Abstract
Previous studies had researched the relationship between rs9642880 gene polymorphism and bladder cancer risk, but the results remained unclear. The comprehensive meta-analysis was performed to clarify this possible association. Relevant articles were searched from Pubmed, Embase and web of science. Odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were calculated to assess the strength of the association. The assessment of publication bias was conducted by Begg’s funnel plots and Egger’s regression test. A total of 7 casecontrol studies involving 4072 cases and 4898 controls were included in our study. Overall, an obvious relationship between rs9642880 polymorphism and increased risk of bladder cancer were detected in all models. Besides, the positive results were observed among both Caucasians and Asians when stratified by ethnicity. Moreover, when stratified by genotyping method, the significant results were detected in all genotyping methods except Sequenom. In addition, in the subgroup analysis by source of control, significant results were detected in both population and hospital based controls. This present meta-analysis with accurate and reliable results indicated that the T allele of SNP rs9642880 confers susceptibility to bladder cancer in both Asian and Caucasian populations.
Keywords: MYC, rs9642880, gene polymorphism, bladder cancer, meta-analysis
Introduction
Bladder cancer, as one of the most common malignant cancers, has a large amount of estimated cases and deaths of approximately 386,300 and 150,200 respectively worldwide [1]. The etiology of bladder cancer is complicated, while two major known risk factors are smoking and occupational exposures [2]. Besides, it has been clarified that genetic polymorphisms are likely to play an important role in the occurrence of bladder cancer [3]. Recently, single-nucleotide polymorphisms (SNPs) have been demonstrated to be associated with bladder cancer risk by genome-wide association studies (GWAS) in European populations [4-8].
As an oncogene, MYC is implicated in the carcinogenesis and tumor progression [9-11]. MYC is a nuclear protein which connects with a small protein called MAX to act as a sequence-specific, DNA-binding transcription factor that regulates various genes involved in cell cell growth, proliferation and apoptosis [12]. Rs9642880 polymorphism is located 30 kb upstream of MYC gene at the 8q24 region [13]. Recently, it has been clarified that rs9642880 GT/TT polymorphism was associated with the enhanced expression of MYC in both mRNA and protein levels in bladder tissues [14], which might play an important role in bladder carcinogenesis.
So far, a growing number of studies have investigated the relevance between rs9642880 polymorphism and bladder cancer susceptibility [15-23]. However, the results are still unclear with limited sample size. Furthermore, lack of stratified analysis prevented comprehensive understanding of the association in current studies. We thereby conducted a meta-analysis to clarify the real association.
Materials and methods
The database PubMed, EMbase and Web of Science were searched for relevant studies until March 31, 2015. The following keywords were utilized for searching: “MYC”, “polymorphism” or “rs9642880” and “bladder caner”. Only the latest or the largest sample size was included in studies with partly overlapping. Additional eligible studies were hand-searched from reference of original studies or reviews.
Relevant studies were selected by the following inclusion criteria: (1) Case-control studies were designed; (2) The number of each genotype was available for estimating the odds ratio (OR) and the corresponding 95% confidence interval (95% CI); (3) The diagnosis of patients was confirmed pathologically and the controls were confirmed as free from any cancer. In addition, the major exclusion criterion was as follows: (1) without control series; (2) not available of genotype frequency data; (3) review articles; (4) duplicates of previous publication.
Data extraction
Two authors (Tang J and Li X) reviewed the studies carefully and independently identify the eligible ones. A consensus was reached on all items and the disagreement would be solved by a discussion. The data was extracted independently and the following data was extracted from each study: first author’s name, year of publication, ethnicity, source of controls (SOC), genotyping method, number of cases and controls, genotype frequency of rs9642880 polymorphism in cases and controls respectively and the results of the Hardy-Weinberg equilibrium (HWE) test.
Statistical analysis
To evaluate the strength of association between rs9642880 polymorphism and bladder cancer risk, the pooled odds ratios (ORs) with 95% confidence intervals (CIs) were calculated. The fixed-effects model (a Mantel-Haenszel method) and the random-effects model (a DerSimonian-Laird method) were respectively utilized to pool the data [24]. If the significant heterogeneity was observed, the random-effects model was applied preferentially. Sensitivity analysis was carried out to evaluate the stability of the results by omitting each study at a time. Then, subgroups analysis was further conducted by ethnicity, genotyping methods and source of control (SOC).
Publication bias between the studies was examined by Begg’s funnel plots and Egger’s linear regression test. It was considered statistically significant when P<0.05 [25]. HWE was estimated by the goodness-of-fit chi-square test and it was regarded as a significantly selective bias when P<0.05 [26]. All statistical analysis was carried out by the STATA software (version 12.0; StataCorp LP, College Station, TX). All tests were two-sided and it was considered statistically significant when P<0.05.
