Abstract
Activation of hepatic stellate cells (HSC) plays a pivotal role in the development of hepatic fibrosis. Transforming growth factor-β1 (TGF-β1) is considered to be the main stimuli factor responsible for the activation of HSC. Diosgenin is a steroidal saponin found in several plants including Solanum and Dioscorea species, and it inhibited high glucose-induced renal tubular fibrosis. However, the effects of diosgenin against hepatic fibrosis remain elusive. Therefore, in this study, we investigated the effects of diosgenin on TGF-β1-induced HSCs and elucidate the possible mechanism of its anti-fibrotic effect. Our results demonstrated that diosgenin inhibited TGF-β1-induced HSC proliferation, reduced the expression of collagen I and α-smooth muscle actin (α-SMA), as well as the expression of TGF-β receptor I (TGF-β RI) and II. Moreover, diosgenin suppressed TGF-β1-induced phosphorylation of Smad3 in HSCs. In conclusion, our data demonstrate that diosgenin inhibited HSC-T6 cell proliferation and activation, at least in part, via the TGF-β1/Smad signaling pathway. These results provide that diosgenin may have potential to treat liver fibrosis.
Keywords: Diosgenin, hepatic stellate cells (HSC), transforming growth factor-β1 (TGF-β1)
Introduction
Liver fibrosis is a major cause of morbidity and mortality from hepatic diseases. It is the result of wound-healing after repeat injury by alcohol, cholestasis, chronic hepatitis, or drugs, and is characterized by the excessive accumulation of extracellular matrix (ECM), which is mainly composed of collagens [1]. Hepatic stellate cells (HSCs) are the most relevant cell type for the development of liver fibrosis, and the activation of HSCs plays an important role during the initiation and development of liver fibrosis [2]. In the healthy liver, HSCs have a quiescent phenotype, but, after liver injury, these quiescent HSCs are exposed to profibrogenic factors, and undergo a process of activation to a myofibroblastic phenotype, finally resulting in the excess production and deposition of ECM components [3]. Therefore, the inactivation of HSCs is the main approach for preventing the progression of liver fibrosis [4]. However, the therapeutic drugs of liver fibrosis are still insufficient. Thus, there is a need to find new agents which can reverse HSC activation.
Diosgenin is a steroidal saponin found in several plants including Solanum and Dioscorea species. A growing body of evidence indicates that diosgenin possesses important pharmacological properties, such as anti-inflammatory [5], anti-atherosclerosis [6], anti-tumor [7] and antioxidant activities [8]. Most recently, one study showed that diosgenin inhibited high glucose-induced renal tubular fibrosis [9]. However, there are no studies focusing on the effects of diosgenin against hepatic fibrosis remain elusive. Therefore, in this study, we investigated the effects of diosgenin on TGF-β1-induced HSCs and elucidate the possible mechanism of its anti-fibrotic effect.
Materials and methods
Cell culture
The human HSC cell line, HSC-T6, was purchased from the American Type Culture Collection (Manassas, VA, USA). HSC-T6 cells were grown in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere.
MTT assay
The cell proliferation assay was carried out using the 3-[4,5-dimethylthiazol-2-ly]-2,5-diphenyltetrazolium bromide (MTT; Sigma-Aldrich, St. Louis, MO, USA) method. The cells were seeded in a 96-well (1×104 cells/well) and treated with TGF-β1 or TGF-β1 + diosgenin (25, 50 and 100 μM) for 24 h. Then MTT was added to the cells at a final concentration of 0.5 mg/ml before the end of the experiment and incubated at 37°C for 4 h. The supernatant was removed, and the crystals were dissolved in 150 μl dimethyl sulfoxide. The absorbance was measured at 570 nm in a microplate reader (Thermo Lab systems, Waltham, MA, USA).
