Abstract
Objectives: Published studies on the association between methylenetetrahydrofolate reductase (MTHFR) gene A1298C polymorphisms and breast cancer risk among Chinese population have yielded conflicting results. The purpose of this study was to clarify the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population. Methods: Systematic searches were performed through the database of Medline/PubMed, Science Direct, Elsevier, CNKI and Wanfang Medical Online. Results: Overall, a significantly increased risk of breast cancer was observed among the subjects carrying MTHFR gene A1298C AC+CC genotype (odds ratio [OR]=1.05 with 95% confidence interval [CI]: 1.01-1.10) as compared to those carrying AA genotype among total Chinese population. We did not observe any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer under the additional genetic models of AC vs. AA, CC vs. AA and C-allele vs. A-allele (OR=1.00 with 95% CI: 0.97-1.02, OR=1.01 with 95% CI: 1.00-1.02 and OR=1.00 with 95% CI: 0.99-1.02, respectively). The cumulative meta-analysis showed similar results. In subgroup analysis, we observed subjects carrying AC+CC genotype had an increased breast cancer risk compared with those carrying AA genotype among the studies of sample size less than 1000. We did not observe any significant association between MTHFR gene A1298C polymorphisms and breast cancer risk in additional subgroup analyses. Conclusions: Our results suggest that MTHFR gene A1298C AC+CC genotype may be a risk factor for the development of breast cancer among Chinese population. Well-designed studies with a large sample size are needed to further confirm our findings.
Keywords: MTHFR, polymorphism, breast cancer, risk, Chinese
Introduction
Methylenetetrahydrofolate reductase (MTHFR) is a folate-dependent enzyme that plays a pivotal role in the conversion of homocysteine to methionine by converting 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate [1,2]. The MTHFR gene is mapped to chromosome 1 location p36.3 in humans. To date, more than forty single nucleotide polymorphisms (SNPs) have been identified in the MTHFR gene. Among them, the polymorphic site of rs1801131 in exon 7 (A1298C, Glu429Ala) is one of the most extensively studied polymorphisms, which is related to the reduction of MTHFR activity [3,4].
Breast cancer is the most commonly diagnosed cancer in women worldwide, and it has become the second leading cause of cancer related death in Chinese females [5]. The etiology of breast cancer is complicated and poorly understood. Some recognized risk factors for breast cancer include age, ethnicity, lifestyle, reproductive events, exogenous hormones, bone density, as well as genetic factors [6]. It has been proposed that low-penetrance susceptibility genes combining with environmental factors may be of importance in the development of breast cancer.
To the best of our knowledge, a growing number of epidemiological studies have focused on to explore the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population [7-20]. However, the results remained conflicting rather than consistent. The discrepancies among these studies might attribute to the relatively small sample size in each study. Meta-analysis is considered as a powerful tool for summarizing the contradicting results from different studies with more statistical power. We set out to clarify the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population by performing a systematic meta-analysis on the most updated data in the literatures.
Materials and methods
Search strategy, inclusion criteria and exclusion criteria
A comprehensive search was carried out through the database of Medline/PubMed, Science Direct, Elsevier, China National Knowledge Infrastructure (CNKI) and Wanfang Medical Online with a combination of the following terms: “breast cancer”, “breast neoplasm” or “breast carcinoma” and “Methylenetetrahydrofolate reductase” or “MTHFR” or “rs180-1131”. Last search was updated on April 30, 2015. The references cited in the publications and review articles were also manually searched.
Data inclusion criteria: (a) The papers should include breast cancer risk and MTHFR gene A1298C polymorphisms; (b) Case-control studies or cohort studies; (c) Sufficient data to estimate the odds ratio (OR) and 95% confidence interval (95% CI). For overlapping or repeated studies, the publication including more information was selected. Accordingly, reviews and papers lacking essential information were excluded.
Data extraction
Data were independently extracted and tabulated by two investigators and an electronic database was established. The following extracted information from each eligible study was included in the database: authors’ name, publishing year, region, source of controls, genotype frequency of cases and controls. Quality of the studies was evaluated according to the “methodological quality assessment scale” [21].
Statistical analysis
The Cochrane Q statistics test was used for the assessment of heterogeneity among studies. A fixed-effects model or a random-effects model was used to estimate the combined effects depending on the results of the heterogeneity test [22]. The fixed-effects model is used while the effects are assumed to be homogenous; otherwise, the random-effects model is used. Begg’s test and Egger’s test were applied to assess the publication bias [23,24]. The χ2 test was applied to check whether the genotype frequencies of the controls were in agreement with Hardy-Weinberg equilibrium (HWE) or not. A sensitivity analysis was performed by deleting one study each time. A cumulative meta-analysis was performed, which accumulated the studies in the light of the publishing year. All of the statistical analyses were performed by using STATA10.0 software package (STATA Corporation, College Station, TX, USA). Statistical significance was determined as a 2-sided P-value less than 0.05.