Results
Characteristics of the studies
Finally, a total of 7 case-control studies including 4072 cases and 4898 controls were involved in the current meta-analysis [15-21] as listed in Table 1. All studies showed that the genotype distribution of control groups was consistent with HWE. The flowchart of literature search and identification process is presented in Figure 1.
Table 1.
Characteristics of individual studies included in the meta-analysis
| rs9642880 | Case (n) | Control (n) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| Year | Surname | Ethnicity | SOC | Genotyping | Case | Control | GG | GT | TT | GG | GT | TT |
| 2014 | Wang | Asian | PB | Taqman | 1210 | 1008 | 550 | 536 | 124 | 514 | 389 | 105 |
| 2013 | Yates | Caucasian | HB | Taqman | 231 | 264 | 64 | 114 | 53 | 84 | 130 | 50 |
| 2013 | Ma | Asian | PB | Sequenom | 171 | 962 | 74 | 74 | 23 | 489 | 371 | 102 |
| 2013 | Ali | Asian | PB | PCR-RFLP | 200 | 200 | 33 | 84 | 83 | 48 | 90 | 62 |
| 2012 | Roupret | Caucasian | HB | Taqman | 261 | 261 | 69 | 119 | 73 | 81 | 130 | 50 |
| 2012 | Schwender | Caucasian | HB | Taqman | 1584 | 1738 | 391 | 767 | 426 | 486 | 876 | 376 |
| 2009 | Wang | Asian | HB | PCR-RFLP | 415 | 465 | 149 | 203 | 63 | 223 | 192 | 50 |
Figure 1.
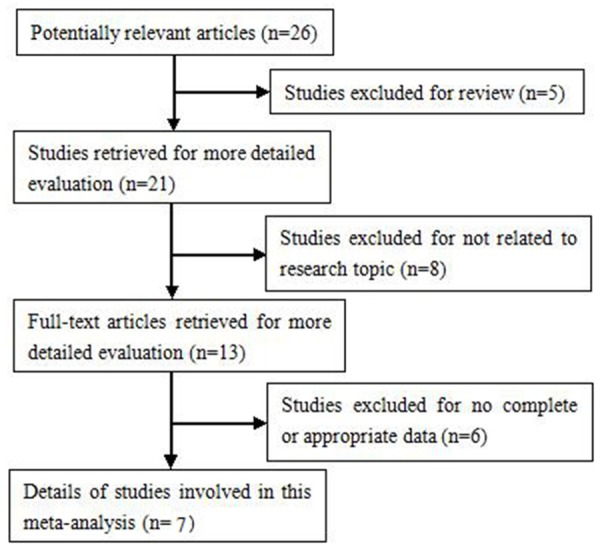
Flow diagram of literature search and selection process.
Quantitative synthesis
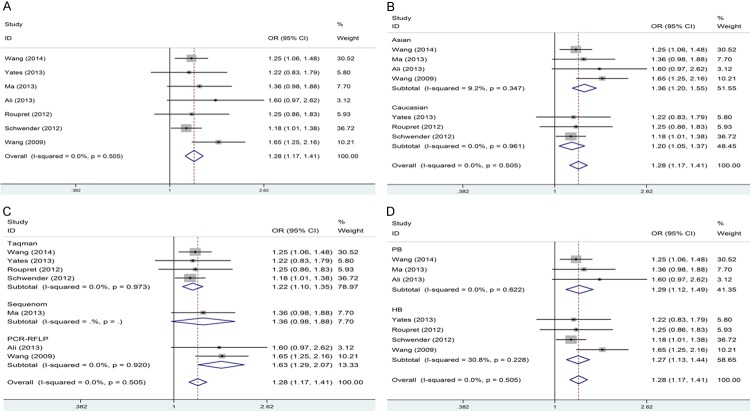
The results of the relationship between rs9642880 polymorphism and bladder cancer risk is listed in Table 2. In general, the pooled OR was 1.23 (95% CI: 1.11-1.35) for heterozygote model, 1.42 (95% CI: 1.25-1.62) for homozygote model, 1.28 (95% CI: 1.17-1.41) for dominant model and 1.31 (95% CI: 1.17-1.46) for recessive model (Figure 2A). Overall, there was an obvious connection between rs9642880 polymorphism and increased risk of bladder cancer. Moreover, the positive results were observed among both Caucasians and Asians when stratified by ethnicity (Figure 2B). Furthermore, when stratified by genotyping method, the significant results were detected in all genotyping methods except Sequenom (Figure 2C). In addition, in the subgroup analysis by SOC, significant results were detected in both population and hospital based controls (Figure 2D).
