Abstract
Objective: The TGFB1 gene is among the most studied genes in high myopia due to its role in scleral remodeling. But reported findings of association on TGFB1 and high myopia are inconsistent. This present study is to evaluate the association of TGFB1 polymorphisms and high myopia. Methods: A comprehensive literature search was conducted on studies published up to April 5, 2015. Summary odds ratios (ORs) and 95% confidence intervals were analyzed. Heterogeneity across studies was evaluated by Cochran Q statistic test and the I2 index. Sensitivity analyses were conducted by the approach of one-study remove to assess the influence of single study on the combined effect. Results: Eight studies were included in this study for meta-analysis. Rs1982073 was associated with high myopia in dominant model (OR=1.64; 95% CI=1.04~2.58; P<0.05), heterozygous model (OR=1.54; 95% CI=1.02~2.33; P<0.05), homozygous model (OR=1.90; 95% CI=1.01~3.55; P=0.05) and allelic model (OR=1.36; 95% CI=1.01~1.84; P=0.05). However, there was no statistical significance when Bonferroni correction was considered. Rs4803455 was associated with high myopia in recessive model (OR=0.40; 95% CI=0.25~0.64; P<0.01) and homozygous model (OR=0.42; 95% CI=0.26~0.68; P<0.01). Rs1800469 was associated with high myopia in allelic model (OR=0.78; 95% CI=0.64~0.96; P<0.05). And the associations can withstand Bonferroni correction in models mentioned above when referring to rs4803455 (P<0.01) and rs1800469 (P<0.05). Conclusions: Meta-analysis of existing data revealed a suggestive association of TGFB1 rs1982073 and rs4803455 with high myopia.
Keywords: Transforming growth factor beta1, single nucleotide polymorphism, high myopia
Introduction
Myopia is a refractive status which focuses image in front of the retina and results in blurred distant vision. Although myopia is often regarded as a benign disorder that can be corrected with optical modalities, the advanced form of myopia, that is high myopia, can greatly affect life quality because of its vision-threatening complications such as glaucoma, macular degeneration, retinal detachment, myopic foveoschisis, choroidal neovascularization and so on [1-3]. Myopia has emerged as a major public health concern worldwide in recent years. In China, Singapore, Taiwan, the prevalence of myopic subjects aged 13-39 years has rapidly increased up to 71-96% in these years [4,5]. Among myopic children, 10-20% have high myopia when they complete high school [6].
Many researches suggest that genetic factors play important roles in myopia [7,8]. As of April 5, 2015, the Online Mendelian Inheritance in Man (OMIM) database has listed 341 genetic factors associated with myopia. Among 24 MYP (MYP1-24) gene regions, MYP1-5, MYP11-13 and MYP15-16 were reported to be associated with high myopia [9-14] and MYP6-10 and MYP14 were associated with common myopia [15-17].
In 2009, Nakanishi et al. [16] reported the first genome-wide association study (GWAS) related with myopia, in which SNP rs577948 was proven to have association with high myopia and the BLID gene 44 kb downstream of this SNP was speculated to play some roles in myopia progression. Since then, GWAS led to the identification of many susceptibility and causative genes for myopia. In Europe, teams of Rotterdam and Twins UK found chromosomal regions 15q14 and 15q25 were myopia related gene mutations [18,19]. Genes GJD2 and RASGRF1 nearby were reported to be associated with myopia [20]. In 2013, the CREAM consortium conducted multicenter genome-wide meta-analyses and identified susceptibility genes of diverse biological pathways [20]. These genes were enriched for certain functional annotations such as neurotransmitter functions (GRIA4), ion channel activity (KCNQ5, CD55, CACNA1D), retinoic acid metabolism (RDH5, CYP26A1, RORB), extracellular matrix remodeling (LAMA2, BMP2) and ocular development (SIX4, CHD7, PRSS56) [21].
The TGFB1 gene locates in 19q13.1-q13.3 of human genome and contains seven exons [22]. This gene encodes a multi-functional peptide that regulates proliferation, differentiation, migration, adhesion, and other functions in many cell types [23,24]. Cloned TGFB1 from a genomic library derived from human term placenta mRNA [25]. TGFB1 was expressed in scleral tissue and can increase collagen production in scleral fibroblasts in a dose-dependent manner [26,27]. Changes in TGFB1 expression has been observed during the development of experimental myopia in animals [28,29]. Besides, Mucida et al. [30] identified the myopia related factor vitamin A metabolite retinoic acid as a key regulator of TGFB dependent immune responses. These evidences suggest that TGFB1 may play an important role in the process of myopia development.