Results
Study description
In total, nineteen published studies were identified. All of the papers were reviewed using the criteria defined above; two reviews and three overlapping articles were excluded. Figure 1 showed the flow diagram of selection process. At last, a total of fourteen studies with 6125 cases and 6936 controls were included in this study. Table 1 listed the essential information including first author, year of publication, source of controls, region, number of cases and controls, quality scores and P value of HWE.
Figure 1.
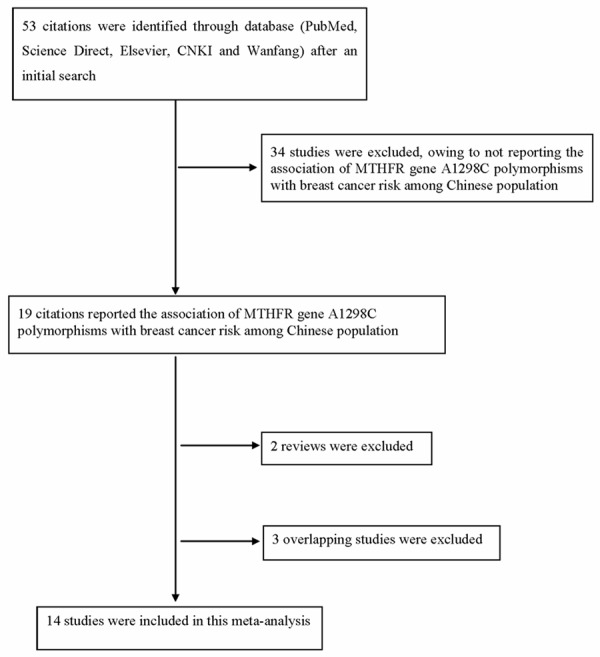
Flow diagram of articles selection process.
Table 1.
General information of the selected articles in this study
| Author | Year | Region | Source of controls | Cases | Controls | P value of HWE | Quality scores | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||
| Total | AA | AC | CC | Total | AA | AC | CC | ||||||
| Qi J [7] | 2004 | Beijing | PB | 217 | 155 | 58 | 4 | 218 | 144 | 71 | 3 | 0.076801 | 6 |
| Shrubsole M [8] | 2004 | Shanghai | PB | 1121 | 768 | 311 | 42 | 1208 | 824 | 344 | 40 | 0.578416 | 8 |
| Kan X [13] | 2007 | Yunnan | PB | 125 | 70 | 41 | 14 | 101 | 61 | 32 | 8 | 0.207067 | 6 |
| Cheng C [9] | 2008 | Taiwan | HB | 351 | 207 | 125 | 19 | 534 | 310 | 207 | 17 | 0.011603 | 7 |
| Inoue M [12] | 2008 | Singapore | PB | 380 | 225 | 139 | 16 | 662 | 387 | 234 | 41 | 0.481608 | 7 |
| Gao C [10] | 2009 | Nanjing | PB | 624 | 446 | 169 | 9 | 624 | 425 | 188 | 11 | 0.056474 | 7 |
| Lin J [14] | 2010 | Guangdong | PB | 65 | 45 | 14 | 6 | 143 | 98 | 35 | 10 | 0.011193 | 5 |
| Hua Z [11] | 2011 | Yunnan | PB | 95 | 50 | 42 | 3 | 90 | 55 | 32 | 3 | 0.522201 | 6 |
| Wu X [15] | 2012 | Yunnan | HB | 75 | 37 | 32 | 6 | 75 | 42 | 28 | 5 | 0.909277 | 5 |
| Liu Y [18] | 2013 | Guangdong | HB | 435 | 206 | 176 | 53 | 435 | 214 | 172 | 49 | 0.111545 | 6 |
| Qiao J [20] | 2014 | Henan | HB | 535 | 258 | 235 | 42 | 673 | 351 | 280 | 42 | 0.158451 | 7 |
| He J [16] | 2014 | Henan | HB | 310 | 138 | 132 | 40 | 381 | 173 | 155 | 53 | 0.058963 | 6 |
| Huang C [17] | 2014 | Taiwan | HB | 1232 | 787 | 386 | 59 | 1232 | 796 | 391 | 45 | 0.723857 | 8 |
| Lu [19] | 2015 | Shanghai | HB | 560 | 369 | 172 | 19 | 560 | 352 | 185 | 23 | 0.831991 | 8 |
PB: Population-based control; HB: Hospital-based control; HWE: Hardy-Weinberg equilibrium.
