Abstract
α-Synuclein gene (SNCA) multiplications cause familial parkinsonism and allele-length polymorphisms within the SNCA dinucleotide repeat REP1 increase the risk for developing Parkinson’s disease (PD). Since SNCA multiplications increase SNCA expression, and REP1-genotypes that increase the risk of developing PD show increased SNCA expression in cell-culture systems, animal models, and human blood and brain, PD therapies seek to reduce SNCA expression. We conducted an observational study of 1,098 PD cases to test the hypothesis that REP1 genotypes correlated with reduced SNCA expression are associated with better motor and cognitive outcomes. We evaluated the association of REP1 genotypes with survival free of Hoehn and Yahr stages 4 or 5 (motor outcome) and of Modified Telephone Interview for Cognitive Status score ≤27 or Alzheimer’s Disease-8 score ≥2 (cognitive outcome). Median disease duration at baseline was 3.3 years and median lag time from baseline to follow-up was 7.8 years. Paradoxically, REP1 genotypes associated with increased risk of developing PD and increased SNCA expression were associated with better motor (HR=0.87, p=0.046 covariate-adjusted age-scale analysis; HR=0.85, p=0.020, covariate-adjusted time-scale analysis) and cognitive outcomes (HR=0.90, p=0.12, covariate-adjusted age-scale analysis; HR=0.85, p=0.023, covariate-adjusted time-scale analysis). Our findings raise the possibility that SNCA has a dual, opposing, and time-dependent role. This may have implications for the development of therapies that target SNCA expression.
Keywords: Parkinson’s disease, α-synuclein gene, outcomes
INTRODUCTION
α-Synuclein gene (SNCA) multiplications cause familial PD via an overexpression mechanism [1]. PD susceptibility is influenced by allele-length polymorphisms in the mixed dinucleotide repeat REP1 (D4S3481, ~10 kb upstream of the SNCA transcription start site) [2], which are correlated with altered SNCA expression in cell cultures [3], transgenic mouse brain [4], and human blood and brain [5–6]. Specifically, longer REP1 allele lengths (263 bp) that correlate with increased SNCA expression are associated with increased risk of developing PD and shorter REP1 allele lengths (259 bp) that correlate with reduced SNCA expression are associated with reduced risk of developing PD [2]. These genetic findings, together with α-synuclein immunostaining of Lewy bodies [7], provide proof of principle for therapies aiming to reduce SNCA expression in PD [8–9].
Recently, Ritz et al. [10] reported the association of SNCA REP1 263 bp alleles (correlated with increased expression) with more rapid motor symptoms progression in PD as assessed by the rate of decline of Unified Parkinson’s Disease Rating Scale (UPDRS) motor scores over a 5-year interval. They interpreted their findings to support therapies targeting reduction of SNCA expression in PD. Here we present a study of the association of twenty SNCA variants (REP1 and 19 additional haplotype-tagging SNPs) and survival free of motor and cognitive outcomes, for 1,098 PD subjects with up to 13 years of follow-up. Since increased SNCA expression is associated with increased risk of developing PD, we hypothesized that REP1 genotypes correlated with reduced SNCA expression (one or two 259 bp alleles) would be associated with greater survival free of developing motor and cognitive outcomes in PD. Surprisingly, the findings of our studies are opposite to this hypothesis and contrast with those of Ritz et al. and suggest a possible time-dependent, dual and opposing effect of SNCA in PD.
MATERIALS AND METHODS
Subjects
Clinical information and biological samples were collected with written informed consent following a protocol approved by the Mayo Clinic IRB (Rochester, MN). Study subjects were 1,098 PD cases from the Molecular Epidemiology of Parkinson’s Disease study (“MEPD study”, NIH 2R01ES10751) referred sequentially to the Department of Neurology of the Mayo Clinic in Rochester, MN, from June 1, 1996 through June 30, 2007. They resided in Minnesota or a neighboring state. All PD cases were examined in a standardized fashion by neurologists specializing in Movement Disorders, and employing a comprehensive protocol for clinical assessment. Cases fulfilled criteria for clinically definite or probable PD [11].
Molecular analyses
Blood was collected and genomic DNA was obtained. Allele length of SNCA REP1 was assessed using an Applied Biosystems sequencing platform (Genotyping Shared Resources, Mayo Clinic, Rochester, MN) and 19 haplotype-tagging SNCA SNPs were assessed using an Illumina GoldenGate genotyping platform [12].
Outcome measurements
Motor and cognitive outcome data for 1,098 PD cases were collected by telephone interview questionnaires directly with the cases or via proxy when incapacitated or deceased. The direct interview questionnaire included the Telephone Interview of Cognitive Status-Modified (TICS-M) [13] and questions regarding motor milestones such as inability to stand or walk unassisted (and dates). The proxy questionnaires collected the same information, with the exception of screening for dementia using the Alzheimer’s Disease Dementia Screening Interview (AD-8) [14] because AD-8 is a brief informant-based measure that reliably differentiates between non-demented and demented individuals and is sensitive to the earliest signs of cognitive change as reported by a proxy informant.