There were also epidemiological evidences supporting the point of view. Lin et al. [31] reported that rs1982073 was the risk of Taiwan Han population susceptible to myopia through restriction fragment length polymorphism (RFLP). Later, Zha et al. [32] found that rs1800469, rs1982070, rs2241716 and rs4803455 were associated with high myopia. In 2010, Khor et al. [33] found that rs4803455 was risk factors of myopia in Singapore Chinese children. However, most SNPs of TGFB1 reported to be associated with high myopia were heterogeneous in different study populations, which might be due to varaitions in small sample sizes and diversities in ethnic backgrounds. For example, the study by Zhou et al. [34] did not support the association of high myopia with allele of rs4803455 in TGFB1, which was inconsistent with the studies by Khor et al. [33] and Zha et al. [32]. Thus, the association of these SNPs in TGFB1 with high myopia remains uncertain.
In the present study, we present a systematic review and meta-analysis of all association studies on TGFB1 and high myopia to evaluate the effects of TGFB1 polymorphisms on high myopia.
Methods
Literature and search strategy
We searched in PubMed, EMBASE, Cochrane Library and some Chinese databases such as chinese biomedical literature database (CBM), China National Knowledge Infrastructure (CNKI), WANFANG DATA and VIP database from their inception to April 5, 2015. The following keywords were used as free words, truncation as well as MeSH terms, “TGF-B”, “transforming growth factor-B”, “polymorphism”, “variant”, “mutation”, “myopia”, “nearsighted”, “refractive error”, “near sight”, “short sight”, “shortsighted”. Detailed search strategy was shown in supplementary data file. Corresponding Chinese terms were used to search in Chinese databases. The reference lists from the retrieved articles were manually screened for potential articles, if any, that had not been captured by electronic search. No language restrictions were applied during the searching process.
Inclusion and exclusion criteria
Inclusion criteria were as follows: (1) Original case-control or family-based studies evaluating the association between polymorphisms of TGFB1 and high myopia; (2) Numbers or frequencies in case and control groups reported for each genotype or allele; (3) If the study was reported in duplicate, the version having comprehensive contents was included. (4) Studies including normal individuals with spherical equivalent refraction ranged from -2.0 to 2.0 diopters (D) and free from any complications. High myopia was defined as having the axial length of not less than 26 mm or having a refractive error of -6 D or less.
Exclusion criteria were as follows: (1) Animal studies, reviews, conference proceedings, case reports, editorials; (2) Articles providing incomplete data.
Data extraction
Two independent authors (M.B. and Y.Y.) screened all retrieved records. Data were extracted with customized data form. Disagreements were resolved by discussion. Further, uncertainties were resolved by consensus with a third author (SYZ). The information of first author, year of publication, ethnicity, sample size, polymorphisms studied, allelic and genotypic counts, minor allele and conclusions on high myopia association were collected. If allele or genotypic data were not available in the original reports, we would calculate the corresponding one based on another.
Assessment of study quality
Study quality was assessed by the following revised criteria according to Little’s recommendations [35] for gene-disease associations, aiming at investigating potential bias and effect on summary results. These criteria included: (1) genotyping method used; (2) definition of cases and method of ascertainment; (3) socio-demographic characteristics of subjects; (4) confounding mentioned in articles; (5) confidence intervals of genotype frequency. An overall quality scoring was generated, and studies with score ≥3 were considered to have high quality. Disagreement was settled as mentioned above.
Statistical analysis
Meta-analysis was performed for SNPs evaluated in at least two studies. Five genetic models, i.e. dominant, recessive, homozygous, heterozygous and allelic model were applied in the investigation of the disease association. Association of each SNP with high myopia in pooled samples, along with the pooled odds ratios (ORs) and 95% confidence intervals (95% CIs) were evaluated using both fixed-effect and random-effect models. Heterogeneity across studies was evaluated by Cochran Q statistic test and the I2 index. The Q statistic was considered significant if P<0.1 and I2 above 50% indicated large heterogeneity. The regression test was used to assess the potential publication bias. If significant heterogeneity was detected, results from the random-effects model should be adopted, if not, the fixed-effects model. Review Manager software (RevMan, version 5.2) was used for data analysis. Sensitivity analyses were conducted by the approach of one-study remove to assess the influence of single study on the combined effect. The Bonferroni correction was used to account for multiple testing in association analyses. When five genetic models were tested for each SNP, a P<0.01 was considered statistivally significant.
Results
A total of 128 articles in English databases and 15 publications in Chinese databases referring to TGFB1 and myopia were identified. Among them, 14 articles (11 in English and 3 in Chinese) were duplicates and 85 had irrelevant titles (81 in English and 5 in Chinese). In the process of abstract evaluation, 26 in English were excluded, including 2 reviews, 13 animal studies and 11 irrelevant articles. Four in Chinese were excluded, including 2 conference proceedings and 2 irrelevant articles. Further, 4 irrelevant articles (3 in English and 1 in Chinese) were excluded after detailed full-text evaluation. Eventually, eight studies in nine articles that met all the criteria were included for meta-analysis. Figure 1 denotes the workflow of study selection.
Figure 1.

Flowchart of study inclusion.