Test of heterogeneity
There are three MTHFR gene A1298C genotypes/polymorphisms present, namely, AA, AC and CC. The heterogeneity was analyzed for MTHFR gene A1298C AC vs. AA, CC vs. AA, AC+CC vs. AA and C-allele vs. A-allele. There was heterogeneity identified in the group of MTHFR gene A1298C AC+CC vs. AA for the analysis of total population study, population-based study, hospital-based study, HWE in controls, publishing year before 2010 and sample size >1000. Therefore, we calculated the pooled odds ratios for the analyses using the random-effects model. The fixed-effects model was used to calculate the pooled odds ratios for the rest.
Quantitative data synthesis
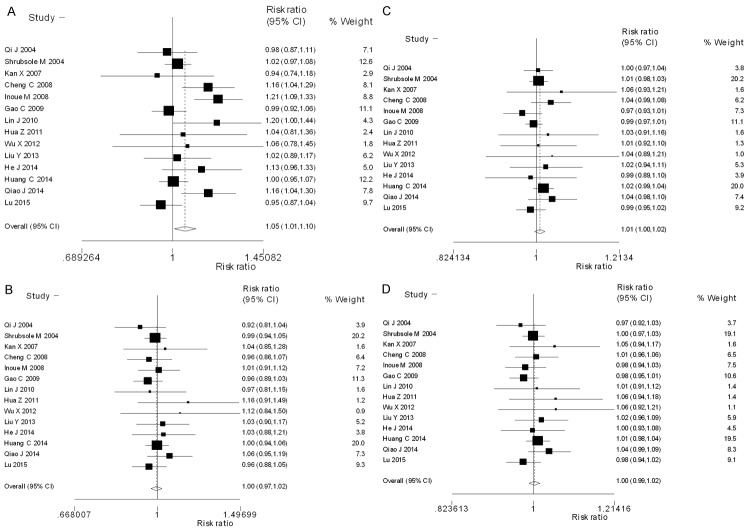
The combined odds ratios of the association between MTHFR gene A1298C polymorphisms and the risk of breast cancer among Chinese population were listed in Table 2. Overall, a significantly increased risk of breast cancer was observed among the subjects carrying MTHFR gene A1298C AC+CC genotype (OR=1.05 with 95% CI: 1.01-1.10) as compared to those carrying AA genotype among the total Chinese population (Figure 2A). We did not observe any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer under the additional genetic models of AC vs. AA, CC vs. AA and C-allele vs. A-allele (OR=1.00 with 95% CI: 0.97-1.02, OR=1.01 with 95% CI: 1.00-1.02 and OR=1.00 with 95% CI: 0.99-1.02, respectively) (Figure 2B-D). Limiting the analysis to the studies with controls in agreement with HWE, we did not observe any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer (OR=1.00 with 95% CI: 0.97-1.03 for AC vs. AA, OR=1.01 with 95% CI: 0.99-1.02 for CC vs. AA, OR=1.04 with 95% CI: 0.99-1.06 for AC+CC vs. AA and OR=1.00 with 95% CI: 0.99-1.02 for C-allele vs. A-allele, respectively) (Table 2).
Table 2.
The summary odds ratio for the association of MTHFR gene A1298C polymorphisms with breast cancer risk among Chinese population
| Genetic model | Cases/controls | Heterogeneity test | Analysis model | Summary OR (95% CI) | Hypothesis test | df | Begg’s test | Egger’s test | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| Q | P | Z | P | Z | P | t | P | |||||
| Total | ||||||||||||
| CA vs. AA | 5793/6586 | 8.15 | 0.833 | Fixed-effects model | 1.00 (0.97-1.02) | 0.37 | 0.710 | 13 | 0.66 | 0.511 | 0.69 | 0.506 |
| CC vs. AA | 4093/4582 | 10.29 | 0.670 | Fixed-effects model | 1.01 (1.00-1.02) | 1.35 | 0.178 | 13 | 0.33 | 0.743 | 0.07 | 0.949 |
| CA+CC vs. AA | 6125/6936 | 28.85 | 0.007 | Random-effects model | 1.06 (1.01-1.10) | 2.24 | 0.025 | 13 | 1.20 | 0.228 | 0.82 | 0.429 |
| C vs. A | 12250/13872 | 10.24 | 0.674 | Fixed-effects model | 1.00 (0.99-1.02) | 0.46 | 0.644 | 13 | 0.22 | 0.827 | 0.36 | 0.723 |
| Stratification by HWE | ||||||||||||
| Yes | ||||||||||||
