Abstract
AIM: To investigate the significance and mechanism of cx-32 mRNA, cx-43 mRNA and their proteins in hepatocarcinogenesis.
METHODS: Sixty-one cases of HCC and 14 cases of normal liver tissues were detected by immunohistochemical and in situ hybridization (ISH) methods.
RESULTS: In HCC grades I, II, III and normal liver tissues, the positive rates of Cx32 protein were 55.6%, 42.1%, 18.2% and 92.9%, respectively. The detection rates of Cx43 protein were 44%, 26.3%, 12.1% and 78.6%, respectively. There was significant difference in Cx32 and Cx43 protein between HCC and normal liver tissues (P < 0.01). ISH the positive rates of cx 32 mRNA shown by ISH in HCC grades I, II, III and normal liver tissues were 88.9%, 84.2%, 87.9% and 92.9%, respectively. Those of cx43 mRNA were 77.8%, 78.6%, 78. 8% and 85.7%, respectively. There was no statistical difference in the positive rates of cx32 mRNA and cx43 mRNA between HCC and normal liver tissue (P > 0.05).
CONCLUSION: The aberrant location of Cx32 and Cx43 proteins could be responsible for progression of hepatocarcinogenesis, and the defect of cx genes in post-translational processing might be the possible mechanism.
Keywords: connexin; gap junction; liver neoplasm; immunohistochemistry; in situ hybridization; carcinoma, hepatocellular; gene expression
INTRODUCTION
Gap junctions are clusters of intercytoplasmic channels connecting neighboring cells which are composed of proteins called connexins (cx). Gap junction intercellular communication (GJIC) mediated by gap junction channels has been believed to be an important mechanism for the maintenance of tissues homeostasis and metabolic cooperation[1]. Carcinogenesis is one of the pathological proc esses in which disorders of GJIC may play an important role[2]. This stu dy was designed to investigate the significance and mechanism of cx32 and cx43 genes and their proteins in hepatocellular carcinoma (HCC).
MATERIALS AND METHODS
Clinical data
The specimens of surgically removed 61 cases of HCC and 14 cases of normal liver tissues were collected at Xijing Hospital during the period of 1996-1998. The samples had not been treated with chemotherapy or radiotherapy before tumor excision. The pathological diagnosis was verifed on the respective paraffin embedded material by histologic examination (HE). The specimens were derived from 9 cases of grade I HCC, 19 of grade II and 33 of grade III, and these were 4 μm thick sections.
SP immunochemistry
SP immunostaining was performed as described by SP immunochemistry kit and DAB kit with mouse anti-cx32 mAb, mouse anti-cx43 mAb of Zymed Lab. Inc., USA. Diagnosis was made by brown or yellow coloration with varied intensities. Negative (-): stained cells < 5%, positive ( + ): stained cells 5%-50%, strongly positive ( ++ ): stained cells > 50%.
Probe labeling
pGEM3-cx32 and pSG5-cx43 plasmids were kindly given by Prof. Gui Yuan Li in Hunan Medical University. After amplification, isolation and purification were done, pGEM3-cx32 plasmid was digested by Ecor-I (Gibco BRL, USA) and pSG5-cx43 by BamH-I (Gibco BRL, USA ). Electroporate the digested plasmids on 7 g/L agarose gel with λ DNA/Hind-III+-EcoR-I marker. Extract and purify the cx32 cDNA and cx43 cDNA from the gel as the protocol of AdvantageTM PCR-Pure Kit from Clontech Lab. Inc., USA. Label the cx32 cDNA and cx43 cDNA using Dig DNA Labeling and detection kit of Boehringer Mannheim, Germany.
mRNA in situ hybridization
Slides were incubated in 0.2 mL/L DEPC at RT for 10 min, in 0.2 mL/L HCl for 10 min and in 5 mg/L PK at 37 °C for 10 min and in 0.1 mol/L glycine to stop the digestion reaction, and then fixed in 40 g/L PFA for 10 min in sequence. After being washed in PBS, the sections were dehydrated in ethanol and air dried. Prehybridized at 42 °C for 30 min, the labeled cDNA probes were denatured in hybridization buffer at 100 °C for 10 min, then -20 °C for 3 min, added on tissues and coverslipped at 42 °C overnight. Sections were washed with 2 × SSC, 1 × SSC, 0.5 × SSC and buffer I, incubated in NSS at 37 °C for 30 min, and then Dig-Ap (1:500) for 2 h, and finally detected with NBT/ BCIP of Dig DNA labeling and detection kit. Diagnosis was made by blue colorat ion with varied intensities and compared with control sections. All results were analyzed by χ2 test.