Caspase 3 activity assay
HSC-T6 cells cultured in 100-mm-diameter dishes were harvested and pelleted by centrifugation. The medium supernatant was discarded, and the cell pellet was washed in 1 mL of ice-cooled PBS. Caspase 3 (DEVDase) activity was determined by a colorimetic caspase 3 activity kit (Abcam) following the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Trizol Reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Then, 2 μg of total RNA was transcribed to first-strand cDNA using TaqMan reverse transcription reagents (Applied Biosystems, Foster City, CA). The following primers were used: collagen I, 5’-TGACTGGAAGAGCGGAGAGTACT-3’ (sense), 5’-GCTGTGGGCATATTGCACAA-3’ (antisense); α-SMA, CCGAGATCTCACCGACTACC (sense), 5’-TCCAGAGCGACATAGCACAG-3’ (antisense); and β-actin 5’-CCGTGAAAAGATGACCCAGATC-3’ (sense), 5’-CACAGCCTGGATGGCTACGT-3’ (antisense). These primers were all synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The PCR procedure was as followed: 95°C for 3 min, followed by 40 cycles of 94°C for 30 s, 59°C for 20 s, and 72°C for 40 s, and finally a single cycle at 72°C for 5 min. For relative quantification, the levels of individual gene mRNA transcripts were firstly normalized to the control β-actin. Subsequently, the differential expression of these genes was analyzed according to the 2-ΔΔCt method.
Western blot
HTC-6 cells were washed with ice-cold PBS and lysed with RIPA lysis buffer (Beyotime, China). The lysates were sonicated for 5 s on ice and centrifuged at 10,000× g for 5 min. The supernatants were collected and the protein concentration was determined using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA). Protein (30 μg) was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membrane was incubated with the primary antibody for collagen α type I, α-smooth muscle actin (α-SMA), TGF-β RI, TGF-β RII, phospho-Smad 2, Smad 2, phospho-Smad3, Smad3, GAPDH (Santa Cruz Biotechnology, CA, USA) at 4°C overnight. After washing with TBST, blots were then incubated with horseradish peroxidase-linked secondary antibodies (Invitrogen, Carlsbad, CA, USA) at room temperature for 1 h. The specific protein bands were developed using a chemiluminescent substrate and imaged using a gel scanner. β-actin was used as the internal control.
Statistical analysis
Results were presented as mean ± S.D. Statistical analysis was performed using One-way ANOVA analysis. Statistical significance was determined at the level of P<0.05.
Results
Diosgenin treatment suppressed TGF-β1-induced HSC proliferation
We first examined the effect of diosgenin on TGF-β1-induced HSC proliferation. As shown in Figure 1A, treatment with diosgenin alone did not obviously affect cell viability. In addition, TGF-β1 significantly reduced the cell viability, however, pretreatment with diosgenin reversed this effect, exhibiting a dose-dependent manner (Figure 1B).
Figure 1.
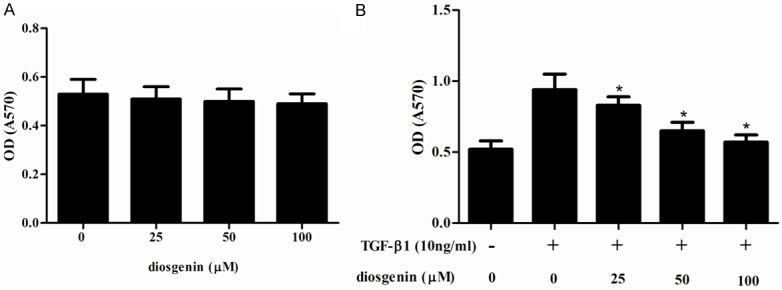
Diosgenin inhibits TGF-β1-induced proliferation in HSC-T6 cells. HSC-T6 cells were stimulated with TGF-β1 after treatment with various concentrations of diosgenin for 24 h. The cell proliferation was determined with MTT assay. Values were expressed as mean ± SD of three independent experiments. *P<0.05 compared with the control group.
Effect of diosgenin on TGF-β1-induced HSC apoptosis
To investigate the effect of diosgenin on TGF-β1-induced HSC apoptosis, we measured caspase-3 activity using colorimetric analysis. As shown in Figure 2, diosgenin treatment has no affect caspase-3 activity. These results suggest that the anti-proliferative effect of diosgenin was not mediated by HSC apoptosis.
Figure 2.