| CA vs. AA | 5402/5936 | 7.77 | 0.734 | Fixed-effects model | 1.00 (0.97-1.03) | 0.15 | 0.879 | 11 | 1.17 | 0.244 | 1.01 | 0.335 |
| CC vs. AA | 3816/4147 | 8.30 | 0.687 | Fixed-effects model | 1.01 (0.99-1.02) | 0.96 | 0.335 | 11 | 0.62 | 0.537 | 0.41 | 0.690 |
| CA+CC vs. AA | 5709/6259 | 21.82 | 0.026 | Random-effects model | 1.04 (0.99-1.02) | 1.54 | 0.123 | 11 | 0.75 | 0.451 | 0.14 | 0.893 |
| C vs. A | 11418/12518 | 10.11 | 0.520 | Fixed-effects model | 1.00 (0.99-1.02) | 0.37 | 0.709 | 11 | 0.07 | 0.945 | 0.29 | 0.780 |
| Stratification by source of control | ||||||||||||
| Population-based control | ||||||||||||
| CA vs. AA | 2533/2930 | 4.12 | 0.660 | Fixed-effects model | 0.98 (0.95-1.02) | 0.84 | 0.402 | 6 | 0.30 | 0.764 | 0.46 | 0.664 |
| CC vs. AA | 1853/2110 | 3.58 | 0.733 | Fixed-effects model | 1.00 (0.99-1.01) | 0.02 | 0.984 | 6 | 0.00 | 1.000 | 0.45 | 0.670 |
| CA+CC vs. AA | 2627/3046 | 15.16 | 0.019 | Random-effects model | 1.05 (0.98-1.12) | 1.36 | 0.173 | 6 | 0.60 | 0.548 | 0.39 | 0.715 |
| C vs. A | 5254/6092 | 4.19 | 0.651 | Fixed-effects model | 0.99 (0.98-1.01) | 0.68 | 0.499 | 6 | 0.30 | 0.764 | 0.82 | 0.450 |
| Hospital-based control | ||||||||||||
| CA vs. AA | 3260/3656 | 3.47 | 0.745 | Fixed-effects model | 1.00 (0.97-1.04) | 0.22 | 0.823 | 6 | 0.60 | 0.548 | 0.73 | 0.498 |
| CC vs. AA | 2240/2472 | 4.28 | 0.639 | Fixed-effects model | 1.02 (1.00-1.04) | 1.64 | 0.102 | 6 | 0.00 | 1.000 | 0.06 | 0.952 |
| CA+CC vs. AA | 3498/3890 | 13.68 | 0.033 | Random-effects model | 1.06 (0.99-1.13) | 1.67 | 0.095 | 6 | 0.60 | 0.548 | 0.74 | 0.491 |
| C vs. A | 6996/7780 | 4.38 | 0.625 | Fixed-effects model | 1.01 (0.99-1.03) | 1.12 | 0.261 | 6 | 0.30 | 0.764 | 0.06 | 0.952 |
| Stratification by publishing year | ||||||||||||
| Before 2010 | ||||||||||||
| CA vs. AA | 2714/3227 | 2.32 | 0.803 | Fixed-effects model | 0.98 (0.94-1.01) | 1.23 | 0.217 | 5 | 0.00 | 1.000 | 0.25 | 0.814 |
| CC vs. AA | 1975/2271 | 5.52 | 0.356 | Fixed-effects model | 1.00 (0.99-1.02) | 0.44 | 0.658 | 5 | 0.00 | 1.000 | 0.07 | 0.949 |
| CA+CC vs. AA | 2818/3347 | 16.71 | 0.005 | Random-effects model | 1.05 (0.98-1.13) | 1.40 | 0.161 | 5 | 0.00 | 1.000 | 0.48 | 0.656 |
| C vs. A | 5636/6694 | 3.28 | 0.656 | Fixed-effects model | 0.99 (0.98-1.01) | 0.72 | 0.469 | 5 | 0.38 | 0.707 | 0.30 | 0.779 |
| After 2010 | ||||||||||||
| CA vs. AA | 3079/3359 | 4.50 | 0.721 | Fixed-effects model | 1.01 (0.97-1.05) | 0.63 | 0.526 | 7 | 0.62 | 0.536 | 0.37 | 0.587 |
| CC vs. AA | 2118/2311 | 3.56 | 0.829 | Fixed-effects model | 1.01 (0.99-1.03) | 1.34 | 0.181 | 7 | 0.62 | 0.536 | 0.91 | 0.399 |
| CA+CC vs. AA | 3307/3589 | 11.97 | 0.101 | Fixed-effects model | 1.04 (1.00-1.08) | 1.72 | 0.085 | 7 | 1.11 | 0.266 | 0.53 | 0.615 |
| C vs. A | 6614/7178 | 5.00 | 0.660 | Fixed-effects model | 1.01 (0.99-1.03) | 1.23 | 0.217 | 7 | 0.37 | 0.711 | 0.11 | 0.916 |