Table 2.
Meta-analysis results of association between rs9642880 polymorphism and bladder cancer risk
| GT vs. GG | TT vs. GG | GT/TT vs. GG | TT vs. GT/GG | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| Na | Sample Size | OR (95% CI)* | Pb | OR (95% CI)* | Pb | OR (95% CI)* | Pb | OR (95% CI)* | Pb | |
| Total | 7 | 8970 | 1.23 (1.11-1.35) | 0.406 | 1.42 (1.25-1.62) | 0.362 | 1.28 (1.17-1.41) | 0.505 | 1.31 (1.17-1.46) | 0.358 |
| Ethnicity | ||||||||||
| Caucasian | 3 | 4339 | 1.36 (1.19-1.55) | 0.690 | 1.44 (1.22-1.70) | 0.752 | 1.20 (1.05-1.37) | 0.961 | 1.36 (1.18-1.56) | 0.624 |
| Asian | 4 | 4631 | 1.09 (0.95-1.26) | 0.966 | 1.40 (1.15-1.71) | 0.113 | 1.36 (1.20-1.55) | 0.347 | 1.23 (1.02-1.47) | 0.177 |
| Genotyping | ||||||||||
| Taqman | 4 | 6557 | 1.17 (1.04-1.30) | 0.565 | 1.35 (1.16-1.56) | 0.386 | 1.22 (1.10-1.35) | 0.973 | 1.27 (1.12-1.44) | 0.157 |
| Sequenom | 1 | 1133 | 1.32 (0.93-1.87) | -- | 1.49 (0.89-2.49) | -- | 1.36 (0.98-1.88) | -- | 1.31 (0.81-2.13) | -- |
| PCR-RFLP | 2 | 1280 | 1.53 (1.19-1.97) | 0.620 | 1.91 (1.36-2.67) | 0.928 | 1.63 (1.29-2.07) | 0.920 | 1.53 (1.15-2.04) | 0.834 |
| SOC | ||||||||||
| PB | 3 | 3751 | 1.30 (1.12-1.51) | 0.979 | 1.29 (1.02-1.62) | 0.167 | 1.29 (1.12-1.49) | 0.622 | 1.17 (0.95-1.43) | 0.148 |
| HB | 4 | 5219 | 1.18 (1.04-1.34) | 0.159 | 1.49 (1.28-1.75) | 0.593 | 1.27 (1.13-1.44) | 0.228 | 1.37 (1.20-1.57) | 0.774 |
Number of studies;
P value of Q test for heterogeneity;
Fixed-effects model was used because all P value for heterogeneity test >0.1.
Figure 2.
Forest plots of association between rs9642880 polymorphism and bladder cancer risk in dominant model. A: Overall results; B: Stratified by ethnicity; C: Stratified by genotyping method; D: Stratified by SOC.
Test of heterogeneity and sensitivity
No significant heterogeneity was observed in all kinds of models, which was confirmed by the analysis of Galbraith (Figure 3). Sensitivity analysis was performed to evaluate the effect of each study on the pooled OR by omitting one single study at a time. The results of the sensitivity analysis demonstrated that no individual study significantly affected the pooled OR (Figure 4). All these indicated the stability of our results.
Figure 3.
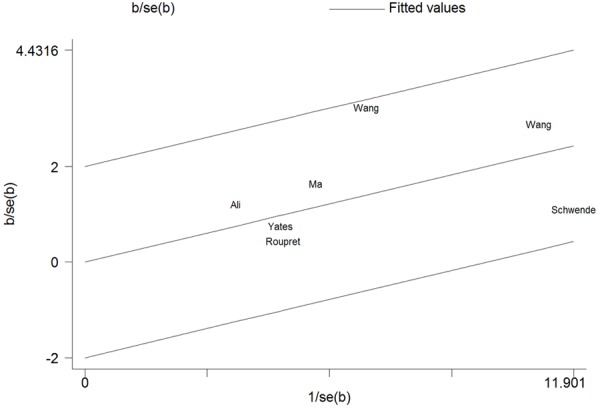
The analysis of Galbraith to assess the heterogeneity.
Figure 4.