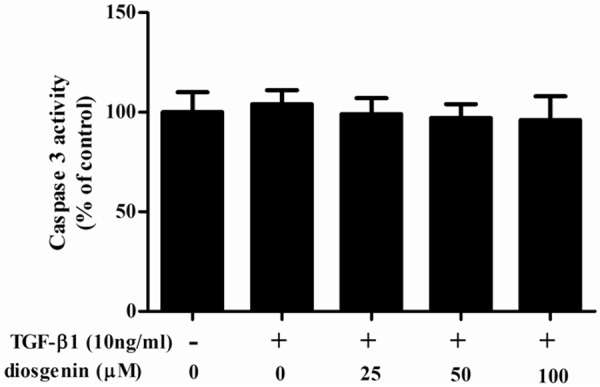
Effect of diosgenin on TGF-β1-induced apoptosis in HSC-T6 cells. HSC-T6 cells were stimulated with TGF-β1 after treatment with various concentrations of diosgenin for 24 h. Caspase 3 activity was measured using Caspase 3 colorimetric assay kits. Values were expressed as mean ± SD of three independent experiments.
Diosgenin treatment suppressed TGF-β1-induced production of type I collagen and α-SMA
We examined whether diosgenin is able to inhibit the fibrotic effects of TGF-β1 on ECM expression in HSCs. As shown in Figure 3, TGF-β1 significantly increased the protein expression of induced a considerable increase in collagen I and α-SMA. However, diosgenin reserved TGF-β1-induced collagen I and α-SMA protein level in a dose dependent manner.
Figure 3.
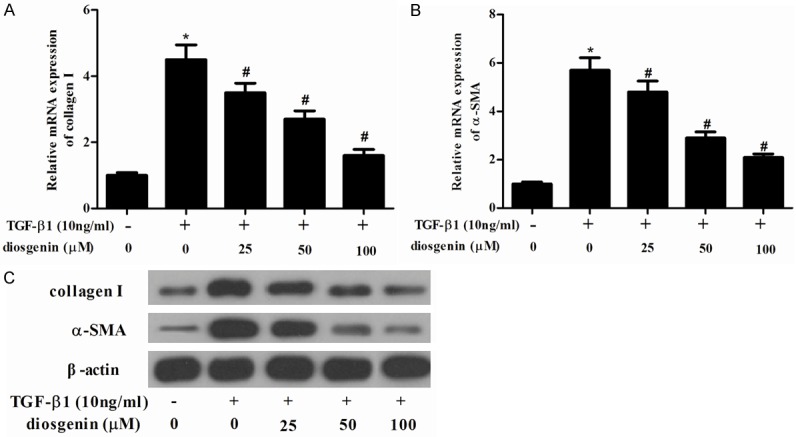
Diosgenin inhibits the expression of α-SMA and collagen type I in TGF-β1-induced HSC-T6 cells. HSC-T6 cells were stimulated with TGF-β1 after treatment with various concentrations of diosgenin for 24 h. RT-PCR was performed to detect the mRNA expression levels of collagen type I (A) α-SMA (B); (C) Western blot analysis was performed to detect the protein expression levels of α-SMA and collagen type I using the antibodies indicated. Values were expressed as mean ± SD of three independent experiments. *P<0.05 compared with the control group. #P<0.05 compared with the TGF-β1 group.
Diosgenin inhibited TGF-β RI and II expression in hepatic stellate cells
Because TGF-β/Smad signaling pathways perform a significant role in liver fibrosis and TGF-β receptor binding initiates the signaling cascade, therefore, we examined the effect of diosgenin on TGF-β RI and II expression levels in TGF-β1-stimulated HSC-T6 cells. As shown in Figure 4, TGF-β1 significantly increased TGF-β RI and TGF-β RII expression in HSC-T6 cells. However, diosgenin treatments dramatically suppressed the TGF-β1-enhanced TGF-β RI and TGF-β RII expression in HSC-T6 cells.
Figure 4.
