRRK2 variants. Ten of the 256 interaction pairs were associated with PD susceptibility at the uncorrected p < 0.05 level (Table 3). This included six SNCA-LRRK2 pairs, three SNCA-MAPT pairs, and one MAPT-LRRK2 pair. None of these interaction effects were significantly associated with PD susceptibility after Bonferroni correction for multiple comparisons. Supplementary Table 1 provides detailed results of the interaction tests for the entire sample.
Table 3.
SNP-SNP Interactions with Marginal Evidence of Association with PD Susceptibility (uncorrected p < 0.05)
| SNP1 | Gene1 | SNP1 OR (95% CI) | SNP1 P value | SNP2 | Gene2 | SNP2 OR (95% CI) | SNP2 P value | Interaction P value |
|---|---|---|---|---|---|---|---|---|
| rs16940758 | MAPT | 1.21 (0.99 – 1.47) | 0.0595 | rs2583959 | SNCA | 1.18 (1.00 – 1.39) | 0.0492 | 0.0049 |
| rs2435211 | MAPT | 1.16 (0.99 – 1.35) | 0.0677 | rs3775423 | SNCA | 1.38 (1.07 – 1.79) | 0.0136 | 0.0105 |
| rs17484286 | LRRK2 | 0.73 (0.57 – 0.94) | 0.0143 | rs3775423 | SNCA | 1.42 (1.10 – 1.85) | 0.0071 | 0.0117 |
| rs17484286 | LRRK2 | 0.72 (0.56 – 0.92) | 0.0093 | rs3775439 | SNCA | 1.21 (0.97 – 1.50) | 0.0914 | 0.0192 |
| rs8079215 | MAPT | 1.16 (0.99 – 1.36) | 0.0713 | rs3775423 | SNCA | 1.41 (1.09 – 1.82) | 0.0083 | 0.0214 |
| rs17484286 | LRRK2 | 0.73 (0.57 – 0.94) | 0.0142 | REP1-263 | SNCA | 1.35 (1.02 – 1.78) | 0.0348 | 0.0233 |
| rs7307562 | LRRK2 | 1.11 (0.96 – 1.29) | 0.1658 | rs9995651 | SNCA | 1.31 (0.92 – 1.86) | 0.1270 | 0.0383 |
| rs17466521 | LRRK2 | 0.88 (0.75 – 1.05) | 0.1522 | rs2572324 | SNCA | 1.23 (1.05 – 1.44) | 0.0120 | 0.0416 |
| rs17484286 | LRRK2 | 0.73 (0.57 – 0.94) | 0.0142 | rs9995651 | SNCA | 1.33 (0.93 – 1.88) | 0.1138 | 0.0445 |
| rs10784486 | LRRK2 | 0.89 (0.76 – 1.05) | 0.1660 | rs2435211 | MAPT | 1.15 (0.98 – 1.35) | 0.0770 | 0.0490 |
P-values are not corrected for multiple testing. For consistency, all results in a given row are based on the same sample that contributed to the interaction analysis, with complete data at both SNPs under consideration.
The 10 nominally significant interactions included interactions of LRRK2 SNP rs17484286 with four SNCA variants, including REP1 coded based on the number of 263 bp alleles, and the three SNPs rs3775423, rs3775439, and rs9995651. LD between the SNCA SNPs rs3775423 and rs3775439 is r2=0.65, while for rs3775423 and rs9995651 r2=0.53. The SNCA SNP rs3775423 showed nominal evidence of interaction with one LRRK2 SNP, as well as two MAPT SNPs (rs2435211 and rs8079215). These two MAPT SNPs are not in very strong LD (r2 = 0.18). There was also nominal evidence of interaction between the MAPT SNP rs2435211 and both a SNCA SNP as well as a LRRK2 SNP.
Stratified analyses also did not identify any significant interactions after adjustment for multiple comparisons. Supplementary Table 2 shows results for all interaction pairs for subgroups defined by type of control, gender, and age at onset. Analysis of 653 cases with matched sibling controls detected 19 nominally significant interactions, while analysis of the 445 cases with unrelated controls detected eight nominally significant interactions. Analyses of 553 men-men pairs detected 11 nominally significant interactions, while analyses of 329 women-women pairs detected five nominally significant interactions. Finally, analysis of 548 early age at onset cases (age at onset ≤median) with their matched controls detected 17 nominally significant interactions, while analysis of 548 late age at onset cases (age at onset >median) with their matched controls detected 16 nominally significant interactions. Median age at onset for cases was 62.16 years. Note that none of the interactions that were nominally significant in the overall sample were also significant in both of two complimentary strata (e.g. in both men and women strata, or in both case-unrelated control and case sibling strata, or in both early and late age at onset strata). However, several of the interactions were significant in more than one of the non-complimentary strata. For example, the interaction between the MAPT SNP rs2435211 and the SNCA SNP rs3775423 was nominally significant in the stratum of men, in the case-sib stratum, and in the stratum containing cases with early age at onset.