| Stratification by sample size | ||||||||||||
| <1000 | ||||||||||||
| CA vs. AA | 1528/1829 | 4.86 | 0.677 | Fixed-effects model | 1.00 (0.95-1.06) | 0.05 | 0.960 | 7 | 0.37 | 0.711 | 0.71 | 0.505 |
| CC vs. AA | 1053/1245 | 2.00 | 0.960 | Fixed-effects model | 1.02 (0.99-1.05) | 1.25 | 0.211 | 7 | 0.12 | 0.902 | 0.08 | 0.942 |
| CA+CC vs. AA | 1673/1977 | 7.70 | 0.360 | Fixed-effects model | 1.07 (1.01-1.14) | 2.36 | 0.018 | 7 | 0.12 | 0.902 | 0.57 | 0.589 |
| C vs. A | 3346/3954 | 3.49 | 0.837 | Fixed-effects model | 1.01 (0.98-1.04) | 0.81 | 0.416 | 7 | 0.62 | 0.536 | 0.42 | 0.688 |
| >1000 | ||||||||||||
| CA vs. AA | 4265/4757 | 3.29 | 0.656 | Fixed-effects model | 0.99 (0.96-1.02) | 0.47 | 0.639 | 5 | 0.00 | 1.000 | 0.20 | 0.851 |
| CC vs. AA | 3040/3337 | 6.82 | 0.234 | Fixed-effects model | 1.00 (0.99-1.02) | 0.71 | 0.476 | 5 | 1.13 | 0.260 | 2.78 | 0.050 |
| CA+CC vs. AA | 4452/4959 | 19.30 | 0.002 | Random-effects model | 1.04 (0.98-1.11) | 1.33 | 0.183 | 5 | 0.38 | 0.707 | 0.67 | 0.540 |
| C vs. A | 8904/9918 | 6.25 | 0.283 | Fixed-effects model | 1.00 (0.99-1.01) | 0.01 | 0.994 | 5 | 1.50 | 0.133 | 1.87 | 0.135 |
HWE: Hardy-Weinberg equilibrium; OR: odds ratio; 95% CI: 95% confidence interval.
Figure 2.
Forest plots of the odds ratio for the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population (A: AC+CC vs. AA; B: AC vs. AA; C: CC vs. AA and D: C-allele vs. A-allele).
In the subgroup analysis by source of controls, we did not observe any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer among subjects on the basis of population-based controls and hospital-based controls (Table 2). When stratified by publishing year, we did not observe any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer among the studies published before 2010 and after 2010 (Table 2). When stratified by sample size, we observed that subjects carrying AC+CC genotype had an increased breast cancer risk compared with those carrying AA genotype among the studies of sample size less than 1000 (Table 2). We did not observe any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer in additional subgroup analyses stratified by sample size (Table 2).
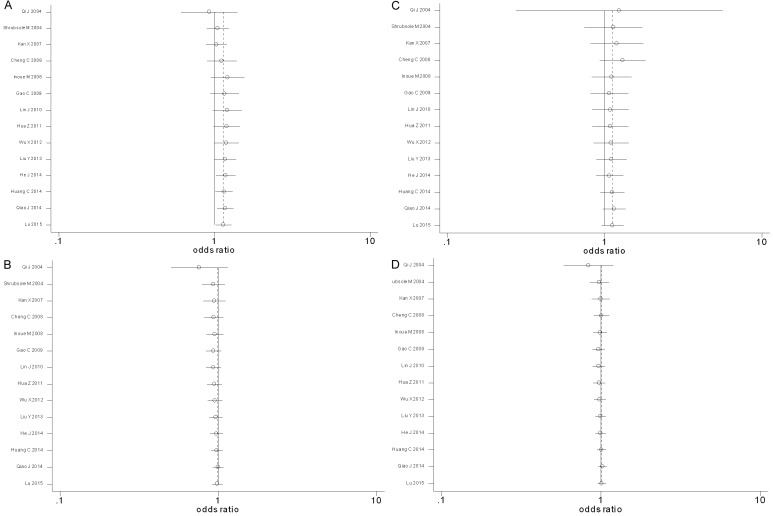
The cumulative meta-analysis showed that there was still a significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer under the genetic model of AC+CC vs. AA, and the cumulative OR was 1.14 with 95% CI: 1.02-1.29 (Figure 3A). Once again, There was still not any significant association between MTHFR gene A1298C polymorphisms and the risk of breast cancer under the additional genetic models of AC vs. AA, CC vs. AA and C-allele vs. A-allele in the cumulative meta-analysis (OR=0.99 with 95% CI: 0.92-1.06, OR=1.12 with 95% CI: 0.95-1.32 and OR=1.01 with 95% CI: 0.96-1.08, respectively) (Figure 3B-D).