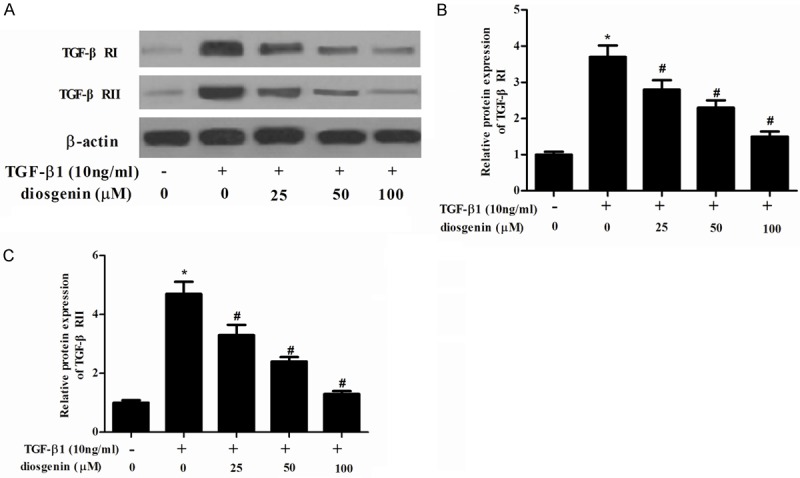
Diosgenin inhibits the expression of TGF-β receptor I and II in TGF-β1-induced HSC-T6 cells. (A) HSC-T6 cells were stimulated with TGF-β1 after treatment with various concentrations of diosgenin for 24 h. Western blot analysis was performed to detect the protein expression levels of TGF-β RI and II using the antibodies indicated. The relative protein expression levels of TGF-β RI (B) and II (C) were quantified using Image-Pro Plus 6.0 software and normalized to β-actin. Values were expressed as mean ± SD of three independent experiments. *P<0.05 compared with the control group. #P<0.05 compared with the TGF-β1 group.
Diosgenin inhibited TGF-β1-induced phosphorylation of Smad3 in hepatic stellate cells
The TGF-β1-mediated signaling pathway depends on the phosphorylation of Smad 2/3. To further evaluate understand the molecular mechanisms responsible for the inhibition of HSCs activation, the protein levels of Smad 2/3 were analyzed. As shown in Figure 5, TGF-β1 treatment stimulated phosphorylation of Smad3, however, diosgenin prevented TGF-β1-induced phosphorylation of Smad3 in HSC-T6 cells.
Figure 5.

Diosgenin attenuates TGF-β1-stimulated phosphorylation of Smad3 in HSC-T6 cells. Serum-starved HSC-T6 cells were treated with diosgenin in the presence or absence of TGF-β1 (5 ng/ml). (A) Phosphorylation level of Smad3 was analyzed by immunoblotting with specific antibodies. The relative protein expression levels of p-Smad3 (B) and Smad3 (C) were quantified using Image-Pro Plus 6.0 software and normalized to β-actin. Values were expressed as mean ± SD of three independent experiments. *P<0.05 compared with the control group. #P<0.05 compared with the TGF-β1 group.
Discussion
Our results demonstrated that diosgenin inhibited TGF-β1-induced HSC proliferation, reduced the expression of collagen I and α-SMA, as well as the expression of TGF-β RI and TGF-β RII. Moreover, diosgenin suppressed TGF-β1-induced phosphorylation of Smad3 in HSCs.
The induction of proliferation is an early step after HSC activation and is stimulated by a variety of cytokines. Among many cytokines, platelet-derived growth factor (PDGF) and TGF-β1 are considered the most prominent mediators of this process. PDGF is one of the most potent mitogen for activated HSC [10], while TGF-β1 is widely accepted as the strongest stimulus for the transdifferentiation of HSCs [11]. In this study, we used TGFβ1 to induce HSC activation. Our data showed that TGF-β1 significantly induced HSC proliferation. These findings are in agreement with earlier reports that TGF-β1 promoted HSC proliferation. In addition, we found that diosgenin inhibited TGF-β1-induced HSC proliferation, and diosgenin had no significant effect on cell apoptosis. These data suggested that diosgenin inhibited TGF-β1-induced HSC proliferation without affecting cell apoptosis.
Activated HSCs are the principal cell type promoting synthesis and deposition of ECM proteins. There is extensive evidence demonstrating that TGF-β1 plays an essential role in modulating ECM expression [12-14]. Previous studies showed that TGF-β1 upregulated expression of α-SMA and collagen I in human HSCs [15-17]. In line with these reports, in this study, we found that TGF-β1 increased the expression of collagen I and α-SMA, while, diosgenin inhibited TGF-β1-induced the expression of collagen I and α-SMA in HSC-T6 cells. These data suggested that diosgenin inhibited TGF-β1-induced HSC activation by reducing the expression of collagen I and α-SMA.