The motor outcome was defined as Hoehn and Yahr (H&Y) stage 4 or 5 and assessed at baseline via clinical assessment and imputed at follow-up via telephone interview. The question asked at the telephone interview was: “Are you able to stand or walk without someone else helping you, or without a cane or walker?” A “no” response corresponded to H&Y stages 4 or 5. A “no” response was followed by the question: “At what age were you no longer able to stand or walk without assistance?” Both questions were appropriately reworded for proxy interviews.
Cognitive outcome was assessed at baseline using the Mini Mental State Examination (MMSE). The cognitive outcome was defined as MMSE <26 [15]. Cognitive outcome at follow-up was assessed via telephone interview using the TICS-M (direct interviews) or the AD-8 (proxy interviews). Cognitive outcome was defined as a TICS-M score ≤27 or AD-8 score ≥2. The outcome assessments were identical to those that we reported for a genome-wide association (GWAS) study of motor and cognitive outcomes in PD [16]. That study did not include SNCA-REP1 genotypes and the 19 SNCA haplotype-tagging SNPs that are the focus of this study, and that study included only a subset (n=443) of the cases included in this study (n=1,098).
Statistical analyses
Statistical analyses evaluated the association of each genetic variant with motor and cognitive impairment, with the primary outcome being time-to-event (motor or cognitive outcomes). Cox proportional hazards regression models were used to assess the association of outcomes with genotypes. For cases with no evidence of motor or cognitive outcomes, time-to-censoring was defined as the time between the baseline clinical assessment and the telephone interview, or time between baseline and death if the individual was deceased. For cases with evidence of motor or cognitive outcomes, time-to-event was defined based on the age of onset of motor or cognitive outcomes as determined by direct or proxy telephone interviews. Subjects with H&Y stage 4/5 or with MMSE <26 at baseline, were excluded from the respective motor or cognitive outcomes survival analyses. To limit survival bias, age at enrollment was considered as time 0 (or baseline). Analyses were performed both using the age-scale and the time-on-study scale. While the time-on-study scale is often used in analyses of disease progression, analysis on the age scale is more appropriate when studying outcomes associated with age in an aging population [18]. Log-rank tests were used to determine significance and associations between outcomes and genetic variables using hazard ratios (HR) with 95% confidence intervals. Kaplan-Meier plots were used to visualize the overall survival function, and stratified by genetic variables.
Prior to testing the effects of SNCA variants on motor and cognitive impairment outcomes, analyses were performed to identify relevant covariates associated with the outcomes. Relevant covariates were then included in the Cox proportional hazards models that were used to assess the association of outcomes with genotypes. The age-scale analysis of the motor impairment outcome included sex, disease duration at baseline, and L-DOPA treatment at baseline as covariates, while the cognitive impairment analysis included sex, disease duration at baseline, and education as covariates. The age-scale analyses of motor and cognitive outcomes were also performed without covariate adjustment. Similarly, the analyses on the time-on-study scale were performed without and with covariate adjustment (same covariates as above with the addition of age at enrollment as a covariate in analyses of both motor and cognitive outcomes).
The primary analysis included the association of outcomes with SNCA REP1 genotypes using the REP1 score as described previously [17]. The REP1 score ranged from 0 (lowest PD risk, lowest SNCA expression) to 4 (highest PD risk, highest SNCA expression). Secondary analyses investigated the association of motor and cognitive outcomes with 19 haplotype-tagging SNCA SNPs defined by the minor allele count (0, 1, or 2 copies of the minor frequency allele); and with REP1 genotypes re-defined by minor allele counts (0, 1, or 2 copies of the 259 bp allele; or 0, 1, or 2 copies of the 263 bp allele). As SNCA REP1 was the genetic polymorphism of interest, primary analyses were not corrected for multiple comparisons. The Bonferroni correction for multiple comparisons was applied to the secondary analyses (20 tests for each outcome). Results with uncorrected p-values < 0.05 in the primary analyses or Bonferroni-corrected p-values < 0.05 in the secondary analyses were considered significant.
The statistical packages SAS® (version 9.2; SAS Institute Inc., Cary, NC) and R (version 2.14; R Development Core Team (2011). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org/) were used for all analyses.
RESULTS
Demographic and clinical characteristics
1,098 MEPD cases were included in this study (Supplemental Table 1). 85 cases were lost to follow-up, and 91 PD cases (or proxies) refused follow-up interviews. Thus, the overall participation rate among those contacted was 91.0%. At follow-up, 467 direct interviews, 180 proxy interviews for incapacitated subjects, and 275 proxy interviews for deceased subjects were performed. From the 922 participating cases 604 were men (65.5%) and 318 women (34.5%) primarily of Caucasian race and European ancestry. Median disease duration at baseline was 3.3 years. 652 of the 922 participating cases (70.7%) had been treated with levodopa at baseline. 62 cases (6.7%) that had reached H&Y stages 4/5 at baseline were removed from the motor outcomes analyses. 59 cases (6.4%) that had reached MMSE <26 at baseline were removed from the cognitive analyses.