Overall, 28 SNPs associated with TGFB1 gene were investigated at least once in eight studies. Of these SNPs, three were tested in at least two studies and then were included in the data synthesis: rs4803455, rs1800469 and rs1982073. All study subjects were Asians (Chinese, India and Japanese) with sample size ranging from 109 to 660. The total sample size was 3481 (1694 with high myopia and 1787 controls).
The methods of gene analysis included restriction fragment length polymorphism (RFLP) [31,32,36,37], gene-chip [33], RT-PCR (TaqMan probe) [34,38] and conformation sensitive gel Electrophoresis (CSGE) [39]. One study used three methods for different SNPs, including RFLP, denaturing high performance liquid chromatography (DHPLC) and allele-specific PCR using SYBR Green I [32]. The quality scores of included studies were greater than 3 except one, which indicating a favorable methodological quality [34]. Table 1 summaries the characteristics of included studies.
Table 1.
Characteristics of all studies included in the meta-analysis
| First author | Year | Quality score | Ethnicity | SNP ID | Sample size | Minor allele | Conclusion on high myopia association | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Cases | Controls | |||||||
| Ahmed | 2014 | 4 | India | rs2229333 | 212 | 239 | T | Associated |
| rs4468717 | 212 | 239 | T | Associated | ||||
| Rasool | 2012 | 3 | India | rs1982073 | 247 | 176 | C | Associated |
| rs1800471 | 247 | 176 | C | NS | ||||
| novel | 247 | 176 | A | NS | ||||
| Khor | 2010 | 3 | Chinese | rs4803455 | 107 | 348 | T | Associated |
| Wang | 2009 | 4 | Chinese | rs1982073 | 288 | 208 | T | NS |
| Zha | 2009 | 5 | Chinese | rs1800469 | 300 | 300 | C | Associated |
| rs1800470 | 300 | 300 | T | Associated | ||||
| rs2241716 | 300 | 300 | A | Associated | ||||
| rs4803455 | 300 | 300 | T | Associated | ||||
| rs11466345 | 300 | 300 | G | NS | ||||
| rs12983047 | 300 | 300 | C | NS | ||||
| rs10417924 | 300 | 300 | C | NS | ||||
| rs12981053 | 300 | 300 | T | NS | ||||
| rs1982073 | 300 | 300 | T | Associated | ||||
| Hayashi | 2007 | 3 | Japanese | rs1800469 | 330 | 330 | T | NS |
| rs2241715 | 330 | 330 | T | NS | ||||
| rs41717 | 330 | 330 | T | NS | ||||
| rs2278422 | 330 | 330 | G | NS | ||||
| rs1800820 | 330 | 330 | T | NS | ||||
| rs1054797 | 330 | 330 | T | NS | ||||
| rs1800468 | 330 | 330 | T | NS | ||||
| rs11466324 | 330 | 330 | A | NS | ||||
| rs11672143 | 330 | 330 | T | NS | ||||
| rs11466334 | 330 | 330 | A | NS | ||||
| Zhou | 2007 | 2 | Chinese | rs4803455 | 9 | 100 | A | NS |
| Lin | 2006 | 4 | Chinese | rs1982073 | 201 | 86 | T | Associated |
NS: Nonassociated.
Meta-analysis under five genetic models is shown in Table 2. Rs1982073 was tested in 4 tudies with 1036 cases and 770 controls. The dominant model (CC+CT vs. TT; OR=1.64; 95% CI=1.04~2.58; P=0.03; Figure 2A), the heterozygous model (CT vs. TT; OR=1.54; 95% CI=1.02~2.33; P=0.04; Figure 2B), the homozygous model (CC vs. TT; OR=1.90; 95% CI=1.01~3.55; P=0.05; Figure 2C) and the allelic model (C vs. T; OR=1.36; 95% CI=1.01~1.84; P=0.05; Figure 3A) best explained its effects and indicated a significant association with high myopia.
Table 2.