Analyses of interaction pairs within each of the three genes of interest showed that seven of the 119 interaction pairs were associated with PD susceptibility at the uncorrected p < 0.05 level (Supplemental Table 3). This included two LRRK2 pairs, three MAPT pairs, and two SNCA pairs. None of these interaction effects were significantly associated with PD susceptibility after Bonferroni correction for multiple comparisons.
Discussion
Previous studies have provided compelling evidence that polymorphisms in SNCA, MAPT, and LRRK2 genes confer susceptibility to PD (main effects). Our study indicates that pairwise interactions between common variations in these genes have limited association with PD susceptibility.
Few prior studies have investigated the joint effects of PARK locus genes and related PD susceptibility genes[26–29]. One study observed significant joint effects on PD susceptibility for the MAPT H1 haplotype and SNCA 3′ SNP variants, with a synergistic interaction of the risk alleles[26]. We previously investigated joint effects for MAPT H1 haplotype and SNCA REP1 variants with PD susceptibility and found that the main effects of the variants were separate and equal with no significant pairwise interactions[27]. Here we extended those analyses of joint effects to include multiple SNPs in SNCA, MAPT, and LRRK2, but again we did not observe any significant pairwise interactions. Recently, a whole-genome conditional two-locus analysis identified SNPs that interacted with SNPs in PARK locus genes[30]. In agreement with our results, that study also failed to detect significant pairwise interactions for SNCA, LRRK2, and MAPT SNPs.
Although none of the interactions were significant after correction for multiple testing in our study, the pairs with suggestive evidence of interaction could be investigated in independent samples. In particular, several of the investigated SNPs or VNTRs were involved in multiple interactions that were significant at the nominal 0.05 significance level. These interactions are good candidates for further investigation.
We focused on the three genes that have been confirmed as playing a role in PD susceptibility by candidate gene and genome-wide association studies (SNCA, MAPT, and LRRK2). Rather than restricting the analyses to the few SNPs with the strongest evidence for association in prior GWAS, we studied a number of variants in each gene (including VNTRs)[21]. Restricting our interaction analyses to the three genes that have recently been confirmed as PD susceptibility loci allowed us to investigate the joint effects of these variants while limiting the multiple testing burden. However, focusing exclusively on these three genes also limited our ability to discover novel genetic risk factors for PD. Nevertheless an assessment of interactions for the three established PD susceptibility genes was an important step towards understanding their joint effects and their full contribution to PD.
Although we had a large sample size of 1,098 PD cases and 1,098 controls, this sample size may still be inadequate for detecting interaction effects, particularly involving rare alleles. We estimated power to detect the interaction of the two genetic variants of primary interest (SNCA REP1 and MAPT H1/H2 variation). The power calculations were performed using the software Quanto [31] (http://hydra.usc.edu/GxE/), with an alpha level of 0.0002, which takes into account the fact that 256 interactions were tested. We used the observed allele frequencies in our calculations, and assumed dominant effects of the 259 allele of SNCA REP1 and the H2 haplotype of MAPT. We considered models with marginal effect sizes consistent with those observed in our data (with both the 259 REP1 allele and the H2 MAPT haplotype having protective effects, with marginal odds ratios of approximately 0.80 and 0.75, respectively). Under these assumptions the power was estimated to be 80% to detect an interaction effect size (which represents a ratio of odds ratios) of 0.40 for the combination of protective genotypes at the two loci, or 2.5 for the combination of high-risk genotypes. Smaller interaction effects may exist, and samples sizes that are currently being used for association studies of main effects may not provide adequate power for analyses of joint effects. Design of gene-gene interaction studies is complicated by the fact that sample size requirements depend on the true effect sizes of interactions, which at this point remain unknown. Collaborative analyses of our suggestive interaction findings within a global genetics consortium may yield more definitive evidence of interactions[8].
