TEXT
Hepatitis B virus (HBV) is a hepadnavirus which can cause systemic infection and hepatocellular damage in humans. Infection is acquired through contact with the blood of a person carrying HBV. The passive administration of anti-HBV surface antigen (HBsAg) antibodies protects against a subsequent infection and vaccination with HBsAg has proved an effective means of protection against HBV infection.
HBsAg purified from the plasma of HBsAg-carriers has been used for many years as an effective vaccine but its major limitation is the availability of the raw material and the cost. Since the late 1980s, genetic engineering has allowed the development of yeast-derived recombinant, very safe and efficient DNA vaccines[1] and compulsory vaccination was introduced in many countries to progressively eradicate HBV infection. Recombinant vaccines contain only the sequence 1-226 of HBsAg (rHBsAg), representing the small nonglycosylated HBs protein (S-domain), with the exclusion of the preS1 and preS2 domains (Figure 1).
Figure 1.
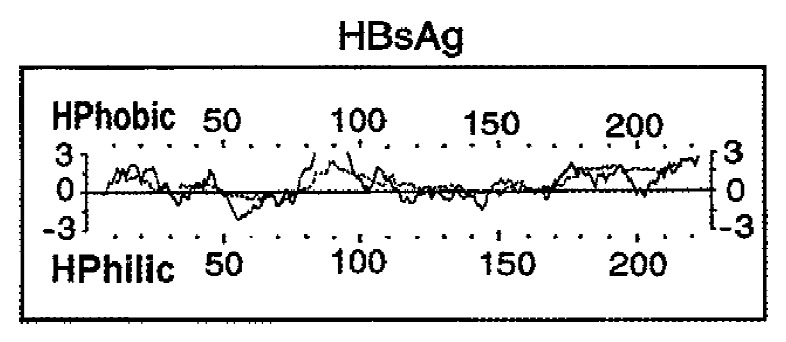
Structure of the HBsAg molecule. The hydropathy of the sequence 1-226 is predicted according to Kyte and Doolittle 1982, J Mol Biol (—) and Engelman et al. 1986, Annu Rev Biophys Biophys Chem (… …).
A serum level of at least 10 mIU/mL of anti-HBs antibodies reached after vaccination has been proposed to be the lowest limit for protection[2]. Several studies were conducted to examine the duration of protection after immunization with recombinant vaccines, but these were related to seroconversion in selected clusters of vaccine recipients, introducing parameters (age, gender, smoking, obesity, etc.) which are known to influence the immune response[3,4]. Recently, we and others proposed a mathematical model to calculate the time after the vaccination with rHBsAg for the loss of protection due to an antibody fall to below 10 mIU/mL[5,6]. This model, which will beavailable on the web-6, showed that the decay in the level of specific immunoglobulins reached after last booster dose is well described by the log time-log titre linear function. In practice, anti-HBs level falls quickly after the peak and then slowly, and the time to reach very low levels depends on the serum level reached at the end of the vaccination protocol.
The analysis of specificity of raised circulating antibodies after inoculation with rHBsAg showed a binding region for one B-cell epitope represented by a peptide located within the HBsAg sequence 110-168 (Table 1). The same specificity has been described after immunization with plasma-derived vaccines or natural infection. This epitope represents the《a》determinant of HBsAg, that may be considered the conserved sequence among different HBV subtypes (adr, adw, ayr, ayw) and is able to stimulate cross-protection after immunisation[7]. This dominant B-cell epitope is conformational, discontinuous, characterized by disulphide bonds and corresponds to a hydrophilic, external region of HBsAg. Although antibodies directed against this region protect against HBV infection, an escape mutant of HBV has been isolated from an infant vaccinated with rHBsAg and later became positive for markers of viral replication. The mutant presents an amino acid substitution of glycine to arginine at position 147[8]. This point mutation is stable, allows the attachment of the virus to hepatocytes and represents a new specificity not cross-reacting with normal virus subtypes. The mutation of one amino acid can be responsible for different conformations of《a》determinant, with varying abilities to elicit and bind anti-HBs antibodies.