Pooled measures for the associations between TGFB1 SNPs and high myopia
| SNPs | Models Tested | Number of. study | Location | Events | Pooled OR | (95% CI) | P | Heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| Cases | Controls | FEM | REM | FEM | REM | Q | PQ | I2 | ||||
| rs4803455 | TT+TG vs. GG | 2 | Intron | 226/407 | 378/648 | 0.89 (0.68-1.15) | 0.97 (0.48-1.93) | 0.36 | 0.92 | 6.26 | 0.01 | 84% |
| TT vs. TG+GG | 2 | 26/407 | 96/648 | 0.40 (0.25-0.64) | 0.40 (0.25-0.64) | 0.0001 | 0.0001 | 0.19 | 0.66 | 0% | ||
| TG vs. GG | 2 | 201/381 | 282/552 | 1.06 (0.81-1.39) | 1.17 (0.54-2.53) | 0.67 | 0.69 | 7.16 | 0.007 | 86% | ||
| TT vs. GG | 2 | 26/206 | 96/366 | 0.42 (0.26-0.68) | 0.43 (0.23-0.79) | 0.0004 | 0.007 | 1.51 | 0.22 | 34% | ||
| T vs. G | 3 | 254/819 | 495/1364 | 0.78 (0.65-0.95) | 0.81 (0.56-1.17) | 0.01 | 0.27 | 4.54 | 0.1 | 56% | ||
| rs1800469 | C vs. T | 2 | Promoter region | 326/790 | 372/788 | 0.78 (0.64-0.96) | 0.78 (0.64-0.96) | 0.02 | 0.02 | 0.59 | 0.44 | 0% |
| rs1982073 | CC+CT vs. TT | 4 | exon | 819/1035 | 546/770 | 1.56 (1.26-1.95) | 1.64 (1.04-2.58) | <0.0001 | 0.03 | 12.07 | 0.007 | 75% |
| CC vs. CT+TT | 4 | 299/1035 | 190/770 | 1.31 (1.05-1.63) | 1.39 (0.92-2.09) | 0.01 | 0.12 | 9.15 | 0.03 | 67% | ||
| CT vs. TT | 4 | 520/736 | 356/580 | 1.48 (1.17-1.86) | 1.54 (1.02-2.33) | 0.001 | 0.04 | 8.9 | 0.03 | 66% | ||
| CC vs. TT | 4 | 299/515 | 190/414 | 1.71 (1.30-2.24) | 1.90 (1.01-3.55) | 0.0001 | 0.05 | 14.11 | 0.003 | 79% | ||
| C vs. T | 4 | 1118/2070 | 736/1540 | 1.31 (1.15-1.50) | 1.36 (1.01-1.84) | <0.0001 | 0.05 | 14.21 | 0.003 | 79% | ||
FEM, fixed-effects model; REM, random-effects model.
Figure 2.
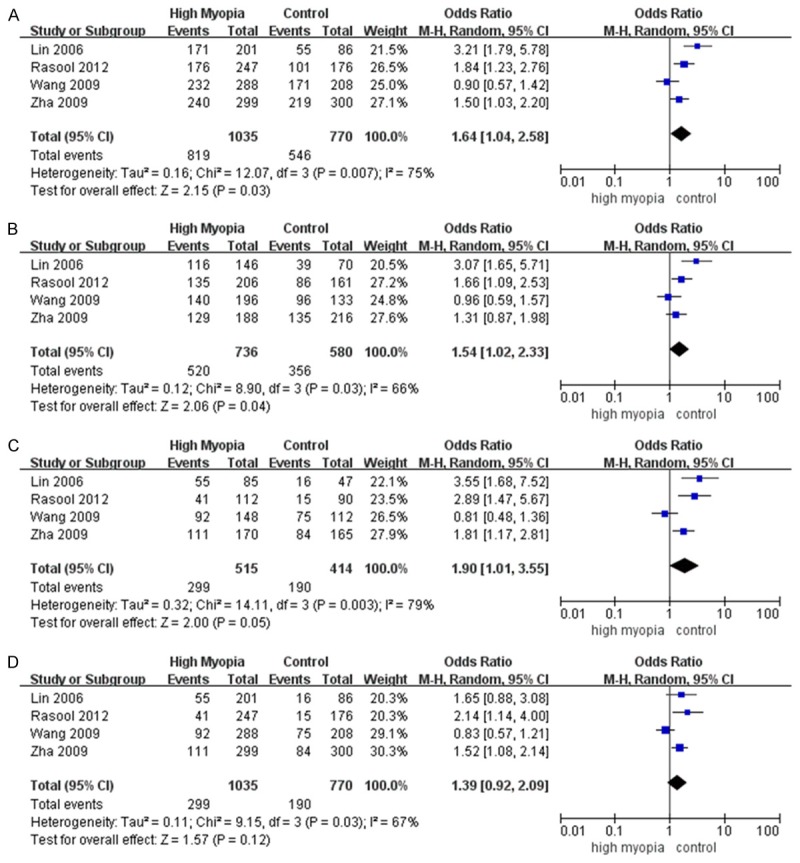
Meta-analysis of the association of TGFB1 rs1982073 with high myopia. The bars with squares in the middle represent 95% confidence intervals (95% CIs) and odds ratios (ORs). The central vertical solid line indicates the ORs for the null hypothesis. Diamond indicates summary OR with its corresponding 95% CI. A. Dominant model (CC+CT vs. TT). B. Heterozygous model (CT vs. TT). C. Homozygous model (CC vs. TT). D. Recessive model (CC vs. CT+TT).
Figure 3.