RESULTS
SP Immunochemical results
Cx32 and Cx43 proteins appeared as numerous individual spots intracytoplasmically ( Figures 1 and 2), and in some as parts of the plasma membrane. There was fairly intense immunoreactivity in nearly all of the 14 normal liver samples ( Cx32 13/14, 92.9%, Cx43 11/14, 78.6%).
Figure 1.
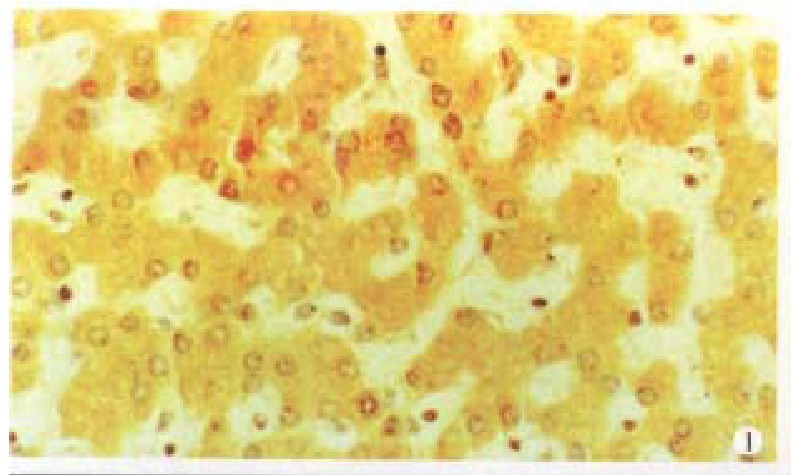
Cx32 protein is positive in normal liver. SP × 400
Figure 2.
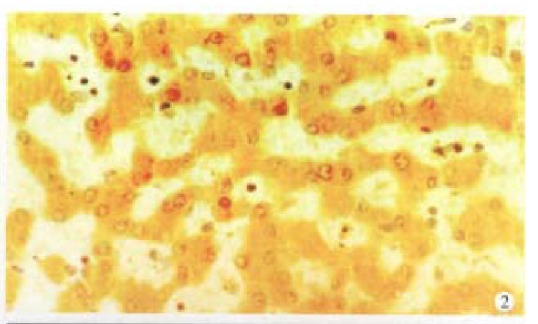
Cx43 protein is positive in normal liver. SP × 400
In contrast, a clear difference was noted between that in normal tissue and in the HCC, the Cx32 and Cx43 positive spots decreased in the latter especially in grade III HCC, only 18.2% (Cx32) and 12.1% (Cx43) specimens exhibited very weak stainings. The expression of Cx32 and Cx43 proteins in normal liver tissue and I, II and III HCC grades were significantly different (P < 0.01) (Table 1).
Table 1.
Positive rates of Cx32 and Cx43 proteins in HCC and normal liver
| Group | n |
Cx32 |
Cx43 |
||||
| + | ++ | % | + | ++ | % | ||
| HCCI | 9 | 2 | 3 | 55.6a | 2 | 2 | 44.4a |
| HCCII | 19 | 6 | 2 | 42.1a | 3 | 2 | 26.3a |
| HCCIII | 33 | 5 | 1 | 18.2a | 4 | 0 | 12.1 |
| Normal liver | 14 | 4 | 9 | 92.9 | 4 | 7 | 78.6 |
P < 0.01 vs normal liver.
mRNA in situ hybridization results
After restriction digestion, 1.5 kb cx32 cDNA probe was obtained from pGEM3-cx32 plasmid (Figure 3), and 1.11 kb cx43 cDNA probe from pSG5- cx43 plasmid (Figure 4).
Figure 3.