Figure 3.
Forest plots of the cumulative odds ratio for the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population (A: AC+CC vs. AA; B: AC vs. AA; C: CC vs. AA and D: C-allele vs. A-allele).
Bias diagnosis
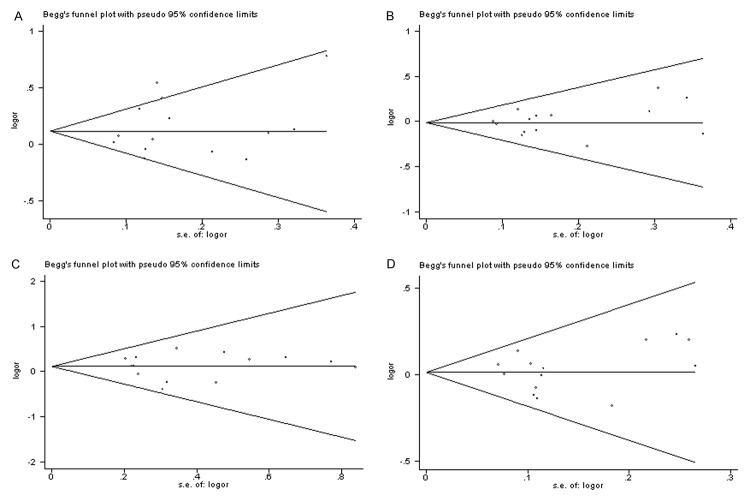
The shape of the funnel plots did not reveal any evidence of obvious asymmetry among the total population (Figure 4A-D), suggesting that no potential publication bias exists. Begg’s test and Egger’s test showed that publication bias might not have significant effects on the pooled results (Table 2).
Figure 4.
Funnel plot analysis to detect publication bias for the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population (A: AC+CC vs. AA, B: AC vs. AA, C: CC vs. AA and D: C-allele vs. A-allele).
Sensitivity analysis
A sensitivity analysis was done to test the effects of the individual dataset on the pooled odds ratio by sequentially removing each eligible study. The pooled effects were not significantly altered with such manipulation for AC+CC vs. AA among the total population (Figure 5).
Figure 5.
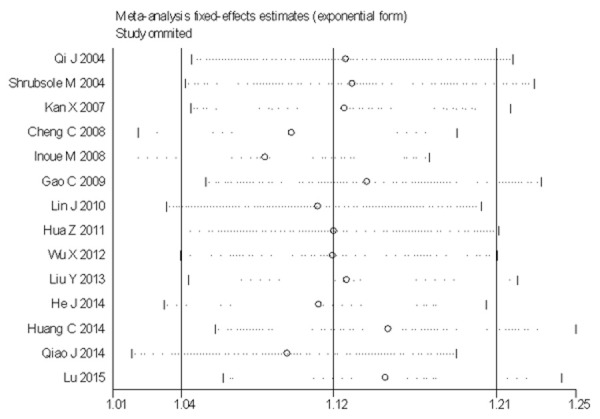
Sensitivity analysis on the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population (AC+CC vs. AA).
Discussion
In 2004, Shrubsole et al., for the first time, investigated the association of MTHFR gene A1298C polymorphisms with breast cancer risk among Chinese population [8]. Thereafter, a number of papers have tested the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population (Data listed in Table 1). The results remained inconsistent. To the best of our knowledge, two meta-analysis studies have been done regarding the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population [25,26]. However, careful examinations of the data reported by the two previous meta-analyses reveal several key issues that are worth noticing. For example, the data reported by Liang et al. [25] for Gao et al.’s study [10] did not seem in line with the data reported by Gao et al. [10]. For overlapping data [9,27], one study reported by Chou et al. [27] with smaller sample size of cases and controls was included in Liang et al.’s study [25] and Song et al.’s study [26] instead of an eligible study reported by Cheng et al. [9] with larger sample size of cases and controls. Three eligible publications [12-14] were missing in Song et al.’s study [26]. Therefore, the conclusions by the previous studies are not entirely credible. Moreover, five studies [16-20] with large sample size have emerged since then. This urged us to clarify the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population.
By carefully combing through the data from fourteen eligible studies, including 6125 cases and 6936 controls, we provided here a more comprehensive and precise assessment of the association between MTHFR gene A1298C polymorphisms and breast cancer risk among Chinese population using up to date published data with the largest case number thus far. We found that the subjects carrying MTHFR gene A1298C AC+CC genotype had an increased risk of breast cancer among Chinese population as comparing to those bearing AA genotype, which is highly statistically significant. Our results are not consistent with these of the previous studies that did not observe any significant association. The discrepancies between our results and theirs may be owing to that our study included comprehensive and precise data. Moreover, a cumulative meta-analysis was carried out to test the tendency by accumulating studies year by year to determine the necessity for new relevant studies. The results remained stable when the studies were accumulated.