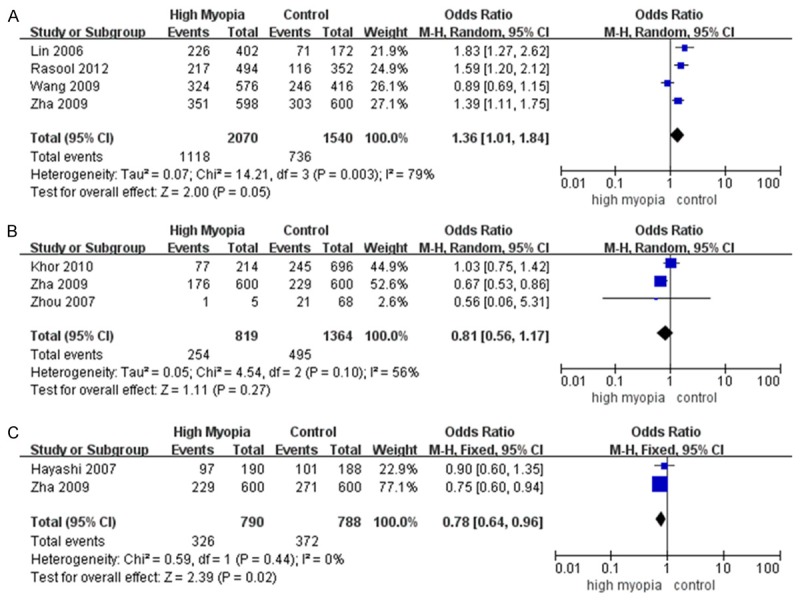
Meta-analysis of the associations of SNPs with high myopia in the allelic model. The bars with squares in the middle represent 95% confidence intervals (95% CIs) and odds ratios (ORs). The central vertical solid line indicates the ORs for the null hypothesis. Diamond indicates summary OR with its corresponding 95% CI. A. rs1982073. B. rs4803455. C. rs1800469.
Rs1800469 was tested in 2 studies with 630 cases and 630 controls. In the allelic model (C vs. T), rs1800469 was associated with high myopia risk (OR=0.78; 95% CI=0.64~0.96; P=0.02; Figure 3C).
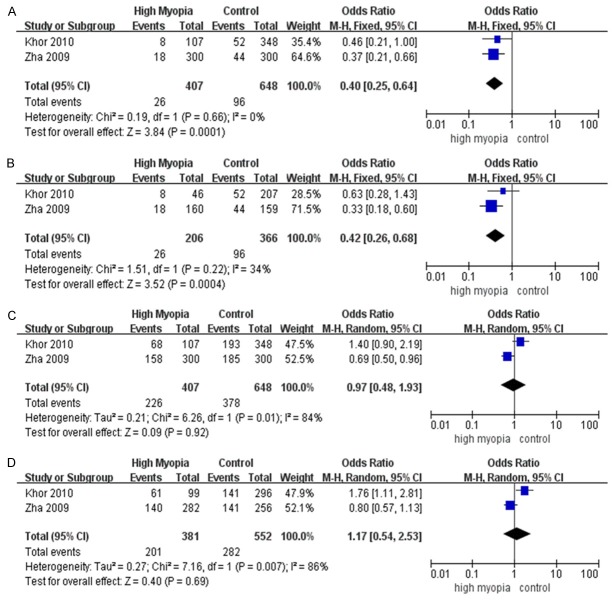
SNP rs4803455 was investigated in 3 studies with 416 cases and 748 controls. The recessive model (TT vs. TG+GG) and the homozygous model (TT vs. GG) showed significant associations with high myopia in a fixed-effects model respectively (OR=0.40; 95% CI=0.25~0.64; P=0.0001; Figure 4A; OR=0.42; 95% CI=0.26~0.68; P=0.0004; Figure 4B). The allelic model (T vs. G) showed a significant association with high myopia (OR=0.78; 95% CI=0.65~0.95; P=0.01; Table 2) in a fixed-effects model. However, as it met the criteria for heterogeneity, the random-effects model was adopted (OR=0.81; 95% CI=0.56~1.17; P=0.27; Figure 3B). The pooled ORs in other genetic models from both fixed-effects model and random-effects model were not significant (P>0.30) (Figure 4C, 4D).
Figure 4.
Meta-analysis of the association of TGFB1 rs4803455 with high myopia. The bars with squares in the middle represent 95% confidence intervals (95% CIs) and odds ratios (ORs). The central vertical solid line indicates the ORs for the null hypothesis. Diamond indicates summary OR with its corresponding 95% CI. A. Recessive model (TT vs. TG+GG). B. Homozygous model (TT vs. GG). C. Dominant model (TT+TG vs. GG). D. Heterozygous model (TG vs. GG).
In our sensitivity analysis of rs1982073 in high myopia, the heterogeneity was significantly diminished after excluding the study of Wang et al. In this condition, we found evidences supporting the association of rs1982073 with high myopia in all genetic models. However, due to the small number of studies (<10) included in each analysis, publication bias was not assessed.
Discussion
In this meta-analysis involving eight studies, rs4803455 in the intron of the TGFB1 gene was found to have association with high myopia in both recessive and homozygous models. Besides, rs1982073 in the exon of the TGFB1 gene was found to have association with high myopia in all models except the recessive one. In the allelic model, there was association between rs1800469 and high myopia. After Boneferroni correction, the association between rs1982073 and high myopia could not withstand P<0.01.