Table 1.
B and T cells epitopes of HBsAg
| Sequence | Determinants | Cell subsets | HLA restriction |
| 110-168[6] | Conformational《a》determinant | B | Not HLA restricted |
| 21-28[11] | Minimal epitope | CD4 T | Class II |
| 124-147[11] | Immunodominant zone | CD4 T | Class II |
| 165-172[11] | Minimal epitope | CD4 T | Class II |
| 215-223[11] | Minimal epitope | CD4 T | Class II |
| 80-98[11] | Containing predicted epitope | CD4 T | Class II |
| 171-179[16] | Minimal epitope | CD8 T | Class I |
Although the immunogenicity and protective effect of rHBsAg are comparable with that of HBV natural infection, differences have been described in IgG subclass distribution after infection or vaccination. The serological response to HBsAg has been well characterized in patients with acute or chronic hepatitis B, who presented circulating immunocomplexes containing mostly IgG1 and IgG4 antibodies, or in convalescent sera where specific antibodies were predominantly IgG1 and IgG3. The analysis of IgG subclass distribution induced by vaccination with rHBsAg shows that the IgG2 subclass is mainly synthesized with IgG1. The presence of IgG2 with IgG1 subclass following vaccination against HBV was also described using plasma-derived vaccines, where HBsAg is glycosylated. Although the synthesis of IgG2 subclass antibodies is generally associated with polysaccharide antigens, the absence of glycosylated residues in the rHBsAg cannot explain the switch to IgG2 production. A hypothesis to explain a comparable immunogenicity from both vaccines is that the network of cytokines secreted by T lymphocytes recognising natural or recombinant HBsAg, could induce the B lymphocyte switch to this IgG subclass synthesis[9].
Anti-HBS production is the consequence of T cell-mediated immunity stimulated by the vaccination, which also plays a fundamental role in preventing HBV infection. The use of genetic engineering to synthesize recombinant HBV envelope protein increased investigation into the immunising activity of HBsAg. Some early studies in vitro disclosed that CD4 + T cells help HBsAg specific B cell for the synthesis of anti-HBs antibodies in subjects vaccinated with rHBsAg[10,11].
The isolation of T cell clones specific for HBsAg from subjects vaccinated with recombinant vaccine provided an important tool for investigating the immunizing activity of this protein. It has been observed that the response of antigen-specific T lymphocytes to HBsAg depends on the sequence of the antigen molecule. In an attempt to identify the sequences recognized by HBsAg-primed T cells from vaccine recipients, synthetic peptides representing short sequences of HBsAg protein have been used in vitro to investigate specific cellular proliferation. Recently, immunodominant HBsAg epitopes have been confirmed or identified analysing the proliferation of CD4 + T cell clones (helper/inducer) by peptides spanning the whole HBsAg protein[12]. The clones belong to the Th0/Th2 subset, expressing and secreting in vitro interleukin 4, 5 and interferon-γ. Three minimal T-cell epitopes have been described: sequences 21-28, 165-172 and 215-223. There is an evidence that T-cell epitopes are also present within the 124-147 sequence, corresponding to that zone on HBsAg containing the《a》determinant. The difficulty in identifying a minimal epitope within this sequence could be due to the low level of immunogenicity of this HBsAg region. We cannot exclude the presence of other important epitopes with in the most hydrophobic region of HBsAg, that is very difficult to study because of this characteristic, which prevents the solubility of peptides in culture media (Table 1). These findings are in agreement with the Berzofsky algorithm[13], which predicts for different protein zones the probability of representing Thepitopes on the basis of the sequential and conformational structure. The pathway of exogenous antigen processing needs the cooperation of antigen presenting cells (APC) and the recognition of short aminoacid sequences (epitopes) by Th cells in association with HLA class II determinants. Recently it has been shown that: ① DR molecules are mostly involved in the presentation of immunodominant HBsAg peptides, but at least DP determinants can participate in antigen presentation; and ② an epitope can be efficiently presented by different MHC loci, suggesting that a short sequence of HBsAg can be immunogenically dominant, because of its specificity for Th cells from subjects presenting different HLA class II alleles[12]. On the other hand, studies on the contribution of HLA class II determinants to the regulation of antibody production suggested that MHC loci are involved in regulation of the immunological response after rHBsAg immunization[14].