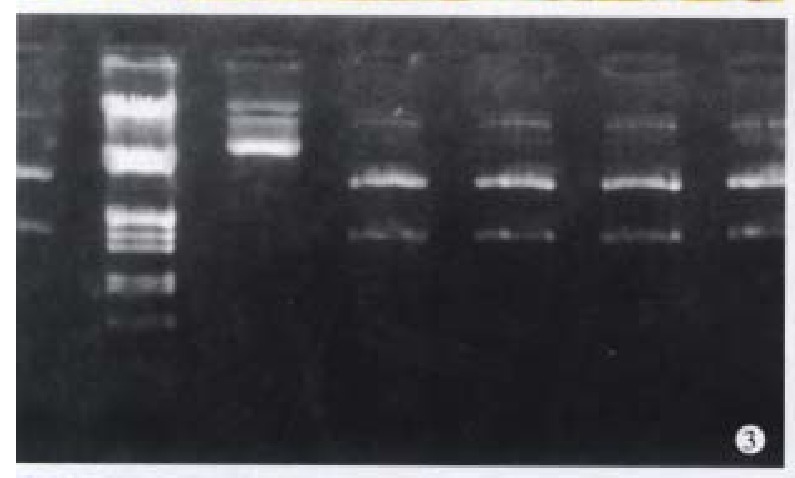
Digestion of pGEM3-cx32 by EcoR-I. A. λ DNA/Hind-III+-EcoR-I marker; B. pGEM3-cx32 plasmid; C. restric tion digestion of 1.5 kb cx32 cDNA.
Figure 4.
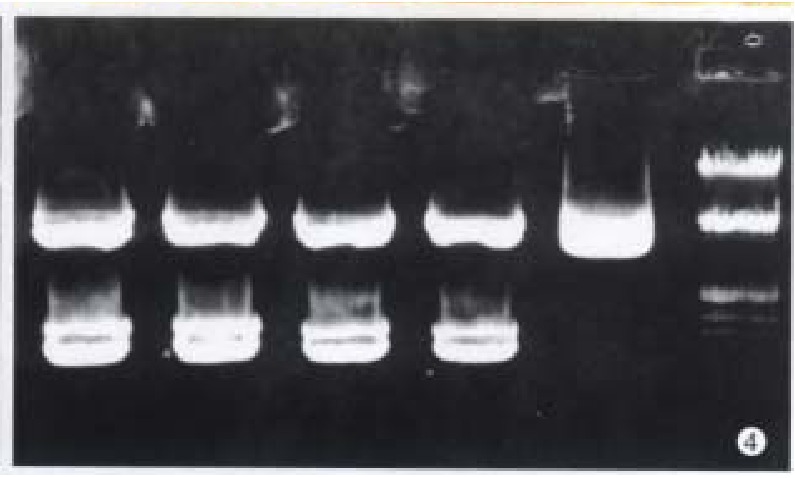
Digestion of pSG5-cx43 by BamH-I. A. λ DNA/Hind-III+-EcoR-I marker; B. pSG5-cx-43 plasmid; C. restricti on digestion of 1.11 kb cx43 cDNA.
Bright blue specific hybridization appearanc of cx32 mRNA and cx43 mRNA were observed intracytoplasmically in normal liver or HCC tissues (Figures 5, 6, 7 and 8). The detection rates of cx32 mRNA and cx43 mRNA in normal liver an dI, II and III HCC grades tissues were not significantly different (P > 0.05).
Figure 5.
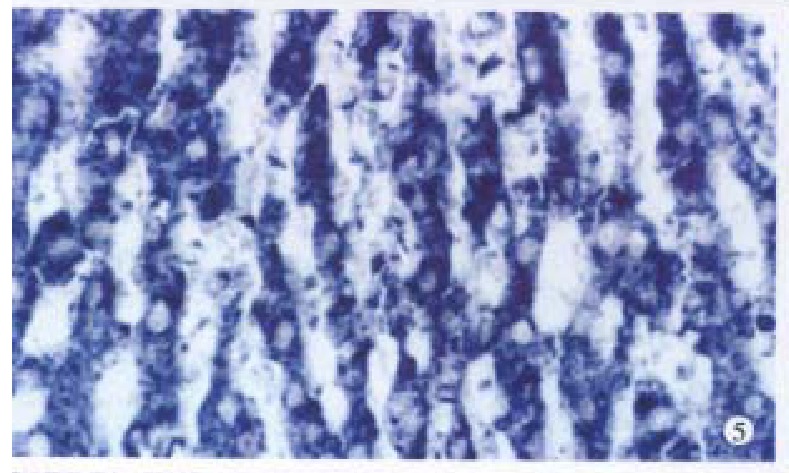
cx32 mRNA is positive in normal liver. ISH × 400
Figure 6.