It has been well-known that some complex diseases such as tumor, diabetes and myopia are due to subtle changes in multiple genes caused by environmental factors [5,40-42]. However, investigating the genetics of complex disorders such as myopia remains to be one great challenge. Myopia involves several overlapping signaling pathways which are mediated by groups of genetic profiles. Studies on the relations between genetic polymorphisms and myopia can provide evidence of etiology and help us to treat myopia. In addition, extened axial length is one important characteristic of high myopia, which is associated with scleral remodeling [43-45]. So it is important to keep eyes on the genes in the pathway of scleral remodeling.
There are three conserved TGFB isoforms found in Homo sapiens: TGFB1, TGFB2 and TGFB3. Shehata et al. [46] found that TGFB1 was directly involved in the pathogenesis of bone marrow reticulin fibrosis in hairy cell leukemia. Besides, Zeisberg et al. [47] considered that cardiac fibrosis was associated with fibroblasts originating from endothelial cells, which suggested an endothelialmesenchymal transition similar to events that occured during the formation of the atrio-ventricular cushion in the embryonic heart. In addition, TGFB1 induced endothelial cells to undergo endothelial-mesenchymal transition. It has been well-known that proliferative vitreoretinopathy is characterized by development of epiretinal and subretinal fibrocellular membranes which contain modified retinal pigment epithelial (RPE) cells.
Moreover, Jobling et al. [26] concluded that TGFB1, TGFB2 and TGFB3 were expressed in scleral tissue and scleral fibroblasts of tree shrew pups. All three isoforms increased collagen production in scleral fibroblasts in a dose-dependent manner. Changes in TGFB1 expression had been observed during development of experimental myopia in these animals. In addition, Awad et al. [48] described a polymorphism of the TGFB1 gene that increased the production of TGFB1 and is associated with the development of fibrotic lung disease, which suggests that SNPs of TGFB1 may also play some roles in myopia.
Rs1982073, which has merged into rs1800470, locates within the exon of TGFB1. Early in 2006, Lin et al. [31] analyzed the association between rs1982073 and high myopia in a Chinese population living in Taiwan. They found that the frequency of the CC homozygote in the high myopia group was much higher than in the control group. Later, the association of the coding SNP rs1800470 with high myopia was successfully replicated by Zha et al. [32]. In this study, six hundred adults were recruited, including 300 subjects with high myopia (-8.0 diopters or worse) and 300 control subjects (within ± 1.0 diopters). In 2012, Rasool et al. [39] confirmed the association above in an ethnic population from Kashmir, India. Our data suggest an association between rs1982073 and high myopia in most models. To eliminate false-positive findings, the Bonferroni corrections were used. After correction, the association could not withstand P>0.01. While Wang et al. [36] provided a view contrary to those in previous reports. Besides, we found evidences supporting the association of rs1982073 with high myopia in all genetic models after excluding the study of Wang et al. [36] in the process of sensitivity analysis. We speculate that the sample recruitment scheme difference may contribute to this. Therefore, further studies are needed to confirm the role of rs1982073.
Rs4803455 locates in the intron 2 of TGFB1. In 2009, Zha et al. [32] found The minor allele T of rs4803455 was protective against high myopia with an odds ratio of 0.67 (95% confidence interval, 0.53-0.86; P=0.001). In addition, Khor et al. [33] observed the association at TGFB1 rs4803455 when children with high myopia vs. non-myopic children were compared (n=348 controls, 107 cases; P=0.007). In our study, we did not include the data of Zhou et al. in models except allelic model due to low study quality and data missing. After Bonferroni corrections, associations between rs4803455 and high myopia in recessive and homogenous model were found. And our results are consistent with the findings of Khor base on the data of an genomewide association study using the Illumina HumanHap 550 Beadchips.
With respect to rs1800469, it resides in the promoter region of TGFB1, which may be the binding site of transcriptional factor. In our study, we included only the data of allelic because of data missing. Our data suggest an association between rs1800469 and high myopia in the allelic model. As there were multiple SNPs associated with TGFB1 gene being investigated, a LD map for this gene region would be very useful so as to see more clearly the relationship between the different associations. The LD map based on 1000 genome data provides potential evidence of haplotypic effect between SNP rs1982073 and rs1800469 (r2>0.8).
Additionally, there were a group of other SNPs that had been studied and three of them showed a significant association with high myopia. For example, rs2229333 and rs4468717 were found to be related with high myopia in one study. This study was conducted by Ahmed et al. [37] in cases with high myopia with a spherical equivalent of ≥6 diopters and emmetropic controls with spherical equivalent within ±0.5 D in one or both eyes of 212 ethnic Kashmiri subjects and 239 controls. Besides, Zha et al. [32] had detected the association between rs2241716 and high myopia. However, meta-analysis of these SNPs was impossible due to the limited numbers of studies. Therefore, whether they are high myopia associated SNPs has yet to be further investigated.