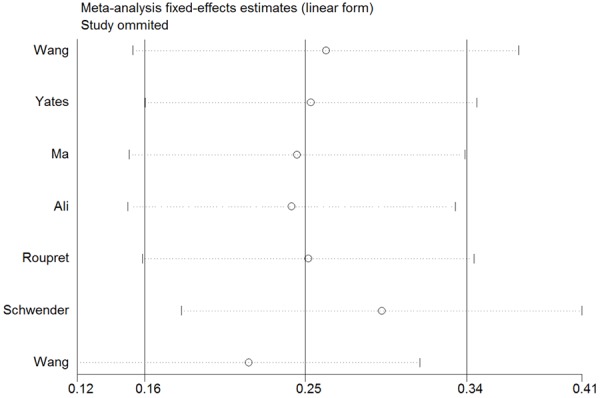
Sensitivity analysis under the dominant model.
Publication bias
Begg’s funnel plot and Egger’s test were utilized to assess the publication bias of the literature. No obvious asymmetry was seen from the shape of the Begg’s funnel plot (Figure 5), indicating there was no significant publication bias and our results were robust. The same result was found in the Egger’s test.
Figure 5.
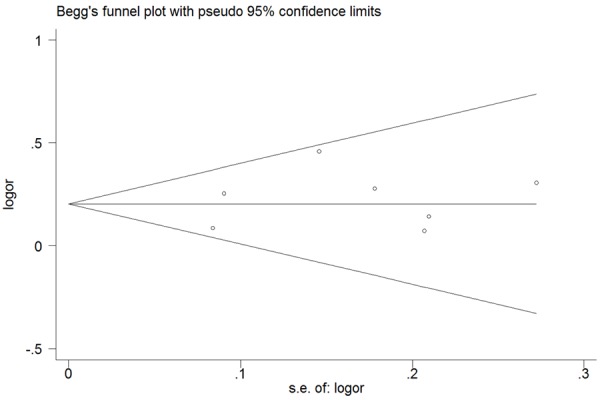
Begg’s funnel plot for publication bias test.
Discussion
The 8q24 region, where rs9642880 polymorphism located at, is a gene desert. Various enhancers regulate transcription are found in this region that can regulate the transcription of MYC [27-29]. Moreover, this region interacts with the MYC protooncogene by a chromatin loop [30]. Meanwhile, it has been demonstrated that T allele of rs9642880 polymorphism was related with the overexpession of MYC mRNA and protein in bladder tissues [14], implying this polymorphism may contribute to the occurrence of bladder cancer through regulating the expression of MYC.
MYC is overexpressed in various tumors, which confers a proliferative advantage to tumor cells both in vitro and in vivo [31,32], regulating cell function associated with tumorgenesis. In addition, Fu et al. [13] found that synthetic artificial microRNAs targeting MYC could inhibit the malignant phenotypes of bladder cancer. Furthermore, it has been shown down-regulating the MYC expression could increase the antitumor activity in gemcitabine-resistant bladder cancer [33], indicating MYC is a potential regulator of chemotherapy resistance.
In 2008, Kiemeney et al. [34] identified rs9642880 polymorphism as the risk locus for bladder cancer. Since then, increasing studies have investigated the association between this polymorphism and bladder cancer risk [15-21]. Nevertheless, the outcomes remained unclear without sufficient support from larger population. Moreover, further studies by different stratified analysis were not carried out. As a powerful tool, meta-analysis can provide more reliable results than individual study and comprehensive information through different subgroup analysis [35]. Therefore, we utilized meta-analysis to illustrate the possible association between rs9642880 polymorphism and bladder cancer risk. In the present meta-analysis, our results indicated the T allele of rs9642880 could increase bladder cancer susceptibility.
Due to various genetic backgrounds, different ethnic populations may have different incidence of gene polymorphisms [36]. As a result, subgroup analysis by ethnic was performed and the positive results were observed among both Asian and Caucasian. When stratified by SOC, both the population and hospital based controls showed significant results. However, in subgroup analysis by genotyping method, the positive results were detected in Taqman and PCR-RFLP method, instead of sequenom. The difference may be caused as only one study take sequenom method, which increased the inaccuracy of the results. In addition, different genotyping methods have specialty in different aspects, so the results would be more reliable and accurate if the same appropriate genotyping method was applied in different studies. In this meta-analysis, no significant heterogeneity was detected and the sensitivity analysis showed the results were stable. In summary, all these analysis suggested rs9642880 polymorphism played an important role in the risk of bladder cancer.
Though the results of this meta-analysis were identified by sufficient statistical evidence, some limitations must be noticed. Firstly, only 7 researches included in the meta-analysis met inclusion criterion, which required more high-quality studies to provide more sufficient evidence. Secondly, the number in some subgroups is relatively small, which limits the statistical power to reveal the real association. Furthermore, bladder cancer susceptibility might result from interactions of various genetic and environmental factors instead of one single gene. Thereby, future studies should focus on the combined effects of different factors.