Publication bias is always a concern when dealing with pooled data from the literatures. To address this issue, Egger’s test and Begg’s test were performed. Our results showed that the likelihood of key publication bias was negligible in this current study. In addition, each study had different eligibility criteria for participants and source of controls, which should be taken into consideration when expounding the pooled estimates. When the subgroup analysis was performed by source of controls, we did not observe any association between MTHFR gene A1298C polymorphisms and breast cancer risk based on population-based study and hospital-based study.
It is widely acknowledged that the results of genetic association studies might be spurious if the distribution frequency of genotypes in the controls deviated from HWE [28]. In order to address this issue, we performed a subgroup analysis by HWE in controls. When the studies being not in agreement with HWE were excluded from this meta-analysis, we did not observe the association between MTHFR gene A1298C AC+CC vs. AA polymorphism and breast cancer risk among Chinese population.
In summary, our results strongly suggest that MTHFR gene A1298C AC+CC vs. AA polymorphism is associated with an increased risk of breast cancer among Chinese population. Well-designed studies with large sample size are needed to further confirm our results.
Acknowledgements
This work was supported by a grant from the National Natural Science Foundation of China (No. U1404815).
Disclosure of conflict of interest
None.
Abbreviations
- CI
confidence interval
- OR
odds ratio
- HWE
Hardy-Weinberg equilibrium
- SNP
single nucleotide polymorphism
- MTHFR
methylenetetrahydrofolate reductase
- CNKI
China National Knowledge Infrastructure
References
- 1.Fodinger M, Horl WH, Sunder-Plassmann G. Molecular biology of 5,10-methylenetetrahydrofolate reductase. J Nephrol. 2000;13:20–33. [PubMed] [Google Scholar]
- 2.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, Boers GJ, den Heijer M, Kluijtmans LA, van den Heuvel LP, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 3.Weisberg I, Tran P, Christensen B, Sibani S, Rozen R. A second genetic polymorphism in methylenetetrahydrofolate reductase (MTHFR) associated with decreased enzyme activity. Mol Genet Metab. 1998;64:169–172. doi: 10.1006/mgme.1998.2714. [DOI] [PubMed] [Google Scholar]
- 4.van der Put NM, Gabreels F, Stevens EM, Smeitink JA, Trijbels FJ, Eskes TK, van den Heuvel LP, Blom HJ. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neuraltube defects? Am J Hum Genet. 1998;62:1044–1051. doi: 10.1086/301825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng W, Chen L, Li D. Association between dietary intake of folate, vitamin B6, B12 & MTHFR, MTR Genotype and breast cancer risk. Pak J Med Sci. 2014;30:106–110. doi: 10.12669/pjms.301.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumitrescu RG, Cotarla I. Understanding breast cancer risk--where do we stand in 2005? J Cell Mol Med. 2005;9:208–221. doi: 10.1111/j.1582-4934.2005.tb00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi J, Miao XP, Tan W, Yu CY, Liang G, Lu WF, Lin DX. Association between genetic polymorphisms in methylenetetrahydrofolate reductase and risk of breast cancer. Zhonghua Zhong Liu Za Zhi. 2004;26:287–289. [PubMed] [Google Scholar]
- 8.Shrubsole MJ, Gao YT, Cai Q, Shu XO, Dai Q, Hebert JR, Jin F, Zheng W. MTHFR polymorphisms, dietary folate intake, and breast cancer risk: results from the Shanghai Breast Cancer Study. Cancer Epidemiol Biomarkers Prev. 2004;13:190–196. doi: 10.1158/1055-9965.epi-03-0273. [DOI] [PubMed] [Google Scholar]
- 9.Cheng CW, Yu JC, Huang CS, Shieh JC, Fu YP, Wang HW, Wu PE, Shen CY. Polymorphism of cytosolic serine hydroxymethyltransferase, estrogen and breast cancer risk among Chinese women in Taiwan. Breast Cancer Res Treat. 2008;111:145–155. doi: 10.1007/s10549-007-9754-x. [DOI] [PubMed] [Google Scholar]
- 10.Gao CM, Tang JH, Cao HX, Ding JH, Wu JZ, Wang J, Liu YT, Li SP, Su P, Matsuo K, Takezaki T, Tajima K. MTHFR polymorphisms, dietary folate intake and breast cancer risk in Chinese women. J Hum Genet. 2009;54:414–418. doi: 10.1038/jhg.2009.57. [DOI] [PubMed] [Google Scholar]