A growing body of evidence demonstrated that the TGF-β1/Smad signaling pathway is a key mediator of progressive liver fibrosis [18-20]. TGF-β signals through a heteromeric receptor complex of type II and type I receptor serine-threonine kinases, which activates the downstream Smad signal pathway. After TGFβ binding to the receptor complex, Smad 2/3 is phosphorylated and binds with SMAD 4 to form multimers, then activated R-Smads translocate to the nucleus and induce transcriptional modulation of target genes, including ECM proteins [21]. So, inhibition of TGF-β1/Smad signaling may be an important therapeutic approach for liver fibrosis. Liu et al. reported that Smad3 specific inhibitor, Naringenin, decreases the expression of ECM induced by TGF-β1 in cultured rat HSCs [22]. Yang et al. reported that astaxanthin decreased TGFβ1-induced α-SMA and procollagen type 1, alpha 1 (Col1A1) mRNA as well as α-SMA protein levels. It also attenuated TGFβ1-induced Smad3 phosphorylation and nuclear translocation with a concomitant inhibition of Smad3, Smad 7, Tβ RI and Tβ RII expression [23]. In agreement with previous data, in the present study, we found that TGF-β1 significantly increased Tβ RI and Tβ RII expression, as well as the phosphorylation level of Smad3 in HSC-T6 cells; whereas co-treatment of diosgenin eliminated these changes. These results suggest that diosgenin attenuated liver fibrosis by inhibiting the TGF-β1/Smad signaling pathway.
In conclusion, our data demonstrate that diosgenin inhibited HSC-T6 cell proliferation and activation, at least in part, via the TGF-β1/Smad signaling pathway. These results provide that diosgenin may have potential to treat liver fibrosis.
Disclosure of conflict of interest
None.
References
- 1.Adrian JE, Kamps JA, Scherphof GL, Meijer DK, Reker-Smit C, Terpstra P, Poelstra K. A novel lipid-based drug carrier targeted to the non-parenchymal cells, including hepatic stellate cells, in the fibrotic livers of bile duct ligated rats. Biochim Biophys Acta. 2007;1768:1430–1439. doi: 10.1016/j.bbamem.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 2.Tsukada S, Parsons CJ, Rippe RA. Mechanisms of liver fibrosis. Clin Chim Acta. 2006;364:33–60. doi: 10.1016/j.cca.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Rockey DC. The cell and molecular biology of hepatic fibrogenesis: clinical and therapeutic implications. Clin Liver Dis. 2000;4:319–355. doi: 10.1016/s1089-3261(05)70113-6. [DOI] [PubMed] [Google Scholar]
- 4.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao M, Chen L, Yu H, Sun Q, Kou J, Yu B. Diosgenin down-regulates NF-κB p65/p50 and p38MAPK pathways and attenuates acute lung injury induced by lipopolysaccharide in mice. Int Immunopharmacol. 2013;15:240–245. doi: 10.1016/j.intimp.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Liu K, Zhao W, Gao X, Huang F, Kou J, Liu B. Diosgenin ameliorates palmitate-induced endothelial dysfunction and insulin resistance via blocking IKKβ and IRS-1 pathways. Atherosclerosis. 2012;223:350–358. doi: 10.1016/j.atherosclerosis.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 7.Rahmati-Yamchi M, Ghareghomi S, Haddadchi G, Milani M, Aghazadeh M, Daroushnejad H. Fenugreek extract diosgenin and pure diosgenin inhibit the hTERT gene expression in A549 lung cancer cell line. Mol Biol Rep. 2014;41:6247–6252. doi: 10.1007/s11033-014-3505-y. [DOI] [PubMed] [Google Scholar]