The median lag time from baseline to follow-up was 7.8 years (range 3.3–13 years). 44 cases with a TICS-M score ≤27 were interviewed directly and repeated interviews (proxy for incapacitated subjects) were conducted in order to obtain valid dates and other information for the survival analyses. Information regarding levodopa therapy at follow-up or cumulative dose exposures was not available.
Molecular analysis
Twenty SNCA variants were genotyped and their positions are shown in a Linkage Disequilibrium (LD) map for SNCA (Supplemental Figure 1).
REP1 and motor outcomes
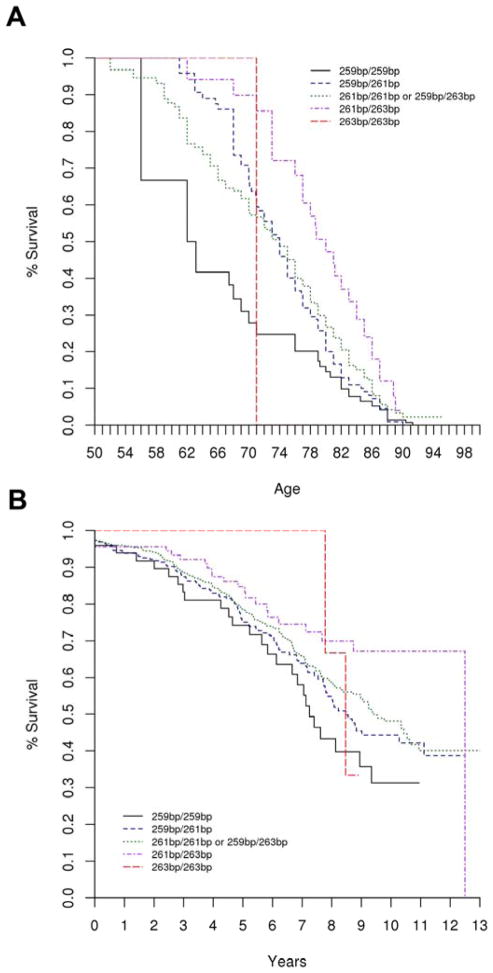
Our initial hypothesis was that PD cases with lower REP1 scores, associated with reduced PD risk and correlated with reduced SNCA expression, would have reduced risk of developing motor and/or cognitive impairment and thus longer survival free of these outcomes. However, we observed the opposite association: PD cases with higher REP1 scores had reduced risk of developing motor impairment and longer survival free of the outcome. This result was statistically significant both in the unadjusted and the covariate-adjusted analyses on the time-on-study scale as well as the unadjusted and covariate-adjusted analyses performed on the age-scale (adjusted analysis on age-scale: HR = 0.87, 95% CI 0.75–1.00, p = 0.046; see Supplemental Table 2). Under the covariate-adjusted age-scale model, having a lower REP1 score conferred a nearly two-fold greater risk for developing the motor outcome than a higher REP1 score (HR=1.78, 95%CI 1.01–3.15, REP1 score of 0 vs. REP1 score of 4). Figure 1 shows the Kaplan-Meier plot for survival free of developing the motor outcome by the REP1 score. Genotype and allele frequencies are shown in Table 1.
Figure 1.
Kaplan Meier plot for SNCA REP1 and survival free of motor impairment (Hoehn and Yahr stages 4 or 5). The genotype 259bp/259bp corresponds to REP1 score = 0, and 259bp/261bp to REP1 score = 1, and 261bp/261bp or 259bp/263bp to REP1 score = 2, and 261bp/263bp to REP1 score = 3, and 263bp/263bp to REP1 score = 4. Figure 1a corresponds to the covariate unadjusted, age-scale analysis (HR = 0.87, 95% CI 0.75–0.99, p = 0.042); and Figure 1b corresponds to the covariate unadjusted, time-on-study analysis (HR = 0.81, 95% CI 0.70–0.93, p = 0.0035).
Table 1.