- 11.Hua Z, Wang Y, Ni J, Ge F, Zou T. Folic acid, Vitamin B12, MTHFR, MS gene polymorphisms associate with the risk of breast cancer. Modern Oncology. 2011;19:428–431. [Google Scholar]
- 12.Inoue M, Robien K, Wang R, Van Den Berg DJ, Koh WP, Yu MC. Green tea intake, MTHFR/TYMS genotype and breast cancer risk: the Singapore Chinese Health Study. Carcinogenesis. 2008;29:1967–1972. doi: 10.1093/carcin/bgn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kan X, Zou T, Wu X, Wang X. Association between MTHFR Genotype Polymorphism and Breast Cancer Susceptibility in Human Population from Yunnan. Cancer Research on Prevention and Treatment. 2007;34:716–718. [Google Scholar]
- 14.Lin J, Chen S, Li W. Single nucleotide polymorphisms in methylenetetrahydrofolate reductase gene and susceptibility to breast cancer. Modern Hospital. 2010;10:15–17. [Google Scholar]
- 15.Wu XY, Ni J, Xu WJ, Zhou T, Wang X. Interactions between MTHFR C677T-A1298C variants and folic acid deficiency affect breast cancer risk in a Chinese population. Asian Pac J Cancer Prev. 2012;13:2199–2206. doi: 10.7314/apjcp.2012.13.5.2199. [DOI] [PubMed] [Google Scholar]
- 16.He JM, Pu YD, Wu YJ, Qin R, Zhang QJ, Sun YS, Zheng WW, Chen LP. Association between dietary intake of folate and MTHFR and MTR genotype with risk of breast cancer. Genet Mol Res. 2014;13:8925–8931. doi: 10.4238/2014.October.31.7. [DOI] [PubMed] [Google Scholar]
- 17.Huang CY, Chang WS, Shui HA, Hsieh YH, Loh CH, Wang HC, Ji HX, Hsiao CL, Hsu CM, Tsai CW, Bau DT. Evaluation of the contribution of methylenetetrahydrofolate reductase genotypes to Taiwan breast cancer. Anticancer Res. 2014;34:4109–4115. [PubMed] [Google Scholar]
- 18.Liu Y, Zhou LS, Xu XM, Deng LQ, Xiao QK. Association of dietary intake of folate, vitamin B6 and B12 and MTHFR genotype with breast cancer risk. Asian Pac J Cancer Prev. 2013;14:5189–5192. doi: 10.7314/apjcp.2013.14.9.5189. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q, Jiang K, Li Q, Ji YJ, Chen WL, Xue XH. Polymorphisms in the MTHFR gene are associated with breast cancer risk and prognosis in a Chinese population. Tumour Biol. 2015;36:3757–3762. doi: 10.1007/s13277-014-3016-4. [DOI] [PubMed] [Google Scholar]
- 20.Qiao J, Jiao D, Lu Z, Cui S, Liu Z. Association of methylenetetrahydrofolate reductase and methionine synthase polymorphisms with breast cancer risk and interaction with folate, vitamin B6, and vitamin B12 intakes. Tumour Biol. 2014;35:11895–11901. doi: 10.1007/s13277-014-2456-1. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Jiang Z, Qiu H, Tang W, Duan T. Associations between CTLA-4 +49 A/G (rs231775) polymorphism and cancer risk: a meta-analysis based on 52 case-control studies. Int J Clin Exp Med. 2015;8:6835–6851. [PMC free article] [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 24.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang H, Yan Y, Li T, Li R, Li M, Li S, Qin X. Methylenetetrahydrofolate reductase polymorphisms and breast cancer risk in Chinese population: a meta-analysis of 22 case-control studies. Tumour Biol. 2014;35:1695–1701. doi: 10.1007/s13277-013-1234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song S, Yang B, Sun G. The correlation between the 5,10-methylenetrahydrofolate reductase C677T or A1298C polymorphisms and breast cancer risk in China: a Metaanalysis. Chin J Prev Contr Chron Dis. 2013;21:560–564. [Google Scholar]
- 27.Chou YC, Wu MH, Yu JC, Lee MS, Yang T, Shih HL, Wu TY, Sun CA. Genetic polymorphisms of the methylenetetrahydrofolate reductase gene, plasma folate levels and breast cancer susceptibility: a case-control study in Taiwan. Carcinogenesis. 2006;27:2295–2300. doi: 10.1093/carcin/bgl108. [DOI] [PubMed] [Google Scholar]
- 28.Salanti G, Amountza G, Ntzani EE, Ioannidis JP. Hardy-Weinberg equilibrium in genetic association studies: an empirical evaluation of reporting, deviations, and power. Eur J Hum Genet. 2005;13:840–848. doi: 10.1038/sj.ejhg.5201410. [DOI] [PubMed] [Google Scholar]