- 8.Kongkaneramit L, Witoonsaridsilp W, Peungvicha P, Ingkaninan K, Waranuch N, Sarisuta N. Antioxidant activity and antiapoptotic effect of Asparagus racemosus root extracts in human lung epithelial H460 cells. Exp Ther Med. 2011;2:143–148. doi: 10.3892/etm.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang WC, Liu SF, Chang WT, Shiue YL, Hsieh PF, Hung TJ, Hung CY, Hung YJ, Chen MF, Yang YL. The effects of diosgenin in the Regulation of renal proximal tubular fibrosis. Exp Cell Res. 2014;323:255–262. doi: 10.1016/j.yexcr.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 10.Pinzani M. PDGF and signal transduction in hepatic stellate cells. Front Biosci. 2002;7:d1720–d1726. doi: 10.2741/A875. [DOI] [PubMed] [Google Scholar]
- 11.Gressner AM, Weiskirchen R, Breitkopf K, Dooley S. Roles of TGF-beta in hepatic fibrosis. Front Biosci. 2002;7:d793–d807. doi: 10.2741/A812. [DOI] [PubMed] [Google Scholar]
- 12.Arnott JA, Nuglozeh E, Rico MC, Arango-Hisijara I, Odgren PR, Safadi FF, Popoff SN. Connective tissue growth factor (CTGF/CCN2) is a downstream mediator for TGF-β1-induced extracellular matrix production in osteoblasts. J Cell Physiol. 2007;210:843–852. doi: 10.1002/jcp.20917. [DOI] [PubMed] [Google Scholar]
- 13.Miralles F, Battelino T, Czernichow P, Scharfmann R. TGF-β plays a key role in morphogenesis of the pancreatic islets of Langerhans by controlling the activity of the matrix metalloproteinase MMP-2. J Cell Biol. 1998;143:827–836. doi: 10.1083/jcb.143.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang HR, Lee CG, Homer RJ, Elias JA. Semaphorin 7A plays a critical role in TGF-β1-induced pulmonary fibrosis. J Exp Med. 2007;204:1083–1093. doi: 10.1084/jem.20061273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman SL. Cytokines and fibrogenesis. Semin Liver Dis. 1999;19:129–40. doi: 10.1055/s-2007-1007105. [DOI] [PubMed] [Google Scholar]
- 16.Wiercinska E, Wickert L, Denecke B, Said HM, Hamzavi J, Gressner AM, Thorikay M, ten Dijke P, Mertens PR, Breitkopf K, Dooley S. Id1 is a critical mediator in TGF-β-induced transdifferentiation of rat hepatic stellate cells. Hepatology. 2006;43:1032–1041. doi: 10.1002/hep.21135. [DOI] [PubMed] [Google Scholar]
- 17.Lindert S, Wickert L, Sawitza I, Wiercinska E, Gressner AM, Dooley S, Breitkopf K. Transdifferentiation-dependent expression of α-SMA in hepatic stellate cells does not involve TGF-β pathways leading to coinduction of collagen type I and thrombospondin-2. Matrix Biol. 2005;24:198–207. doi: 10.1016/j.matbio.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, Zheng J, Wu L, Shi M, Zhang H, Wang X, Xia N, Wang D, Liu X, Yao L. NDRG2 ameliorates hepatic fibrosis by inhibiting the TGF-β1/Smad pathway and altering the MMP2/TIMP2 ratio in rats. PLoS One. 2011;6:e27710. doi: 10.1371/journal.pone.0027710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zong L, Qu Y, Xu MY, Dong YW, Lu LG. 18α-glycyrrhetinic acid down-regulates expression of type I and III collagen via TGF-Β1/Smad signaling pathway in human and rat hepatic stellate cells. Int J Med Sci. 2012;9:370–379. doi: 10.7150/ijms.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yao QY, Xu BL, Wang JY, Liu HC, Zhang SC, Tu CT. Inhibition by curcumin of multiple sites of the transforming growth factor-beta1 signalling pathway ameliorates the progression of liver fibrosis induced by carbon tetrachloride in rats. BMC Complement Altern Med. 2012;12:156–166. doi: 10.1186/1472-6882-12-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X, Wang W, Hu H, Tang N, Zhang C, Liang W, Wang M. Smad3 specific inhibitor, naringenin, decreases the expression of extracellular matrix induced by TGF-β1 in cultured rat hepatic stellate cells. Pharm Res. 2006;23:82–89. doi: 10.1007/s11095-005-9043-5. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y, Kim B, Park YK, Koo SI, Lee JY. Astaxanthin prevents TGFβ1-induced pro-fibrogenic gene expression by inhibiting Smad3 activation in hepatic stellate cells. Biochim Biophys Acta. 2015;1850:178–185. doi: 10.1016/j.bbagen.2014.10.014. [DOI] [PubMed] [Google Scholar]