REP1 genotype frequencies for MEPD Motor outcomes (N=854)
| REP1 genotypes | Genotype Frequency n (%) |
| 257/261 | 2 (0.2%) |
| 257/263 | 1 (0.1%) |
| 259/259 | 49 (5.7%) |
| 259/261 | 257 (30.1%) |
| 259/263 | 25 (2.9%) |
| 259/265 | 1 (0.1%) |
| 261/261 | 422 (49.4%) |
| 261/263 | 90 (10.5%) |
| 261/265 | 1 (0.1%) |
| 263/263 | 6 (0.7%) |
| REP1 score | Genotype Frequency n (%) |
| 0 | 49 (5.8%) |
| 1 | 257 (30.3%) |
| 2 | 447 (52.7%) |
| 3 | 90 (10.6%) |
| 4 | 6 (0.7%) |
| REP1 259 bp | Genotype Frequency n (%) |
| 0 | 522 (61.1%) |
| 1 | 283 (33.1%) |
| 2 | 49 (5.7%) |
| REP1 263 bp | Genotype Frequency n (%) |
| 0 | 732 (85.7%) |
| 1 | 116 (13.6%) |
| 2 | 6 (0.7%) |
| REP1 261 bp | Genotype Frequency n (%) |
| 0 | 82 (9.6%) |
| 1 | 350 (41.0%) |
| 2 | 422 (49.4%) |
REP1 and cognitive outcomes
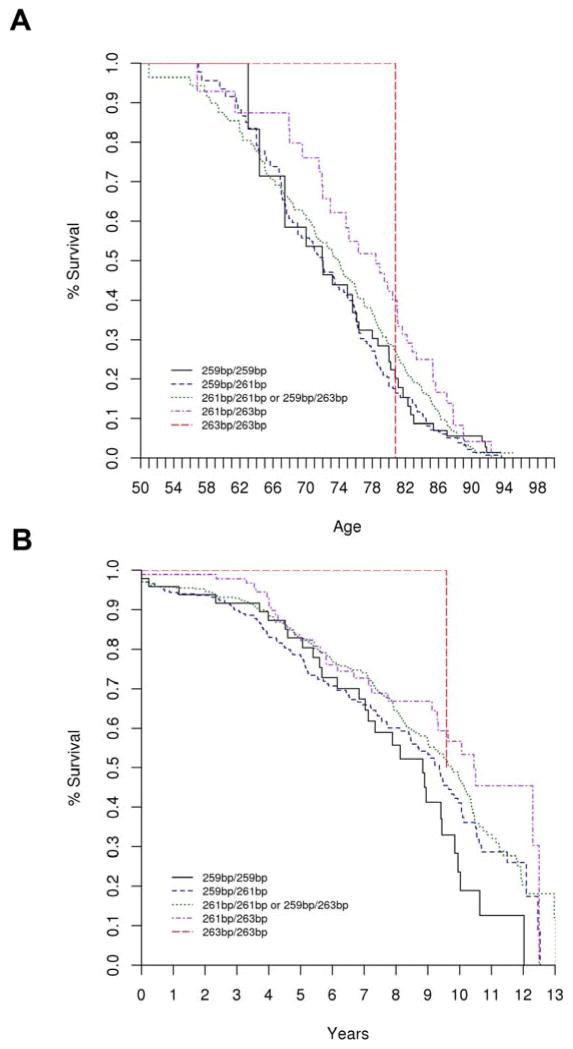
Consistent with our findings for motor outcomes, we observed an opposite-than-expected association for cognitive outcomes. PD cases with higher REP1 scores had reduced risk of developing cognitive impairment and longer survival free of the outcomes. While this result was statistically significant in the unadjusted and covariate-adjusted analysis on the time scale (adjusted analysis on time-scale: HR = 0.85, 95% CI 0.75–0.98, p = 0.023), it just failed to achieve significance (p=0.058) in the unadjusted analysis on the age-scale, and was not significant in the covariate-adjusted analysis on the age-scale (p=0.12). Under the covariate-adjusted age-scale model, having a low REP1 score conferred a 1.5 times greater risk for developing the cognitive outcome than having a high REP1 score (HR=1.53, 95% CI 0.90–2.60, REP1 score of 0 vs. REP1 score of 4). Results of all analyses are shown in Supplemental Table 3. Figure 2 shows the Kaplan-Meier plots for survival free of the cognitive outcome by REP1 score. Genotype and allele frequencies are shown in Table 2.
Figure 2.
Kaplan Meier plot for SNCA REP1 and survival free of cognitive impairment (TICS-M score ≤27 or AD-8 score ≥2). The genotype 259bp/259bp corresponds to REP1 score = 0, and 259bp/261bp to REP1 score = 1, and 261bp/261bp or 259bp/263bp to REP1 score = 2, and 261bp/263bp to REP1 score = 3, and 263bp/263bp to REP1 score = 4. Figure 2a corresponds to the covariate unadjusted, age-scale analysis (HR = 0.88, 95% CI 0.78–1.00, p = 0.058); and Figure 2b corresponds to the covariate unadjusted, time-on-study analysis (HR = 0.81, 95% CI 0.71–0.92, p = 0.0017).
Table 2.
REP1 genotype frequencies in cognitive outcomes (N=858)
| REP1 genotypes | Genotype Frequencies n (%) |
| 257/261 | 2 (0.2%) |
| 257/263 | 1 (0.1%) |
| 259/259 | 48 (5.6%) |
| 259/261 | 265 (30.9%) |
| 259/263 | 25 (2.9%) |
| 259/265 | 1 (0.1%) |
| 261/261 | 416 (48.5%) |
| 261/263 | 94 (11.0%) |
| 261/265 | 1 (0.1%) |
| 263/263 | 5 (0.6%) |
| REP1 score | Genotype Frequency n (%) |
| 0 | 48 (5.6%) |
| 1 | 265 (31.1%) |
| 2 | 441 (51.7%) |
| 3 | 94 (11.0%) |
| 4 | 5 (0.6%) |
| REP1 259 bp | Genotype Frequency n (%) |
| 0 | 519 (60.5%) |
| 1 | 291 (33.9%) |
| 2 | 48 (5.6%) |
| REP1 263 bp | Genotype Frequency n (%) |
| 0 | 733 (85.4%) |
| 1 | 120 (14.0%) |
| 2 | 5 (0.6%) |
| REP1 261 bp | Genotype Frequency n (%) |
| 0 | 80 (9.3%) |
| 1 | 362 (42.2%) |
| 2 | 416 (48.5%) |
SNPs and motor and cognitive outcomes
The association results for all genotyped SNCA variants (19 SNPs, REP1) and developing the motor and cognitive outcomes are included in Supplemental Tables 2 and 3. None of the SNPs were significantly associated with developing motor or cognitive impairment after Bonferroni correction for multiple testing, including those tagging the 3′ LD block.