In vitro studies on CD8 + T cells responsible for cytotoxic activity are hampered by the difficulty of utilising target cells expressing appropriate HLA class II determinants for the association with the antigen. Although the cytotoxic activity of CD8 + T cells stimulated by HBsAg is not well known, one minimal essential epitope on HBsAg has been described in sequence 172-180[15] corresponding to a peptide candidate for the binding to HLA-A2 determinant and this sequence has been described to activate T CD8+ lymphocytes isolated from subjects sharing this HLA class II determinant. It is also noteworthy that this antigen can evoke cytotoxic activity by subsets of CD4 + T cells expressing the CD56 phenotype[16]. We do not exclude that different short sequences of S protein could induce an immunological response involving both CD4+ and CD8 + T lymphocytes. Double pathway for the antigen processing, utilising endosomic and cytosomic cellular districts, has been described for the pre S2 protein sequence 120-134 of HBV, which can be presented by HLA class I and class II determinants, leading to activation of CD4+ (helper/inducer) and CD8+ (cytotoxic) T cells[17]. Identification of dominant epitopes on HBsAg specific B, CD4+ and CD8 + T cells may clarify the effective role of rHBsAg in inducing a protective cellular and humoral response against HBV infection. The major histocompatibility complex (MHC) controls the immune response to protein antigen and allelic variants of MHC can influence the lack of response to the immunisation in a small percentage of vaccine recipients (5%-10%).
More recently some authors have analyzed the possibility of utilising HBsAg-encoding plasmid DNA as vaccine. Good findings have been obtained in vivo following i.m. injection of the plasmid into mice[18]. This route of administration induced muscular cells to synthesize endogenous HBsAg followed by the association of peptides with HLA class I determinants and the secretion of the whole molecule. In the future, DNA vaccine could be considered an alternative method to obtain an active immunization to several pathogens if this approach proves to be as effective and safe as recombinant protein.
Footnotes
Supported in part by Grants from the Italian Ministry of Health, University of Bologna (ex60%) and Ricerca Corrente, Istituti Ortopedici Rizzoli, Bologna, Italy.
References
- 1.Harford N, Cabezon T, Colau B, Delisse AM, Rutgers T, De Wilde M. Construction and characterization of a Saccharomyces cerevisiae strain (RIT4376) expressing hepatitis B surface antigen. Postgrad Med J. 1987;63 Suppl 2:65–70. [PubMed] [Google Scholar]
- 2.Hollinger FB. Hepatitis B vaccines--to switch or not to switch. JAMA. 1987;257:2634–2636. [PubMed] [Google Scholar]
- 3.Honorati MC, Mariani E, Dolzani P, Facchini A. Biological parameters influencing the immunological response to plasma derived and recombinant hepatitis B vaccines. Ann Ist Super Sanita. 1996;32:369–374. [PubMed] [Google Scholar]
- 4.Hollinger FB. Factors influencing the immune response to hepatitis B vaccine, booster dose guidelines, and vaccine protocol recommendations. Am J Med. 1989;87:36S–40S. doi: 10.1016/0002-9343(89)90530-5. [DOI] [PubMed] [Google Scholar]
- 5.Gesemann M, Scheiermann N. Quantification of hepatitis B vaccine-induced antibodies as a predictor of anti-HBs persistence. Vaccine. 1995;13:443–447. doi: 10.1016/0264-410x(94)00010-k. [DOI] [PubMed] [Google Scholar]