There are several limitations in the current meta-analysis. First, the results were pooled from a small number of studies. It is necessary to validate the association of TGFB1 with high myopia in more study cohorts. Second, only SNPs investigated in ≥2 studies were included. However, SNPs that were studied in one study may also be associated with high myopia. Thirdly, heterogeneity in some models was detected and the random-effect model was used, yielding more conservative ORs. Finally, the existing studies were based on Asians.The ethnic background may effect the extrapolation of our results. In conclusion, this meta-analysis suggested an association of TGFB1 SNPs (rs1982073, rs4803455) with high myopia in Asians. Therefore, TGFB1 gene may have some effect on myopia development according to the existing evidence.
Acknowledgements
This work was supported by the Major State Basic Research Development Program of China (“973” Program, 2011CB504601), the Major International (Regional) Joint Research Project of the National Natural Science Foundation of China (81120108807), Beijing Nova Program (Z121107002512055), and the National Natural Science Foundation of China (81300797). Bo Meng and Shi-Ming Li contributed equally to this work.
Disclosure of conflict of interest
None.
References
- 1.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25:381–91. doi: 10.1111/j.1475-1313.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 2.Coppe AM, Ripandelli G, Parisi V, Varano M, Stirpe M. Prevalence of asymptomatic macular holes in highly myopic eyes. Ophthalmology. 2005;112:2103–9. doi: 10.1016/j.ophtha.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Hornbeak DM, Young TL. Myopia genetics: a review of current research and emerging trends. Curr Opin Ophthalmol. 2009;20:356–62. doi: 10.1097/ICU.0b013e32832f8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan IG, Ohno-Matsui K, Saw SM. Myopia. Lancet. 2012;379:1739–48. doi: 10.1016/S0140-6736(12)60272-4. [DOI] [PubMed] [Google Scholar]
- 5.Tang WC, Yap MK, Yip SP. A review of current approaches to identifying human genes involved in myopia. Clin Exp Optom. 2008;91:4–22. doi: 10.1111/j.1444-0938.2007.00181.x. [DOI] [PubMed] [Google Scholar]
- 6.Lin LL, Shih YF, Hsiao CK, Chen CJ. Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore. 2004;33:27–33. [PubMed] [Google Scholar]
- 7.Mackey DA, Hewitt AW. Genome-wide association study success in ophthalmology. Curr Opin Ophthalmol. 2014;25:386–93. doi: 10.1097/ICU.0000000000000090. [DOI] [PubMed] [Google Scholar]
- 8.Stambolian D. Genetic susceptibility and mechanisms for refractive error. Clin Genet. 2013;84:102–8. doi: 10.1111/cge.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz M, Haim M, Skarsholm D. X-linked myopia: Bornholm eye disease. Linkage to DNA markers on the distal part of Xq. Clin Genet. 1990;38:281–6. [PubMed] [Google Scholar]
- 10.Young TL, Ronan SM, Alvear AB, Wildenberg SC, Oetting WS, Atwood LD, Wilkin DJ, King RA. A second locus for familial high myopia maps to chromosome 12q. Am J Hum Genet. 1998;63:1419–24. doi: 10.1086/302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. A new locus for autosomal dominant high myopia maps to 4q22-q27 between D4S1578 and D4S1612. Mol Vis. 2005;11:554–60. [PubMed] [Google Scholar]
- 12.Zhang Q, Guo X, Xiao X, Jia X, Li S, Hejtmancik JF. Novel locus for X linked recessive high myopia maps to Xq23-q25 but outside MYP1. J Med Genet. 2006;43:e20. doi: 10.1136/jmg.2005.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lam CY, Tam PO, Fan DS, Fan BJ, Wang DY, Lee CW, Pang CP, Lam DS. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- 14.Paget S, Julia S, Vitezica ZG, Soler V, Malecaze F, Calvas P. Linkage analysis of high myopia susceptibility locus in 26 families. Mol Vis. 2008;14:2566–74. [PMC free article] [PubMed] [Google Scholar]