DISCUSSION
In contrast to our original hypothesis our results reveal an unexpected association of the SNCA REP1 genotypes correlated with reduced SNCA expression with the development of worse motor and cognitive outcomes in a large and well-characterized cohort of PD cases. Our results are inconsistent with findings from SNCA multiplication mutations in familial parkinsonism, where increased SNCA-genomic dosage and SNCA expression were associated with earlier age at onset, faster progression and increased motor and cognitive severity [1]. However, our results are consistent with clinical and genetic information that two of the authors (KM, BAC) newly obtained from a previously reported Greek family with PD [19]. In affected kindred members, the SNCA c.157G>A (p.A53T) mutation is in phase with the REP1 259-bp allele (Supplemental Figure 2). All affected individuals with the 259-bp REP1 allele had a severe phenotype with poor motor and cognitive outcomes (median duration to H&Y stages 4/5 was 7 years and to dementia was 7 years), while a recombinant lacking the 259-bp REP1 allele remains asymptomatic past the mean age of onset for their generation. This is consistent with the hypothesis that the co-localization of the causal gene mutation with the 259-bp REP1 allele is associated with variable expressivity (worse outcomes) in this family.
Our results are also inconsistent with findings reported by Ritz et al [10], where REP1 alleles associated with increased PD risk and correlated with increased SNCA expression were associated with the development of worse motor outcomes. There are several possible explanations for these conflicting results. First, the difference may be due to sample size. Our sample at study enrollment was 3-fold larger than that of Ritz et al. (1,098 vs. 363 PD cases), and at follow-up was nearly 4-fold larger (854 vs. 233 PD cases). Small and underpowered samples have greater false-positive rates and may provide erroneous or exaggerated estimates of the direction or size of effect. Second, the population characteristics were different. Our study population was racially and ethnically homogeneous with subjects of Caucasian and European descent, whereas in the Ritz study ~19% of the subjects were non-Caucasian. The frequencies and effects of gene variants vary by race and ethnicity, and population stratification can further erode statistical power or bias towards false-positive associations. Third, we defined motor outcome differently. Our outcome was reaching H&Y stages 4/5; inability to stand or walk unassisted represents a major milestone in PD progression and is therefore likely to be recalled reliably by patients or their proxies. Ritz et al. used the rate of decline in motor severity measured by the UPDRS part III scale. The UPDRS is subject to treatment effects and inter-rater variability, and change in the UPDRS score by five points may not result in a meaningful change in H&Y stage. Our findings were internally consistent for the development of both motor and cognitive outcomes, whereas the Ritz study did not assess cognitive measures. Fourth, our follow-up duration was substantially longer, nearly fifteen years vs. five years of Ritz et al. It is conceivable that we are observing differential effects of REP1-allele genotypes at different disease stages.
While a cross-sectional study of SNCA polymorphisms and the development of motor outcomes in a cohort of multiplex families with PD has been reported, it was not representative of sporadic PD [20]. Association studies of SNCA polymorphisms and cognitive outcomes in sporadic PD were small, considered only a few variants, and were mostly cross-sectional [21–23].
Our genetic association study has several strengths: First, we used an observational study design to evaluate the effects of SNCA genotypes on the development of motor and cognitive outcomes in PD. While there have been experimental studies of therapies targeting SNCA expression in model systems [8–9], observational studies of SNCA effects on the development of motor and cognitive outcomes to assess the long term benefits of therapeutic interventions are lacking. Second, all MEPD study cases were recruited using strict enrollment and diagnostic criteria, resided in a defined geographic region, and examined at baseline by Movement Disorders specialists using a standardized and comprehensive clinical assessment protocol. Third, our follow-up telephone interview assessments included validated measures allowing us to determine outcomes directly or by proxy for 84% of the subjects. Fourth, follow-up interval was long (up to 13 years) and the study had sufficient power to detect clinically meaningful effects. Fifth, genotypes were determined for REP1 but also for 19 SNCA SNPs.
Our study also has several weaknesses. First, a sampling bias cannot be excluded as our PD cohort was referral-based. Attempts to limit sampling bias included recruiting cases prospectively from a defined geographic region and by not excluding or enriching for familial PD cases. The frequency of known PARK mutations was very low [24–25]. Second, to limit survival bias we evaluated outcomes in PD cases with variable disease duration at baseline and included disease duration at baseline as an adjustment variable in the statistical models. Third, the measurements at baseline and follow-up employed different methods and the validity of self or proxy reported assessments and imputed H&Y stages are unclear. Fourth, treatment effects cannot be excluded since our PD cohort was not randomized to treatment. L-DOPA therapy at baseline was included as an adjustment variable in some of the models, but cumulative dose exposures to dopaminergic therapy could not be ascertained. Fifth, statistical power may be greater for repeated quantitative measures than for singly performed dichotomous measures and survival analyses.