Another limitation of our study stems from the use of sibling controls for some of the cases, which may reduce power relative to a sample with unrelated controls. Sibling controls were selected when possible to ensure close matching on ethnicity, and thus avoid population stratification effects. The disadvantage of using sibling controls is that because siblings share a considerable proportion of genetic background, the power for detecting genetic associations (both main effects and interactions) is reduced. However, the reduction in power to detect gene-gene interactions is usually quite small, as shown by Gauderman [31]. Gauderman’s power calculations demonstrate that while larger sample sizes are often needed to detect gene-gene interactions in case-sib studies than in case-unrelated-control studies, the sample size requirements are not substantially different, usually requiring no more than a 20% increase in sample size for the case-sib design. Gauderman also showed some scenarios for which the case-sib design had greater power to detect gene-gene interactions than did the case-unrelated-control design.
Finally, we note that our study only considered SNPs and VNTRs. Additional interaction variables to consider in the study of PD include copy number variations, and environmental and epigenetic variations. Studies that include measurement of multiple types of variations in samples from multiple diverse populations may ultimately uncover the complex causes of PD.
Supplementary Material
Results for the association of PD with interactions of each SNP or VNTR variant in two different genes in the sample overall.
Results of tests of association of PD with interactions of each SNP or VNTR in two different genes in the sample stratified by sex, type of control (sibling or unrelated), and age of the case.
Results for the association of PD with interactions of pairs of SNPs within the same gene for the sample overall.
Figure 1. Linkage disequilibrium plots.
Plots of the linkage disequilibrium (LD) structure of the SNCA (Figure 1A), MAPT, (Figure 1B), and LRRK2 (Figure 1C) genes are shown. For the SNCA gene, the multiallelic VNTR REP1 was coded as 259 bp vs. others. The LD values as measured using r2 are given by numbers and the LD values as measured by D′ are given by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data). SNPs used in the interaction analyses are listed in Table 1.
Acknowledgments
The study was funded by the NIH grant 2R01ES10751. We wish to thank the many research personnel who comprised the Molecular Epidemiology of Parkinson’s Disease research team (beyond the authors listed here). We especially wish to thank the many Parkinson’s disease patients, their siblings, and also the unrelated population controls for their participation in the study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pankratz N, Wilk J, Latourelle J, DeStefano A, Halter C, Pugh E, et al. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Human Genetics. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nature genetics. 2009 Dec;41(12):1303–7. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 3.Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nature genetics. 2009 Dec;41(12):1308–12. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards TL, Scott WK, Almonte C, Burt A, Powell EH, Beecham GW, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74(2):97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997 Jun 27;276(5321):2045–7. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 6.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004 Nov 18;44(4):601–7. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, et al. alpha-Synuclein locus triplication causes Parkinson’s disease. Science. 2003 Oct 31;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 8.Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006 Aug 9;296(6):661–70. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 9.Maraganore DM, Hernandez DG, Singleton AB, Farrer MJ, McDonnell SK, Hutton ML, et al. Case-Control study of the extended tau gene haplotype in Parkinson’s disease. Ann Neurol. 2001 Nov;50(5):658–61. doi: 10.1002/ana.1228. [DOI] [PubMed] [Google Scholar]
- 10.Arima K, Hirai S, Sunohara N, Aoto K, Izumiyama Y, Uéda K, et al. Cellular co-localization of phosphorylated tau- and NACP/[alpha]-synuclein-epitopes in Lewy bodies in sporadic Parkinson’s disease and in dementia with Lewy bodies. Brain Research. 1999;843(1–2):53–61. doi: 10.1016/s0006-8993(99)01848-x. [DOI] [PubMed] [Google Scholar]