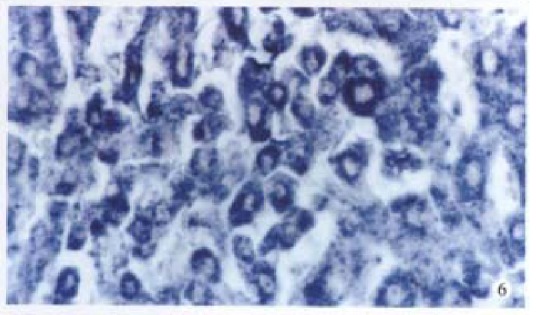
cx32 mRNA is positive in HCC. ISH × 400
Figure 7.
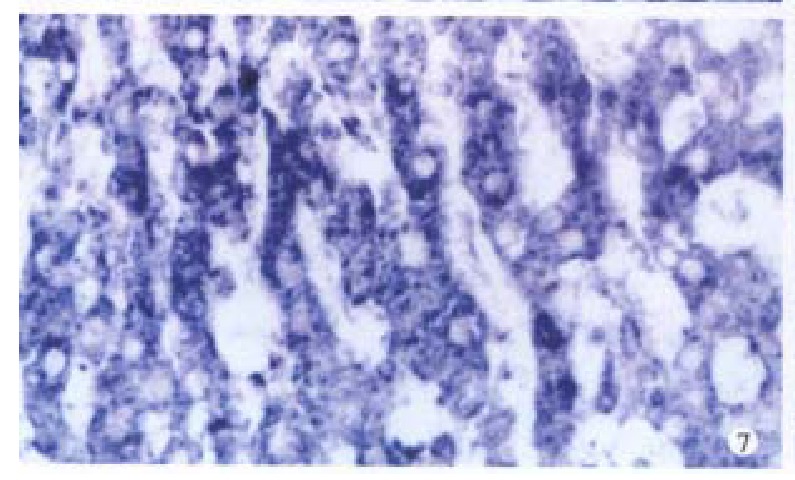
cx43 mRNA is positive in normal liver. ISH × 400
Figure 8.
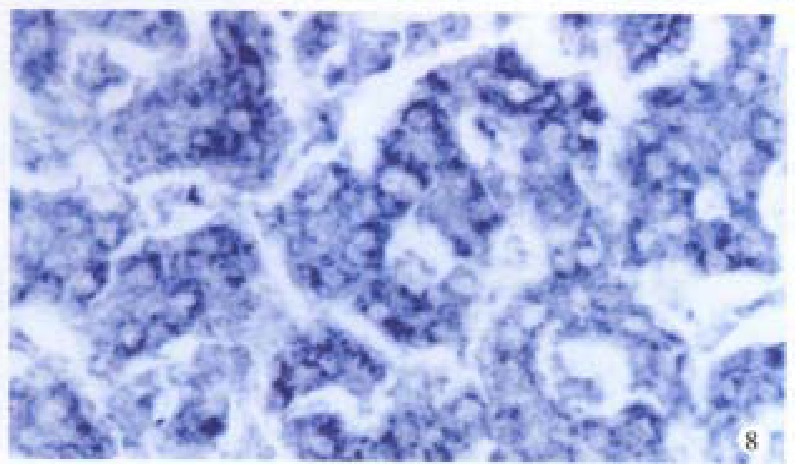
cx43 mRNA is positive in HCC. ISH × 400
DISCUSSION
Since the first cloning of cx gene in 1986, there have been rapid progress in identifying and characterizing a multigene family (including at least 13 members ) which codes the gap junction proteins[3]. The cloning sequence of cDNA shows that there is a highly homogenic and strict gene structure among these family members.
The expressions of cx genes in different tissues are specific. GJIC plays an important role in the rapid progress of cell society. Gap junctions contain channels that connect contacting cells, and have an apparent selectivity based principally on molecular size, allowing the movement of molecules smaller than Mr 1000, such as cAMP, but preventing the movement of proteins or nucleic acids.
Gap junction mediated GJIC has been postulated to be an important mechanism to maintain tissue homeostasis and to control growth and differentiation. The decreased level of GJIC has been found in many kinds of tumor cells, and this can be important in neoplastic progression by allowing tumor cells to escape local control mechanisms[4].