Both a neurotoxic and a neuroprotective role for SNCA expression have been proposed in cell culture and animal models [26]. In addition, both up-regulation and down-regulation of SNCA expression in PD and control brains has been reported [6, 27–29]. It is conceivable that SNCA expression variability may reflect a dynamic state of SNCA regulation over the disease course. SNCA REP1 genotypes associated with reduced expression may function in a neuroprotective manner reducing the risk to develop PD, whereas well after PD symptom-onset these genotypes may exacerbate disease progression. One speculative mechanism is that after disease onset, as α-synuclein and its proteolytic fragments are sequestered in aggregates (e.g., Lewy Bodies), relatively reduced expression fails to support the production of sufficient pools of normal α-synuclein, which in turn contributes to further disease progression. Individuals with short REP1 alleles may be more at-risk for this condition following disease onset. A dual and opposing effect of gene/protein function in neurological disease is not limited to SNCA/α-synuclein. β-Amyloid, which plays a critical role in the pathogenesis of Alzheimer’s disease, has a beneficial effect in animal models of multiple sclerosis [30, 31]. The apolipoprotein E gene ε2 variant (APOE ε2) is associated with a reduced risk for Alzheimer’s disease, but with an increased amyloid neuropathology burden in Alzheimer’s disease patients older than 90 years; and while APOE ε2 reduces the risk for Alzheimer’s disease, it increases the risk for cerebral amyloid angiopathy and associated hemorrhages (which are prevalent in Alzheimer’s disease patients) [32]. Further, APOEε4 alleles have differential effects on amyloid load and glucose metabolism in the frontal and occipital cortices of Alzheimer’s disease patients [33].
Taken together, these observations point to a paradigm for neurodegeneration in which a critical gene’s function depends on its spatial and temporal context, potentially resulting in dual and opposing effects. Our findings suggest that therapies aiming at SNCA reduction may worsen later outcomes. These effects may not be detected by Phase 1 clinical trials that select small samples of early PD cases and employ short follow-up periods. Observational studies using large and representative PD cohorts with long follow-up periods may evaluate this critical issue.
Supplementary Material
Demographic characteristics of cases from the Molecular Epidemiology of Parkinson’s Disease Study
Genetic association with time-to-motor-impairment
Genetic association with time-to-cognitive-impairment
Linkage disequilibrium map for SNCA. The multi-allelic variable number tandem repeat REP1 was coded as 259 bp vs. others. The LD values as measured using r2 are given by numbers and the LD values as measured by D′ are given by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data).
Haplotype analysis in Family H (modified to protect subject confidentiality). Circles correspond to women and squares to men. Diagonal lines through a symbol indicate deceased status. Shaded lower right quadrants identify individuals affected with PD, while clear symbols indicate unaffected status. A “+” sign indicates that DNA was available for genotyping. The other nine rows beneath the pedigree symbols correspond to: individual identifier (top row) and marker genotypes for D4S2460, D4S3481 (REP1), rs2619363, rs2301135, rs104893877 c.157G>A, rs1005233, rs3216147 and D4S423 (bottom row). The black bar signifies the c.157G>A (p.A53T) haplotype; shaded/unfilled bars distinguish four SNCA promoter haplotypes with different D4S3481 (REP1), rs2619363, and rs2301135 alleles. Haplotypes for deceased family members, without DNA, are probabilistically inferred to best fit the available genotype data. The range and mean of the age for disease onset in 20 affected members of this family is 31–71 and 52.9. In generations III, IV, and V, the respective ranges, n, and mean are 50–71, 8, and 61.2; 40–63, 9, and 50.2; and 31–53, 3, and 39.
Acknowledgments
This paper was made possible by the grant 2R01ES10751 from the National Institutes of Health, and by a grant from Alnylam Pharmaceuticals, Inc. and Medtronic, Inc. (to Demetrius M. Maraganore, MD). Matthew J. Farrer, PhD acknowledges support from the Canada Excellence Research Chairs Program and Leading Edge Endowment Funds provided by the Province of British Columbia, Life Labs, and Genome BC. Work on Family H was supported in part by a grant from the National Parkinson Foundation (to Bruce A. Chase, PhD and Katerina Markopoulou MD, PhD), a seed grant from the University of Nebraska Medical Center (to Katerina Markopoulou MD, PhD), and from the National Institutes of Health grant R15NS043162-01A2 (to Bruce A. Chase, PhD).