In summary, this present meta-analysis with accurate and reliable results indicated the T allele of rs9642880 polymorphism confers susceptibility to bladder cancer both in Asian and Caucasian populations. Future studies on combined effects of different gene polymorphisms are required to explore the etiology and provide an early diagnosis of bladder cancer.
Disclosure of conflict of interest
None.
References
- 1.Aly M, Wiklund F, Xu J, Isaacs WB, Eklund M, D’Amato M, Adolfsson J, Gronberg H. Polygenic risk score improves prostate cancer risk prediction: results from the Stockholm-1 cohort study. Eur Urol. 2011;60:21–28. doi: 10.1016/j.eururo.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murta-Nascimento C, Schmitz-Drager BJ, Zeegers MP, Steineck G, Kogevinas M, Real FX, Malats N. Epidemiology of urinary bladder cancer: from tumor development to patient’s death. World J Urol. 2007;25:285–295. doi: 10.1007/s00345-007-0168-5. [DOI] [PubMed] [Google Scholar]
- 3.Murta-Nascimento C, Silverman DT, Kogevinas M, Garcia-Closas M, Rothman N, Tardon A, Garcia-Closas R, Serra C, Carrato A, Villanueva C, Dosemeci M, Real FX, Malats N. Risk of bladder cancer associated with family history of cancer: do low-penetrance polymorphisms account for the increase in risk? Cancer Epidemiol Biomarkers Prev. 2007;16:1595–1600. doi: 10.1158/1055-9965.EPI-06-0743. [DOI] [PubMed] [Google Scholar]
- 4.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, Blondal T, Witjes JA, Vermeulen SH, Hulsbergen-van de Kaa CA, Swinkels DW, Ploeg M, Cornel EB, Vergunst H, Thorgeirsson TE, Gudbjartsson D, Gudjonsson SA, Thorleifsson G, Kristinsson KT, Mouy M, Snorradottir S, Placidi D, Campagna M, Arici C, Koppova K, Gurzau E, Rudnai P, Kellen E, Polidoro S, Guarrera S, Sacerdote C, Sanchez M, Saez B, Valdivia G, Ryk C, de Verdier P, Lindblom A, Golka K, Bishop DT, Knowles MA, Nikulasson S, Petursdottir V, Jonsson E, Geirsson G, Kristjansson B, Mayordomo JI, Steineck G, Porru S, Buntinx F, Zeegers MP, Fletcher T, Kumar R, Matullo G, Vineis P, Kiltie AE, Gulcher JR, Thorsteinsdottir U, Kong A, Rafnar T, Stefansson K. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu X, Ye Y, Kiemeney LA, Sulem P, Rafnar T, Matullo G, Seminara D, Yoshida T, Saeki N, Andrew AS, Dinney CP, Czerniak B, Zhang ZF, Kiltie AE, Bishop DT, Vineis P, Porru S, Buntinx F, Kellen E, Zeegers MP, Kumar R, Rudnai P, Gurzau E, Koppova K, Mayordomo JI, Sanchez M, Saez B, Lindblom A, de Verdier P, Steineck G, Mills GB, Schned A, Guarrera S, Polidoro S, Chang SC, Lin J, Chang DW, Hale KS, Majewski T, Grossman HB, Thorlacius S, Thorsteinsdottir U, Aben KK, Witjes JA, Stefansson K, Amos CI, Karagas MR, Gu J. Genetic variation in the prostate stem cell antigen gene PSCA confers susceptibility to urinary bladder cancer. Nat Genet. 2009;41:991–995. doi: 10.1038/ng.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, Real FX, Van Den Berg D, Matullo G, Baris D, Thun M, Kiemeney LA, Vineis P, De Vivo I, Albanes D, Purdue MP, Rafnar T, Hildebrandt MA, Kiltie AE, Cussenot O, Golka K, Kumar R, Taylor JA, Mayordomo JI, Jacobs KB, Kogevinas M, Hutchinson A, Wang Z, Fu YP, Prokunina-Olsson L, Burdett L, Yeager M, Wheeler W, Tardón A, Serra C, Carrato A, García-Closas R, Lloreta J, Johnson A, Schwenn M, Karagas MR, Schned A, Andriole G Jr, Grubb R 3rd, Black A, Jacobs EJ, Diver WR, Gapstur SM, Weinstein SJ, Virtamo J, Cortessis VK, Gago-Dominguez M, Pike MC, Stern MC, Yuan JM, Hunter DJ, McGrath M, Dinney CP, Czerniak B, Chen M, Yang H, Vermeulen SH, Aben KK, Witjes JA, Makkinje RR, Sulem P, Besenbacher S, Stefansson K, Riboli E, Brennan P, Panico S, Navarro C, Allen NE, Bueno-de-Mesquita HB, Trichopoulos D, Caporaso N, Landi MT, Canzian F, Ljungberg B, Tjonneland A, Clavel-Chapelon F, Bishop DT, Teo MT, Knowles MA, Guarrera S, Polidoro S, Ricceri F, Sacerdote C, Allione A, Cancel-Tassin G, Selinski S, Hengstler JG, Dietrich H, Fletcher T, Rudnai P, Gurzau E, Koppova K, Bolick SC, Godfrey A, Xu Z, Sanz-Velez JI, D García-Prats M, Sanchez M, Valdivia G, Porru S, Benhamou S, Hoover RN, Fraumeni JF Jr, Silverman DT, Chanock SJ. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiemeney LA, Sulem P, Besenbacher S, Vermeulen SH, Sigurdsson A, Thorleifsson G, Gudbjartsson DF, Stacey SN, Gudmundsson J, Zanon C, Kostic J, Masson G, Bjarnason H, Palsson ST, Skarphedinsson OB, Gudjonsson SA, Witjes JA, Grotenhuis AJ, Verhaegh GW, Bishop DT, Sak SC, Choudhury A, Elliott F, Barrett JH, Hurst CD, de Verdier PJ, Ryk C, Rudnai P, Gurzau E, Koppova K, Vineis P, Polidoro S, Guarrera S, Sacerdote C, Campagna M, Placidi D, Arici C, Zeegers MP, Kellen E, Gutierrez BS, Sanz-Velez JI, Sanchez-Zalabardo M, Valdivia G, Garcia-Prats MD, Hengstler JG, Blaszkewicz M, Dietrich H, Ophoff RA, van den Berg LH, Alexiusdottir K, Kristjansson K, Geirsson G, Nikulasson S, Petursdottir V, Kong A, Thorgeirsson T, Mungan NA, Lindblom A, van Es MA, Porru S, Buntinx F, Golka K, Mayordomo JI, Kumar R, Matullo G, Steineck G, Kiltie AE, Aben KK, Jonsson E, Thorsteinsdottir U, Knowles MA, Rafnar T, Stefansson K. A sequence variant at 4p16.3 confers susceptibility to urinary bladder cancer. Nat Genet. 2010;42:415–419. doi: 10.1038/ng.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rafnar T, Vermeulen SH, Sulem P, Thorleifsson G, Aben KK, Witjes JA, Grotenhuis AJ, Verhaegh GW, Hulsbergen-van de Kaa CA, Besenbacher S, Gudbjartsson D, Stacey SN, Gudmundsson J, Johannsdottir H, Bjarnason H, Zanon C, Helgadottir H, Jonasson JG, Tryggvadottir L, Jonsson E, Geirsson G, Nikulasson S, Petursdottir V, Bishop DT, Chung-Sak S, Choudhury A, Elliott F, Barrett JH, Knowles MA, de Verdier PJ, Ryk C, Lindblom A, Rudnai P, Gurzau E, Koppova K, Vineis P, Polidoro S, Guarrera S, Sacerdote C, Panadero A, Sanz-Velez JI, Sanchez M, Valdivia G, Garcia-Prats MD, Hengstler JG, Selinski S, Gerullis H, Ovsiannikov D, Khezri A, Aminsharifi A, Malekzadeh M, van den Berg LH, Ophoff RA, Veldink JH, Zeegers MP, Kellen E, Fostinelli J, Andreoli D, Arici C, Porru S, Buntinx F, Ghaderi A, Golka K, Mayordomo JI, Matullo G, Kumar R, Steineck G, Kiltie AE, Kong A, Thorsteinsdottir U, Stefansson K, Kiemeney LA. European genome-wide association study identifies SLC14A1 as a new urinary bladder cancer susceptibility gene. Hum Mol Genet. 2011;20:4268–4281. doi: 10.1093/hmg/ddr303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-David E, Bester AC, Shifman S, Kerem B. Transcriptional dynamics in colorectal carcinogenesis: new insights into the role of c-Myc and miR17 in benign to cancer transformation. Cancer Res. 2014;74:5532–5540. doi: 10.1158/0008-5472.CAN-14-0932. [DOI] [PubMed] [Google Scholar]
- 10.Fonseca-Alves CE, Rodrigues MM, de Moura VM, Rogatto SR, Laufer-Amorim R. Alterations of C-MYC, NKX3.1, and E-cadherin expression in canine prostate carcinogenesis. Microsc Res Tech. 2013;76:1250–1256. doi: 10.1002/jemt.22292. [DOI] [PubMed] [Google Scholar]
- 11.Xiao W, Wang J, Ou C, Zhang Y, Ma L, Weng W, Pan Q, Sun F. Mutual interaction between YAP and c-Myc is critical for carcinogenesis in liver cancer. Biochem Biophys Res Commun. 2013;439:167–172. doi: 10.1016/j.bbrc.2013.08.071. [DOI] [PubMed] [Google Scholar]
