Abstract
Objective: A number of studies have investigated the relationship between the PON1 gene polymorphisms and breast cancer risk, but the conclusions are not consistent. In this paper, a meta-analysis was conducted to explore the possible reasons for these inconsistencies, expecting to further clarify the correlation between PON1 gene polymorphisms and breast cancer risk. Methods: After searches in the database such as MEDLINE, EBSCO, ProQuest, Google Scholar, High-Wire, SID (Scientific Information Database) and PubMed, 7 literatures were collected. RevMan 5.2 software was used to perform the meta-analysis. Random-effects or fixed-effects model was used to analyze the odds ratio (OR) and 95% confidence intervals. Results: The analysis of L55M single nucleotide polymorphisms (SNPs) showed that M allele frequency was positively correlated with the incidence risk of breast cancer (OR=1.34, 95% CI: 1.03-1.74). While we did not found Q192R polymorphism associated with the risk of breast cancer (OR=1.0, 95% CI: 0.71-1.42). Conclusion: For PON1 gene, the frequencies of M allele were associated with the incidence risk of breast cancer.
Keywords: Paraoxonase, genetics, meta-analysis
Introduction
Paraoxonase (PON) is a class of aromatic esterase catalyzing the hydrolysis of phosphate bonds, which can degrade organic phosphate, aromatic carboxylic acid esters and carbamates [1]. Since the most common substrate of the enzyme for activity test is paraoxon, it is named as paraoxonase. Recent studies have found that it not only plays an important role in the detoxification of organophosphate compounds, but also closely relates with the onset of atherosclerosis by inhibiting the oxidation of LDL [2,3]. PON gene is located on human chromosome 7, mainly including PON1, PON2 and PON3. The present studies [4,5] showed that in the coding region of PON1 gene, there were two common functional SNPs, which were L55M (rs854560) and Q192R (rs662). L55M polymorphism can affect the concentration of paraoxonase and Q192R polymorphism can affect the ability of enzyme to hydrolyze lipid peroxides. Given the important role of PON1 in tumor-induced oxidative stress reactions, some scholars believe that PON1 polymorphisms may be associated with breast cancer risk.
In the past decade, some studies have investigated the relationship between PON1 polymorphisms and breast cancer risk [6-12], but the results of these studies were not consistent. This inconsistency may be due to the small influence of PON1 gene polymorphisms on breast cancer risk and/or the interference of some false-positive results. Therefore, we conducted a meta-analysis of some published literatures, trying to explore the possible reasons for this inconsistency, and expecting to further clarify the correlation between PON1 gene polymorphisms and breast cancer risk.
Methods
The literatures on the correlation between PON1 gene polymorphisms and breast cancer risk published before January 2015 were searched on the databases of MEDLINE, EBSCO, ProQuest, Google Scholar, High-Wire, SID (Scientific Information Database) and PubMed, with “Breast cancer”, “polymorphisms”, “L55M”, “Q192R”, “paraoxonase” and “PON1” as keywords. Meanwhile, the references in the obtained literatures were analyzed for further follow-up analysis of other related researches.
In this study, only English and Chinese literatures were included and a total of 7 literatures complying with the requirements were retrieved, which were shown in Table 1.
Table 1.
The characteristics of the included studies
| Control subjects | Breast cancer Patients | Control subjects | Breast cancer Patients | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||||
| Authors | Publication year | Country | Case/control | LL | LM | MM | LL | LM | MM | QR | RR | QR | RR | HWE | ||
| Stevens et al. | 2006 | USA | 483/483 | 202 | 223 | 58 | 176 | 230 | 77 | 238 | 198 | 47 | 259 | 182 | 42 | |
| Antognelli et al. | 2009 | Italy | 547/544 | 188 | 125 | 231 | 107 | 115 | 325 | 340 | 152 | 52 | 484 | 50 | 13 | |
| Naidu et al. | 2010 | Malaysia | 387/252 | 126 | 109 | 17 | 159 | 178 | 50 | 115 | 115 | 22 | 200 | 158 | 29 | |
| Hussein et al. | 2011 | Egypt | 200/200 | 81 | 65 | 54 | 70 | 62 | 68 | 46 | 42 | 12 | 51 | 41 | 8 | |
| Agachan et al. | 2006 | Turkey | 33/52 | - | - | - | - | - | - | 17 | 29 | 6 | 17 | 4 | 12 | |
| Gallicchio et al. | 2007 | USA | 58/904 | - | - | - | - | - | - | 469 | 353 | 82 | 38 | 15 | 5 | |
| DeRoo et al. | 2014 | USA | 294/646 | HR=0.96 (95% CI: 0.78-1.20) | HR=0.97 (95% CI: 0.77-1.22) | |||||||||||
Data extraction
Standardized forms were used for data extraction of all documents, including the author, publication time, country, experimental design, information of the control group (data from the non-breast cancer patients in the hospital or healthy individuals), race, the specific number of cases of different PON1 genotypes in experimental and control groups.