- 6.Honorati MC, Palareti A, Dolzani P, Busachi CA, Rizzoli R, Facchini A. A mathematical model predicting anti-hepatitis B virus surface antigen (HBs) decay after vaccination against hepatitis B. Clin Exp Immunol. 1999;116:121–126. doi: 10.1046/j.1365-2249.1999.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neurath AR. Chemical synthesis of hepatitis B vaccines. In Zuckerman AJ, ed Recent developments in prophylactic immunization. London: Kluwer Academic; 1989. pp. 210–242. [Google Scholar]
- 8.Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman AJ, Thomas HC. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 9.Honorati MC, Borzì RM, Dolzani P, Toneguzzi S, Facchini A. Distribution of IgG subclasses after anti-hepatitis B virus immunization with a recombinant vaccine. Int J Clin Lab Res. 1997;27:202–206. doi: 10.1007/BF02912459. [DOI] [PubMed] [Google Scholar]
- 10.Degrassi A, Mariani E, Honorati MC, Roda P, Miniero R, Capelli M et al. Cellular response and anti-HBs synthesis in vitro after vaccination with yeast-derived recombinant hepatitis vaccine. Vaccine. 1992;10(9):617–622. doi: 10.1016/0264-410x(92)90443-n. [DOI] [PubMed] [Google Scholar]
- 11.Ducos J, Bianchi-Mondain AM, Pageaux G, Conge AM, Poncet R, Vendrell JP, Segondy M, Serre A. Hepatitis B virus (HBV)-specific in vitro antibody production by peripheral blood mononuclear cells (PBMC) after vaccination by recombinant hepatitis B surface antigen (rHBsAg) Clin Exp Immunol. 1996;103:15–18. doi: 10.1046/j.1365-2249.1996.928621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honorati MC, Dolzani P, Mariani E, Piacentini A, Lisignoli G, Ferrari C, Facchini A. Epitope specificity of Th0/Th2 CD4+ T-lymphocyte clones induced by vaccination with rHBsAg vaccine. Gastroenterology. 1997;112:2017–2027. doi: 10.1053/gast.1997.v112.pm9178695. [DOI] [PubMed] [Google Scholar]
- 13.Margalit H, Spouge JL, Cornette JL, Cease KB, Delisi C, Berzofsky JA. Prediction of immunodominant helper T cell antigenic sites from the primary sequence. J Immunol. 1987;138:2213–2229. [PubMed] [Google Scholar]
- 14.Mineta M, Tanimura M, Tana T, Yssel H, Kashiwagi S, Sasazuki T. Contribution of HLA class I and class II alleles to the regulation of antibody production to hepatitis B surface antigen in humans. Int Immunol. 1996;8:525–531. doi: 10.1093/intimm/8.4.525. [DOI] [PubMed] [Google Scholar]
- 15.Nayersina R, Fowler P, Guilhot S, Missale G, Cerny A, Schlicht HJ, Vitiello A, Chesnut R, Person JL, Redeker AG, et al. HLA A2 restricted cytotoxic T lymphocyte responses to multiple hepatitis B surface antigen epitopes during hepatitis B virus infection. J Immunol. 1993;150:4659–4671. [PubMed] [Google Scholar]
- 16.Barnaba V, Franco A, Paroli M, Benvenuto R, De Petrillo G, Burgio VL, Santilio I, Balsano C, Bonavita MS, Cappelli G. Selective expansion of cytotoxic T lymphocytes with a CD4+CD56+ surface phenotype and a T helper type 1 profile of cytokine secretion in the liver of patients chronically infected with Hepatitis B virus. J Immunol. 1994;152:3074–3087. [PubMed] [Google Scholar]
- 17.Barnaba V, Franco A, Alberti A, Balsano C, Benvenuto R, Balsano F. Recognition of hepatitis B virus envelope proteins by liver-infiltrating T lymphocytes in chronic HBV infection. J Immunol. 1989;143:2650–2655. [PubMed] [Google Scholar]
- 18.Böhm W, Mertens T, Schirmbeck R, Reimann J. Routes of plasmid DNA vaccination that prime murine humoral and cellular immune responses. Vaccine. 1998;16:949–954. doi: 10.1016/s0264-410x(97)00302-2. [DOI] [PubMed] [Google Scholar]