- 12.Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- 13.Fu X, Liu Y, Zhuang C, Liu L, Cai Z, Huang W. Synthetic artificial microRNAs targeting UCA1-MALAT1 or c-Myc inhibit malignant phenotypes of bladder cancer cells T24 and 5637. Mol Biosyst. 2015;11:1285–9. doi: 10.1039/c5mb00127g. [DOI] [PubMed] [Google Scholar]
- 14.Wang M, Wang M, Zhang W, Yuan L, Fu G, Wei Q, Zhang Z. Common genetic variants on 8q24 contribute to susceptibility to bladder cancer in a Chinese population. Carcinogenesis. 2009;30:991–996. doi: 10.1093/carcin/bgp091. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Ye D, Guo J, Liu F, Jiang H, Gong J, Gu C, Shao Q, Sun J, Zheng SL, Yu H, Lin X, Xia G, Fang Z, Zhu Y, Ding Q, Xu J. Genetic score of multiple risk-associated single nucleotide polymorphisms is a marker for genetic susceptibility to bladder cancer. Genes Chromosomes Cancer. 2014;53:98–105. doi: 10.1002/gcc.22121. [DOI] [PubMed] [Google Scholar]
- 16.Yates DR, Roupret M, Drouin SJ, Audouin M, Cancel-Tassin G, Comperat E, Bitker MO, Cussenot O. Genetic polymorphisms on 8q24.1 and 4p16.3 are not linked with urothelial carcinoma of the bladder in contrast to their association with aggressive upper urinary tract tumours. World J Urol. 2013;31:53–59. doi: 10.1007/s00345-012-0954-6. [DOI] [PubMed] [Google Scholar]
- 17.Ma Z, Hu Q, Chen Z, Tao S, Macnamara L, Kim ST, Tian L, Xu K, Ding Q, Zheng SL, Sun J, Xia G, Xu J. Systematic evaluation of bladder cancer risk-associated single-nucleotide polymorphisms in a Chinese population. Mol Carcinog. 2013;52:916–921. doi: 10.1002/mc.21932. [DOI] [PubMed] [Google Scholar]
- 18.Ali SHB, Younis M, Bangash KS, Rauf A, Anwar K, Khurram R, Khawaja MA, Qureshi AA, Akhter S, Azam M, Kiemeney LA, Qamar R. Common Variants at 8q24 Confer Susceptibility to Urothelial Bladder Cancer in the Pakistani Population. PAK J ZOOL. 2013;45:1501–1509. [Google Scholar]
- 19.Roupret M, Drouin SJ, Cancel-Tassin G, Comperat E, Larre S, Cussenot O. Genetic variability in 8q24 confers susceptibility to urothelial carcinoma of the upper urinary tract and is linked with patterns of disease aggressiveness at diagnosis. J Urol. 2012;187:424–428. doi: 10.1016/j.juro.2011.10.038. [DOI] [PubMed] [Google Scholar]
- 20.Schwender H, Selinski S, Blaszkewicz M, Marchan R, Ickstadt K, Golka K, Hengstler JG. Distinct SNP combinations confer susceptibility to urinary bladder cancer in smokers and non-smokers. PLoS One. 2012;7:e51880. doi: 10.1371/journal.pone.0051880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golka K, Hermes M, Selinski S, Blaszkewicz M, Bolt HM, Roth G, Dietrich H, Prager HM, Ickstadt K, Hengstler JG. Susceptibility to urinary bladder cancer: relevance of rs9642880[T] , GSTM1 0/0 and occupational exposure. Pharmacogenet Genomics. 2009;19:903–906. doi: 10.1097/FPC.0b013e328331b554. [DOI] [PubMed] [Google Scholar]
- 22.Figueroa JD, Ye Y, Siddiq A, Garcia-Closas M, Chatterjee N, Prokunina-Olsson L, Cortessis VK, Kooperberg C, Cussenot O, Benhamou S, Prescott J, Porru S, Dinney CP, Malats N, Baris D, Purdue M, Jacobs EJ, Albanes D, Wang Z, Deng X, Chung CC, Tang W, Bas Bueno-de-Mesquita H, Trichopoulos D, Ljungberg B, Clavel-Chapelon F, Weiderpass E, Krogh V, Dorronsoro M, Travis R, Tjønneland A, Brenan P, Chang-Claude J, Riboli E, Conti D, Gago-Dominguez M, Stern MC, Pike MC, Van Den Berg D, Yuan JM, Hohensee C, Rodabough R, Cancel-Tassin G, Roupret M, Comperat E, Chen C, De Vivo I, Giovannucci E, Hunter DJ, Kraft P, Lindstrom S, Carta A, Pavanello S, Arici C, Mastrangelo G, Kamat AM, Lerner SP, Barton Grossman H, Lin J, Gu J, Pu X, Hutchinson A, Burdette L, Wheeler W, Kogevinas M, Tardón A, Serra C, Carrato A, García-Closas R, Lloreta J, Schwenn M, Karagas MR, Johnson A, Schned A, Armenti KR, Hosain GM, Andriole G Jr, Grubb R 3rd, Black A, Ryan Diver W, Gapstur SM, Weinstein SJ, Virtamo J, Haiman CA, Landi MT, Caporaso N, Fraumeni JF Jr, Vineis P, Wu X, Silverman DT, Chanock S, Rothman N. Genome-wide association study identifies multiple loci associated with bladder cancer risk. Hum Mol Genet. 