Literature inclusion criteria
The included literatures for meta-analysis must meet the following criteria: (1) The included studies must be case-control studies about PON1 polymorphisms and susceptibility of breast cancer. (2) The distribution of genotype frequency, or evaluable indicators, such as odds ratios (odd ratios, OR) and 95% confidence intervals (confidential interval, CI) must be specific or can be calculated. (3) The frequency of population genotype must meet the Hardy-Weinberg equilibrium in the control group. (4) If there are duplicated publication or data, we selected the articles with the largest sample size or the latest published literatures.
Exclusion criteria
(1) Review articles. (2) The reports repeated the same population. (3) The frequency distribution of gene loci cannot be obtained. (4) Diagnostic criteria were different from other studies. (5) Genotype distribution in the control group did not meet the Hardy-Weinberg (HW) equilibrium.
Meta-analysis
Meta-analysis of all literatures was performed utilized RevMan 5.2 software. Chi-square test was used for Hardy-Weinberg equilibrium (HWE) analysis of the distribution of PON1 genotypes in control groups. Taking into account the differences that may exist among different studies, Cochran’s Q test was used to analyze the differences among various experiments, and P<0.05 indicated that there was a significant difference; and based on the differences among studies, the random-effects and fixed-effects models were used to analyze the correlation between gene polymorphisms and breast cancer risk. Fixed-effects model assumes that there were no significant differences among various results; the random-effects model is applicable to the studies with significant differences. Statistical analysis of random-effects and fixed-effects model specifically referred to the relevant literatures [13,14].
Results
Baseline characteristics of included studies
As shown in Figure 1, we retrieved 46 literatures, and excluded 39 literatures for duplicated publication and other reason. There are 7 literatures [6-11,14] were selected to perform meta-analysis finally. These 7 studies included a total of 4893 cases (including 1902 cases of breast cancer patients and 2991 cases of healthy control subjects); all the patients were consistent to the diagnostic criteria of breast cancer. Peripheral-blood specimens were used during the investigations of polymorphism in all the studies. All the genotypes distributions were in line with Hardy-Weinberg equilibrium (HWE). The characteristics of included studies were shown in Table 1.
Figure 1.
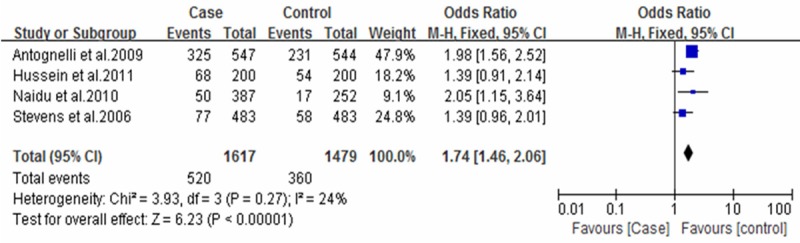
Forest plot of breast cancer and L55M polymorphism (MM vs. LL+LM).
Meta-analysis results
For the SNPs analysis of L55M, five documents were totally included, and Q test showed no significant differences among the studies. Correlation analysis showed that M allele frequency was positively correlated with the incidence risk of breast cancer. MM genotype frequency was higher in the case group than that in the control group (OR=1.74; 95% CI: 1.46-2.06; Figure 1). Also, M allele frequency was positively correlated with the incidence risk of breast cancer (OR=1.34, 95% CI: 1.03-1.74, Figure 2). Q192R SNPs analysis showed that neither RR genotype nor R allele frequency was correlated with the incidence risk of breast cancer (Figures 3 and 4).
Figure 2.
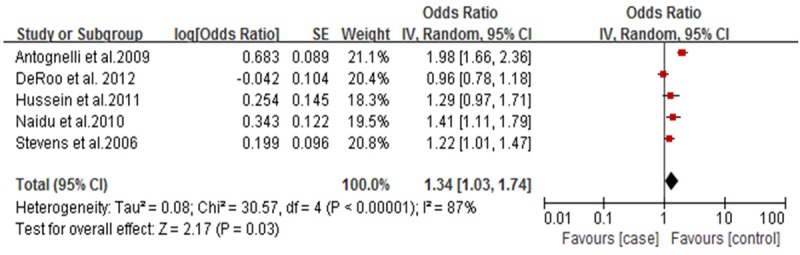
Forest plot of breast cancer and L55M polymorphism (M allele vs. M allele).