Abbreviations
- AD-8
Alzheimer’s Disease-8
- bp
base pair
- H&Y
Hoehn and Yahr
- HR
hazard ratio
- LD
linkage disequilibrium
- MEPD
Molecular Epidemiology of Parkinson’s Disease
- MMSE
Mini Mental State Examination
- SNCA
α-synuclein gene
- SNP
single-nucleotide polymorphism
- TICS-M
Modified Telephone Interview for Cognitive Status
- UPDRS
Unified Parkinson’s Disease Rating Scale
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Farrer M, Kachergus J, Forno L, Lincoln S, Wang DS, Hulihan M, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55:174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 2.Maraganore DM, de AM, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, et al. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296:661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- 3.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10:3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 4.Cronin KD, Ge D, Manninger P, Linnertz C, Rossoshek A, Orrison BM, et al. Expansion of the Parkinson disease-associated SNCA-Rep1 allele upregulates human alpha-synuclein in transgenic mouse brain. Hum Mol Genet. 2009;18:3274–3285. doi: 10.1093/hmg/ddp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs J, Tichopad A, Golub Y, Munz M, Schweitzer KJ, Wolf B, et al. Genetic variability in the SNCA gene influences alpha-synuclein levels in the blood and brain. FASEB J. 2008;22:1327–1334. doi: 10.1096/fj.07-9348com. [DOI] [PubMed] [Google Scholar]
- 6.Linnertz C, Saucier L, Ge D, Cronin KD, Burke JR, Browndyke JN, et al. Genetic regulation of alpha-synuclein mRNA expression in various human brain tissues. PLoS One. 2009;4:e7480. doi: 10.1371/journal.pone.0007480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 8.Lewis J, Melrose H, Bumcrot D, Hope A, Zehr C, Lincoln S, et al. In vivo silencing of alpha-synuclein using naked siRNA. Mol Neurodegener. 2008;3:19. doi: 10.1186/1750-1326-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormack AL, Mak SK, Henderson JM, Bumcrot D, Farrer MJ, Di Monte DA. Alpha-synuclein suppression by targeted small interfering RNA in the primate substantia nigra. PLoS One. 2010;5:e12122. doi: 10.1371/journal.pone.0012122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ritz B, Rhodes SL, Bordelon Y, Bronstein J. alpha-Synuclein genetic variants predict faster motor symptom progression in idiopathic Parkinson disease. PLoS One. 2012;7:e36199. doi: 10.1371/journal.pone.0036199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bower JH, Maraganore DM, McDonnell SK, Rocca WA. Incidence and distribution of parkinsonism in Olmsted County, Minnesota, 1976–1990. Neurology. 1999;52:1214–1220. doi: 10.1212/wnl.52.6.1214. [DOI] [PubMed] [Google Scholar]
- 12.Chung SJ, Armasu SM, Biernacka JM, Lesnick TG, Rider DN, Lincoln SJ, et al. Common variants in PARK loci and related genes and Parkinson’s disease. Mov Disord. 2011;26:280–288. doi: 10.1002/mds.23376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plassman BL, Newman TT, Welsh KA, Helms M, Breitner JCS. Application in Epidemiological and Longitudinal Studies. Cognitive and Behavioral Neurology. 1994;7:235–241. [Google Scholar]
- 14.Galvin JE, Roe CM, Powlishta KK, Coats MA, Muich SJ, Grant E, et al. The AD8: a brief informant interview to detect dementia. Neurology. 2005;65:559–564. doi: 10.1212/01.wnl.0000172958.95282.2a. [DOI] [PubMed] [Google Scholar]
- 15.Dubois B, Burn D, Goetz C, Aarsland D, Brown RG, Broe GA, et al. Diagnostic procedures for Parkinson’s disease dementia: Recommendations from the movement disorder society task force. Mov Disord. 2007;22:2314–2324. doi: 10.1002/mds.21844. [DOI] [PubMed] [Google Scholar]
- 16.Chung SJ, Armasu SM, Biernacka JM, Anderson KJ, Lesnick TG, Rider DN, et al. Genomic determinants of motor and cognitive outcomes in Parkinson’s disease. Parkinsonism Relat Disord. 2012;18:881–886. doi: 10.1016/j.parkreldis.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brighina L, Schneider NK, Lesnick TG, de AM, Cunningham JM, Mrazek D, et al. Alpha-synuclein, alcohol use disorders, and Parkinson disease: a case-control study. Parkinsonism Relat Disord. 2009;15:430–434. doi: 10.1016/j.parkreldis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamarca R, Alonso J, Gomez G, Munoz A. Left-truncated data with age as time scale: an alternative for survival analysis in the elderly population. J Gerontol A Biol Sci Med Sci. 1998;53:M337–M343. doi: 10.1093/gerona/53a.5.m337. [DOI] [PubMed] [Google Scholar]
- 19.Markopoulou K, Wszolek ZK, Pfeiffer RF, Chase BA. Reduced expression of the G209A alpha-synuclein allele in familial Parkinsonism. Ann Neurol. 1999;46:374–381. doi: 10.1002/1531-8249(199909)46:3<374::aid-ana13>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Pankratz N, Nichols WC, Elsaesser VE, Pauciulo MW, Marek DK, Halter CA, et al. Alpha-synuclein and familial Parkinson’s disease. Mov Disord. 2009;24:1125–1131. doi: 10.1002/mds.22524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goris A, Williams-Gray CH, Clark GR, Foltynie T, Lewis SJ, Brown J, et al. Tau and alpha-synuclein in susceptibility to, and dementia in, Parkinson’s disease. Ann Neurol. 2007;62:145–153. doi: 10.1002/ana.21192. [DOI] [PubMed] [Google Scholar]