- 11.Alegre-Abarrategui J, Ansorge O, Esiri M, Wade-Martins R. LRRK2 is a component of granular alpha-synuclein pathology in the brainstem of Parkinson’s disease. Neuropathology and Applied Neurobiology. 2008;34(3):272–83. doi: 10.1111/j.1365-2990.2007.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, et al. Initiation and Synergistic Fibrillization of Tau and Alpha-Synuclein. Science. 2003 Apr 25;300(5619):636–40. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 13.Esposito A, Dohm CP, Kermer P, Bähr M, Wouters FS. [alpha]-Synuclein and its disease-related mutants interact differentially with the microtubule protein tau and associate with the actin cytoskeleton. Neurobiology of Disease. 2007;26(3):521–31. doi: 10.1016/j.nbd.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Lei P, Ayton S, Finkelstein DI, Adlard PA, Masters CL, Bush AI. Tau protein: Relevance to Parkinson’s disease. The International Journal of Biochemistry & Cell Biology. 2010 doi: 10.1016/j.biocel.2010.07.016. In Press, Corrected Proof. [DOI] [PubMed] [Google Scholar]
- 15.Taymans JM, Cookson MR. Mechanisms in dominant parkinsonism: The toxic triangle of LRRK2, α-synuclein, and tau. BioEssays. 2010;32(3):227–35. doi: 10.1002/bies.200900163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carballo-Carbajal I, Weber-Endress S, Rovelli G, Chan D, Wolozin B, Klein CL, et al. Leucine-rich repeat kinase 2 induces [alpha]-synuclein expression via the extracellular signal-regulated kinase pathway. Cellular Signalling. 2010;22(5):821–7. doi: 10.1016/j.cellsig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. Journal of neuroscience research. 2008 Jun;86(8):1711–20. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinton LK, Blurton-Jones M, Myczek K, Trojanowski JQ, LaFerla FM. Synergistic Interactions between A{beta}, Tau, and {alpha}-Synuclein: Acceleration of Neuropathology and Cognitive Decline. J Neurosci. 2010 May 26;30(21):7281–9. doi: 10.1523/JNEUROSCI.0490-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Apaydin H, Ahlskog JE, Parisi JE, Boeve BF, Dickson DW. Parkinson disease neuropathology: later-developing dementia and loss of the levodopa response. Archives of Neurology. 2002;59(1):102–12. doi: 10.1001/archneur.59.1.102. [DOI] [PubMed] [Google Scholar]
- 20.Brighina L, Frigerio R, Schneider NK, Lesnick TG, de Andrade M, Cunningham JM, et al. Alpha-synuclein, pesticides, and Parkinson disease: a case-control study. Neurology. 2008 Apr 15;70(16 Pt 2):1461–9. doi: 10.1212/01.wnl.0000304049.31377.f2. [DOI] [PubMed] [Google Scholar]
- 21.Chung S, Armasu S, Biernacka J, Lesnick TG, Rider D, Lincoln S, et al. Common Variants in PARK Loci and Related Genes and Parkinson’s Disease. 2010 doi: 10.1002/mds.23376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Incidence and distribution of parkinsonism in Olmsted County, Minnesota, 1976–1990. Neurology. 1999 Apr 12;52(6):1214–20. doi: 10.1212/wnl.52.6.1214. [DOI] [PubMed] [Google Scholar]
- 23.Rocca WA, Maraganore DM, McDonnell SK, Schaid DJ. Validation of a telephone questionnaire for Parkinson’s disease. J Clin Epidemiol. 1998 Jun;51(6):517–23. doi: 10.1016/s0895-4356(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 24.Kooperberg C, Leblanc M. Increasing the power of identifying gene x gene interactions in genome-wide association studies. Genet Epidemiol. 2008 Apr;32(3):255–63. doi: 10.1002/gepi.20300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Goris A, Williams-Gray CH, Clark GR, Foltynie T, Lewis SJ, Brown J, et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson’s disease. Ann Neurol. 2007 Aug;62(2):145–53. doi: 10.1002/ana.21192. [DOI] [PubMed] [Google Scholar]
- 27.Mamah CE, Lesnick TG, Lincoln SJ, Strain KJ, de Andrade M, Bower JH, et al. Interaction of alpha-synuclein and tau genotypes in Parkinson’s disease. Ann Neurol. 2005 Mar;57(3):439–43. doi: 10.1002/ana.20387. [DOI] [PubMed] [Google Scholar]
- 28.Maraganore DM, de Andrade M, Lesnick TG, Farrer MJ, Bower JH, Hardy JA, et al. Complex interactions in Parkinson’s disease: a two-phased approach. Mov Disord. 2003 Jun;18(6):631–6. doi: 10.1002/mds.10431. [DOI] [PubMed] [Google Scholar]
- 29.McCulloch CC, Kay DM, Factor SA, Samii A, Nutt JG, Higgins DS, et al. Exploring gene-environment interactions in Parkinson’s disease. Hum Genet. 2008 Apr;123(3):257–65. doi: 10.1007/s00439-008-0466-z. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Perez A, Gayan J, Marin J, Galan JJ, Saez ME, Real LM, et al. Whole-genome conditional two-locus analysis identifies novel candidate genes for late-onset Parkinson’s disease. Neurogenetics. 2009 Jan 21; doi: 10.1007/s10048-009-0170-8. [DOI] [PubMed] [Google Scholar]
- 31.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. American journal of epidemiology. 2002 Mar 1;155(5):478–84. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Results for the association of PD with interactions of each SNP or VNTR variant in two different genes in the sample overall.
Results of tests of association of PD with interactions of each SNP or VNTR in two different genes in the sample stratified by sex, type of control (sibling or unrelated), and age of the case.
Results for the association of PD with interactions of pairs of SNPs within the same gene for the sample overall.