Figure 3.
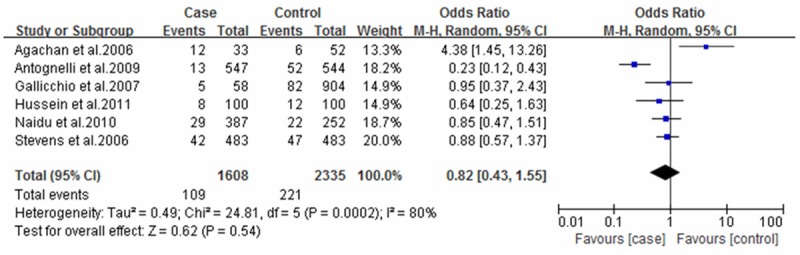
Forest plot of breast cancer and Q192R polymorphism (RR vs. QR+QQ).
Figure 4.
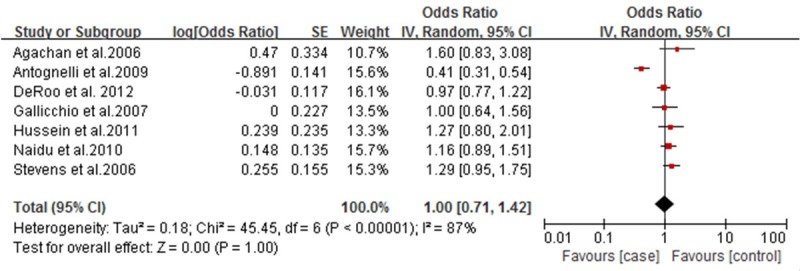
Forest plot of breast cancer and Q192R polymorphism (R allele vs. Q allele).
Publication bias
We strengthened the confidence level in the results by conducting a publication bias analysis. The shape of the funnel plots was symmetrical, suggesting no evidence of publication bias among the studies (Figure 5). The Egger’s test, performed to provide statistical evidence of funnel plot asymmetry, indicated a lack of publication bias in the current meta-analysis (P>0.05).
Figure 5.
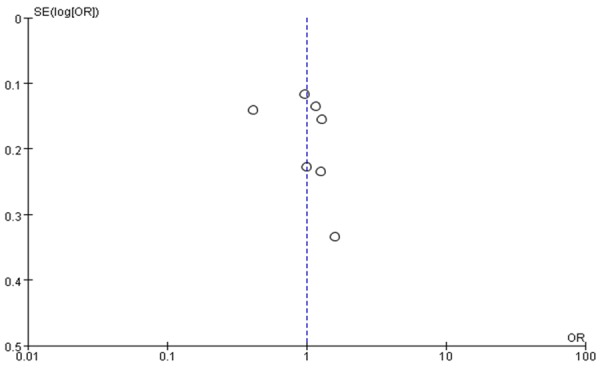
Funnel plot for publication bias tests.
Sensitivity analysis
Sensitivity analysis was used in this study. The statistical results show that there is no single study can significantly influence the results of the existing analysis.
Discussion
Through the meta-analysis of the published literatures, we found that the 55M allele but not 192Q allele in PON1 gene waspositively correlated with the risk of breast cancer. Previous studies have shown [15] that, the changes of 55M allele may reduce the stability of the PON1 enzyme, thereby reducing the serum concentration of PON1 enzyme and ultimately affecting the enzyme activity. The enzyme activity of LM genotype was between that of LL gentype and MM genotype. The studies also showed that the changes of 192R allele promoted the production of PON1 enzyme, thereby enhancing its response to oxidative stress and lipid peroxide reaction which were induced by tumor [16]. Therefore, individuals with MM genotype and QQ genotype were easily influenced by some external factors (such as food, etc.), who were more susceptible to breast cancer, due to the reduced concentration and activity of PON1 enzyme. However, in the present study, we did not find Q192R polymorphism associated with breast cancer. This result was different from the previous two meta-analysis studies [17,18]. Saadat et al.’s meta-analysis included six studies and found PON1 55M and 192Q alleles are associated with a higher risk of breast cancer. Liu et al. found PON1 L55M but not Q192R polymorphism increased breast cancer risk. Our result was in line with the Liu et al’s.
Previous studies have shown thatthere were differences among races in the correlation between polymorphisms and cancer risk. For example, glutathione-S-transferase T1 (GST T1) polymorphisms were associated with the increased risk of gastric cancer, which was limited to Caucasians; the analysis of the XRCC1 gene polymorphisms showed that Arg399Gln polymorphism loci were associated with lung cancer risk, which was only foundin the Asian population and not suitable for the people in Western countries.