Cx32 is widely expressed in many tissues, especially in normal liver, Cx43 is also involved in the gap junctional pathway in liver. This study revealed that Cx32 protein and Cx43 protein expressed at a high level in normal liver and even in pericancerous liver tissues, but at a decreased level in HCC tissues. The lack of Cx32 and Cx43 proteins reduced the number of GJIC between HCC cells and surrounding non-tumorous cells, finally resulting in progression of hepatocar cinogenesis.
In some studies, Cx43 protein was not detectable in normal liver, but could be found in HCC cells. These results suggested that certain tumors or transformed cells have normal levels of gap junction or GJIC and lack of functional gap junctions is not a general feature of maligancy[5], the exact mechanisms remain to be elucidated.
Because there was no decrease in levels of cx32 mRNA and cx43 mRNA in HCC and normal liver tissues, the aberrant localization of Cx32 and Cx43 might be responsible for the reduced GJIC in HCC[6]. It appears that the control mechanisms of cx32 and cx43 genes transcription are not affected during human liver tumorigenesis.
Not any mutation in the coding sequence of cx genes from any of the human tumors has been found. It is likely that the aberrant location of Cx32 and Cx43 in HCC cells is due to disruption of the mechanisms for construction of these proteins into gap junction plaques rather than to structural abnormality of the Cx32 and Cx43 themselves. The possibility is that there is defect in post-translati onal processing of Cx32 and Cx43 proteins, which may be essential for their tran sport to membrane. Post-translational phosphorylation[7] may be the imp ortant factors controlling the GJIC mediated by Cx32 and Cx43 in HCC, it is also responsible for the assembly or function of these proteins.
In conclusion, the aberrant localization of Cx32 and Cx43 proteins in HCC is no teworthy, which has direct biological significance in the process of hepatocarc inogenesis. Recent results demonstrate that the cx gene family is a class of non-mutant tumor-suppressive gene[8], it should be regarded as an effectual marker of early diagnosis or treatment for liver cancer.
Footnotes
Edited by Wu XN and Ma JY
Proofread by Miao QH
References
- 1.Kumar NM, Gilula NB. The gap junction communication channel. Cell. 1996;84:381–388. doi: 10.1016/s0092-8674(00)81282-9. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg TH. Gap junction function: the messenger and the message. Am J Pathol. 1998;152:851–854. [PMC free article] [PubMed] [Google Scholar]
- 3.Haefliger JA, Bruzzone R, Jenkins NA, Gilbert DJ, Copeland NG, Paul DL. Four novel members of the connexin family of gap junction proteins. Molecular cloning, expression, and chromosome mapping. J Biol Chem. 1992;267:2057–2064. [PubMed] [Google Scholar]
- 4.Holder JW, Elmore E, Barrett JC. Gap junction function and cancer. Cancer Res. 1993;53:3475–3485. [PubMed] [Google Scholar]
- 5.Wilgenbus KK, Kirkpatrick CJ, Knuechel R, Willecke K, Traub O. Expression of Cx26, Cx32 and Cx43 gap junction proteins in normal and neoplastic human tissues. Int J Cancer. 1992;51:522–529. doi: 10.1002/ijc.2910510404. [DOI] [PubMed] [Google Scholar]
- 6.Tsuda H, Asamoto M, Baba H, Iwahori Y, Matsumoto K, Iwase T, Nishida Y, Nagao S, Hakoi K, Yamaguchi S. Cell proliferation and advancement of hepatocarcinogenesis in the rat are associated with a decrease in connexin 32 expression. Carcinogenesis. 1995;16:101–105. doi: 10.1093/carcin/16.1.101. [DOI] [PubMed] [Google Scholar]
- 7.Neveu MJ, Hully JR, Babcock KL, Hertzberg EL, Nicholson BJ, Paul DL, Pitot HC. Multiple mechanisms are responsible for altered expression of gap junction genes during oncogenesis in rat liver. J Cell Sci. 1994;107(Pt 1):83–95. doi: 10.1242/jcs.107.1.83. [DOI] [PubMed] [Google Scholar]
- 8.Krutovskikh V, Mazzoleni G, Mironov N, Omori Y, Aguelon AM, Mesnil M, Berger F, Partensky C, Yamasaki H. Altered homologous and heterologous gap-junctional intercellular communication in primary human liver tumors associated with aberrant protein localization but not gene mutation of connexin 32. Int J Cancer. 1994;56:87–94. doi: 10.1002/ijc.2910560116. [DOI] [PubMed] [Google Scholar]