- 22.De Marco EV, Tarantino P, Rocca FE, Provenzano G, Civitelli D, De L, et al. Alpha-synuclein promoter haplotypes and dementia in Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet. 2008;147:403–407. doi: 10.1002/ajmg.b.30611. [DOI] [PubMed] [Google Scholar]
- 23.Keri S, Nagy H, Myers CE, Benedek G, Shohamy D, Gluck MA. Risk and protective haplotypes of the alpha-synuclein gene associated with Parkinson’s disease differentially affect cognitive sequence learning. Genes Brain Behav. 2008;7:31–36. doi: 10.1111/j.1601-183X.2007.00315.x. [DOI] [PubMed] [Google Scholar]
- 24.Farrer M, Stone J, Mata IF, Lincoln S, Kachergus J, Hulihan M, et al. LRRK2 mutations in Parkinson disease. Neurology. 2005;65:738–740. doi: 10.1212/01.wnl.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- 25.Maraganore DM, Wilkes K, Lesnick TG, Strain KJ, de AM, Rocca WA, et al. A limited role for DJ1 in Parkinson disease susceptibility. Neurology. 2004;63:550–553. doi: 10.1212/01.wnl.0000133402.78621.ad. [DOI] [PubMed] [Google Scholar]
- 26.Eschbach J, Danzer KM. Neurodegener Dis. 2013 Sep 24; Epub ahead of print α-Synuclein in Parkinson’s Disease: Pathogenic Function and Translation into Animal Models. [Google Scholar]
- 27.Sutherland GT, Matigian NA, Chalk AM, Anderson MJ, Silburn PA, Mackay-Sim A, et al. A cross-study transcriptional analysis of Parkinson’s disease. PLoS One. 2009;4:e4955. doi: 10.1371/journal.pone.0004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simunovic F, Yi M, Wang Y, Macey L, Brown LT, Krichevsky AM, et al. Gene expression profiling of substantia nigra dopamine neurons: further insights into Parkinson’s disease pathology. Brain. 2009;132:1795–1809. doi: 10.1093/brain/awn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dachsel JC, Lincoln SJ, Gonzalez J, Ross OA, Dickson DW, Farrer MJ. The ups and downs of alpha-synuclein mRNA expression. Mov Disord. 2007;22:293–295. doi: 10.1002/mds.21223. [DOI] [PubMed] [Google Scholar]
- 30.Grant JL, Ghosn EE, Axtell RC, Herges K, Kuipers HF, Woodling NS, et al. Reversal of paralysis and reduced inflammation from peripheral administration of beta-amyloid in TH1 and TH17 versions of experimental autoimmune encephalomyelitis. Sci Transl Med. 2012;4:145ra105. doi: 10.1126/scitranslmed.3004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurnellas MP, Adams CM, Sobel RA, Steinman L, Rothbard JB. Amyloid fibrils composed of hexameric peptides attenuate neuroinflammation. Sci Transl Med. 2013;5:179ra42. doi: 10.1126/scitranslmed.3005681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ossenkoppele R, van der Flier WM, Zwan MD, Adriaanse SF, Boellaard R, Windhorst AD, et al. Differential effect of APOE genotype on amyloid load and glucose metabolism in AD dementia. Neurology. 2013;80:359–365. doi: 10.1212/WNL.0b013e31827f0889. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Demographic characteristics of cases from the Molecular Epidemiology of Parkinson’s Disease Study
Genetic association with time-to-motor-impairment
Genetic association with time-to-cognitive-impairment
Linkage disequilibrium map for SNCA. The multi-allelic variable number tandem repeat REP1 was coded as 259 bp vs. others. The LD values as measured using r2 are given by numbers and the LD values as measured by D′ are given by color intensity (red squares indicate strong LD, pink squares indicate intermediate LD, and white squares indicate low LD, with evidence for ancestral recombination; blue indicates limited data).
Haplotype analysis in Family H (modified to protect subject confidentiality). Circles correspond to women and squares to men. Diagonal lines through a symbol indicate deceased status. Shaded lower right quadrants identify individuals affected with PD, while clear symbols indicate unaffected status. A “+” sign indicates that DNA was available for genotyping. The other nine rows beneath the pedigree symbols correspond to: individual identifier (top row) and marker genotypes for D4S2460, D4S3481 (REP1), rs2619363, rs2301135, rs104893877 c.157G>A, rs1005233, rs3216147 and D4S423 (bottom row). The black bar signifies the c.157G>A (p.A53T) haplotype; shaded/unfilled bars distinguish four SNCA promoter haplotypes with different D4S3481 (REP1), rs2619363, and rs2301135 alleles. Haplotypes for deceased family members, without DNA, are probabilistically inferred to best fit the available genotype data. The range and mean of the age for disease onset in 20 affected members of this family is 31–71 and 52.9. In generations III, IV, and V, the respective ranges, n, and mean are 50–71, 8, and 61.2; 40–63, 9, and 50.2; and 31–53, 3, and 39.