In conclusion, the present study indicated that L55M polymorohism was associated with breast cancer. However, due to the small number of the included studies in the meta-analysis, we cannot identify the differences among various ethnics in the correlation between PON1 polymorphisms and breast cancer risk, which required a combination of more research in the future to clarify and confirm.
Acknowledgements
This work was supported by the Natural Science Foundation of Chongqing (CSTC, 2011BB5032).
Disclosure of conflict of interest
None.
References
- 1.Milnerowicz H, Kowalska K, Socha E. Paraoxonase activity as a marker of exposure to xenobiotics in tobacco smoke. Int J Toxicol. 2015;34:224–32. doi: 10.1177/1091581815584624. [DOI] [PubMed] [Google Scholar]
- 2.Costa LG, Li WF, Richter RJ, Shih DM, Lusis A, Furlong CE. The role of paraoxonase (PON1) in the detoxication of organophosphates and its human polymorphism. Chem Biol Interact. 1999;119-120:429–38. doi: 10.1016/s0009-2797(99)00055-1. [DOI] [PubMed] [Google Scholar]
- 3.Kowalska K, Socha E, Milnerowicz H. Review: The role of paraoxonase in cardiovascular diseases. Ann Clin Lab Sci. 2015;45:226–33. [PubMed] [Google Scholar]
- 4.Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–6. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- 5.Leviev I, Deakin S, James RW. Decreased stability of the M54 isoform of paraoxonase as a contributory factor to variations in human serum paraoxonase concentrations. J Lipid Res. 2001;42:528–35. [PubMed] [Google Scholar]
- 6.Hussein YM, Gharib AF, Etewa RL, ElSawy WH. Association of L55M and Q192R polymorphisms in paraoxonase 1 (PON1) gene with breast cancer risk and their clinical significance. Mol Cell Biochem. 2011;351:117–23. doi: 10.1007/s11010-011-0718-4. [DOI] [PubMed] [Google Scholar]
- 7.Naidu R, Har YC, Taib NA. Genetic Polymorphisms of Paraoxonase 1 (PON1) Gene: Association between L55M or Q192R with Breast Cancer Risk and Clinico-Pathological Parameters. Pathol Oncol Res. 2010 [Epub ahead of print] [Google Scholar]
- 8.Antognelli C, Del Buono C, Ludovini V, Gori S, Talesa VN, Crinò L, Barberini F, Rulli A. CYP17, GSTP1, PON1 and GLO1 gene polymorphisms as risk factors for breast cancer: An Italian case-control study. BMC Cancer. 2009;9:115. doi: 10.1186/1471-2407-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallicchio L, McSorley MA, Newschaffer CJ, Huang HY, Thuita LW, Hoffman SC, Helzlsouer KJ. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detect Prev. 2007;31:95–101. doi: 10.1016/j.cdp.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Stevens VL, Rodriguez C, Pavluck AL, Thun MJ, Calle EE. Association of polymorphisms in the paraoxonase 1 gene with breast cancer incidence in the CPS-II Nutrition Cohort. Cancer Epidemiol Biomarkers Prev. 2006;15:1226–8. doi: 10.1158/1055-9965.EPI-05-0930. [DOI] [PubMed] [Google Scholar]
- 11.Deroo LA, Bolick SC, Xu Z, Umbach DM, Shore D, Weinberg CR, Sandler DP, Taylor JA. Global DNA methylation and one-carbon metabolism gene polymorphisms and the risk of breast cancer in the Sister Study. Carcinogenesis. 2014;35:333–8. doi: 10.1093/carcin/bgt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agachan B, Yaylım I, Ergen HA, Arıkan S, Kucucuk S, Yılmaz H. Is paraoxonase 1 192 BB genotype risk factor for breast cancer? Adv Mol Med. 2006;2:37–40. [Google Scholar]
- 13.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 15.Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat Genet. 1993;3:73–6. doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- 16.Leviev I, Deakin S, James RW. Decreased stability of the M54 isoform of paraoxonase as a contributory factor to variations in human serum paraoxonase concentrations. J Lipid Res. 2001;42:528–35. [PubMed] [Google Scholar]
- 17.Liu C, Liu L. Polymorphisms in three obesityrelated genes (LEP, LEPR, and PON1) and breast cancer risk: Ameta-analysis. Tumour Biol. 2011;32:1233–40. doi: 10.1007/s13277-011-0227-9. [DOI] [PubMed] [Google Scholar]
- 18.Saadat M. Paraoxonase 1 genetic polymorphisms and susceptibility to breast cancer: A meta-analysis. Cancer Epidemiol. 2012;36:e101–3. doi: 10.1016/j.canep.2011.10.015. [DOI] [PubMed] [Google Scholar]