2014;23:1387–1398. doi: 10.1093/hmg/ddt519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortessis VK, Yuan J, Van Den Berg D, Jiang X, Gago-Dominguez M, Stern MC, Castelao JE, Xiang Y, Gao Y, Pike MC, Conti DV. Risk of Urinary Bladder Cancer Is Associated with 8q24 Variant rs9642880[T] in Multiple Racial/Ethnic Groups: Results from the Los Angeles-Shanghai Case-Control Study. Cancer Epidem Biomar. 2010;19:3150–3156. doi: 10.1158/1055-9965.EPI-10-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DerSimonian R, Kacker R. Random-effects model for meta-analysis of clinical trials: an update. Contemp Clin Trials. 2007;28:105–114. doi: 10.1016/j.cct.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 27.Lavenu A, Pournin S, Babinet C, Morello D. The cis-acting elements known to regulate c-myc expression ex vivo are not sufficient for correct transcription in vivo. Oncogene. 1994;9:527–536. [PubMed] [Google Scholar]
- 28.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genomewide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 29.Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, Manak JR, Berman BP, Henderson BE, Frenkel B, Haiman CA, Freedman M, Tanay A, Coetzee GA. Functional enhancers at the genepoor 8q24 cancer-linked locus. PLoS Genet. 2009;5:e1000597. doi: 10.1371/journal.pgen.1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sotelo J, Esposito D, Duhagon MA, Banfield K, Mehalko J, Liao H, Stephens RM, Harris TJ, Munroe DJ, Wu X. Long-range enhancers on 8q24 regulate c-Myc. Proc Natl Acad Sci U S A. 2010;107:3001–3005. doi: 10.1073/pnas.0906067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adams JM, Harris AW, Pinkert CA, Corcoran LM, Alexander WS, Cory S, Palmiter RD, Brinster RL. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 32.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 33.Seo HK, Ahn KO, Jung NR, Shin JS, Park WS, Lee KH, Lee SJ, Jeong KC. Antitumor activity of the c-Myc inhibitor KSI-3716 in gemcitabineresistant bladder cancer. Oncotarget. 2014;5:326–337. doi: 10.18632/oncotarget.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, Stacey SN, Gudmundsson J, Jakobsdottir M, Bergthorsson JT, Sigurdsson A, Blondal T, Witjes JA, Vermeulen SH, Hulsbergenvan de Kaa CA, Swinkels DW, Ploeg M, Cornel EB, Vergunst H, Thorgeirsson TE, Gudbjartsson D, Gudjonsson SA, Thorleifsson G, Kristinsson KT, Mouy M, Snorradottir S, Placidi D, Campagna M, Arici C, Koppova K, Gurzau E, Rudnai P, Kellen E, Polidoro S, Guarrera S, Sacerdote C, Sanchez M, Saez B, Valdivia G, Ryk C, de Verdier P, Lindblom A, Golka K, Bishop DT, Knowles MA, Nikulasson S, Petursdottir V, Jonsson E, Geirsson G, Kristjansson B, Mayordomo JI, Steineck G, Porru S, Buntinx F, Zeegers MP, Fletcher T, Kumar R, Matullo G, Vineis P, Kiltie AE, Gulcher JR, Thorsteinsdottir U, Kong A, Rafnar T, Stefansson K. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munafo MR, Flint J. Meta-analysis of genetic association studies. Trends Genet. 2004;20:439–444. doi: 10.1016/j.tig.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]