- 15.Hammond CJ, Andrew T, Mak YT, Spector TD. A susceptibility locus for myopia in the normal population is linked to the PAX6 gene region on chromosome 11: a genomewide scan of dizygotic twins. Am J Hum Genet. 2004;75:294–304. doi: 10.1086/423148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, Ohno-Matsui K, Mochizuki M, Saito M, Iida T, Matsuo K, Tajima K, Yoshimura N, Matsuda F. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wojciechowski R, Moy C, Ciner E, Ibay G, Reider L, Bailey-Wilson JE, Stambolian D. Genomewide scan in Ashkenazi Jewish families demonstrates evidence of linkage of ocular refraction to a QTL on chromosome 1p36. Hum Genet. 2006;119:389–99. doi: 10.1007/s00439-006-0153-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Solouki AM, Verhoeven VJ, van Duijn CM, Verkerk AJ, Ikram MK, Hysi PG, Despriet DD, van Koolwijk LM, Ho L, Ramdas WD, Czudowska M, Kuijpers RW, Amin N, Struchalin M, Aulchenko YS, van Rij G, Riemslag FC, Young TL, Mackey DA, Spector TD, Gorgels TG, Willemse-Assink JJ, Isaacs A, Kramer R, Swagemakers SM, Bergen AA, van Oosterhout AA, Oostra BA, Rivadeneira F, Uitterlinden AG, Hofman A, de Jong PT, Hammond CJ, Vingerling JR, Klaver CC. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hysi PG, Young TL, Mackey DA, Andrew T, Fernandez-Medarde A, Solouki AM, Hewitt AW, Macgregor S, Vingerling JR, Li YJ, Ikram MK, Fai LY, Sham PC, Manyes L, Porteros A, Lopes MC, Carbonaro F, Fahy SJ, Martin NG, van Duijn CM, Spector TD, Rahi JS, Santos E, Klaver CC, Hammond CJ. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42:902–5. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verhoeven VJ, Hysi PG, Wojciechowski R, Fan Q, Guggenheim JA, Hohn R, MacGregor S, Hewitt AW, Nag A, Cheng CY, Yonova-Doing E, Zhou X, Ikram MK, Buitendijk GH, McMahon G, Kemp JP, Pourcain BS, Simpson CL, Makela KM, Lehtimaki T, Kahonen M, Paterson AD, Hosseini SM, Wong HS, Xu L, Jonas JB, Parssinen O, Wedenoja J, Yip SP, Ho DW, Pang CP, Chen LJ, Burdon KP, Craig JE, Klein BE, Klein R, Haller T, Metspalu A, Khor CC, Tai ES, Aung T, Vithana E, Tay WT, Barathi VA, Chen P, Li R, Liao J, Zheng Y, Ong RT, Doring A, Evans DM, Timpson NJ, Verkerk AJ, Meitinger T, Raitakari O, Hawthorne F, Spector TD, Karssen LC, Pirastu M, Murgia F, Ang W, Mishra A, Montgomery GW, Pennell CE, Cumberland PM, Cotlarciuc I, Mitchell P, Wang JJ, Schache M, Janmahasatian S, Igo RP Jr, Lass JH, Chew E, Iyengar SK, Gorgels TG, Rudan I, Hayward C, Wright AF, Polasek O, Vatavuk Z, Wilson JF, Fleck B, Zeller T, Mirshahi A, Muller C, Uitterlinden AG, Rivadeneira F, Vingerling JR, Hofman A, Oostra BA, Amin N, Bergen AA, Teo YY, Rahi JS, Vitart V, Williams C, Baird PN, Wong TY, Oexle K, Pfeiffer N, Mackey DA, Young TL, van Duijn CM, Saw SM, Bailey-Wilson JE, Stambolian D, Klaver CC, Hammond CJ. Genome-wide meta-analyses of multiancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat Genet. 2013;45:314–8. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hysi PG, Wojciechowski R, Rahi JS, Hammond CJ. Genome-wide association studies of refractive error and myopia, lessons learned, and implications for the future. Invest Ophthalmol Vis Sci. 2014;55:3344–51. doi: 10.1167/iovs.14-14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujii D, Brissenden JE, Derynck R, Francke U. Transforming growth factor beta gene maps to human chromosome 19 long arm and to mouse chromosome 7. Somat Cell Mol Genet. 1986;12:281–8. doi: 10.1007/BF01570787. [DOI] [PubMed] [Google Scholar]
- 23.Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, et al. Transforming growth factor type beta: rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci U S A. 1986;83:4167–71. doi: 10.1073/pnas.83.12.4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–29. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 25.Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goeddel DV. Human transforming growth factor-beta complementary DNA sequence and expression in normal and transformed cells. Nature. 1985;316:701–5. doi: 10.1038/316701a0. [DOI] [PubMed] [Google Scholar]
- 26.Jobling AI, Nguyen M, Gentle A, McBrien NA. Isoform-specific changes in scleral transforming growth factor-beta expression and the regulation of collagen synthesis during myopia progression. J Biol Chem. 2004;279:18121–6. doi: 10.1074/jbc.M400381200. [DOI] [PubMed] [Google Scholar]
- 27.McBrien NA. Regulation of scleral metabolism in myopia and the role of transforming growth factor-beta. Exp Eye Res. 2013;114:128–40. doi: 10.1016/j.exer.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Jobling AI, Gentle A, Metlapally R, McGowan BJ, McBrien NA. Regulation of scleral cell contraction by transforming growth factor-beta and stress: competing roles in myopic eye growth. J Biol Chem. 2009;284:2072–9. doi: 10.1074/jbc.M807521200. [DOI] [PubMed] [Google Scholar]
- 29.Jobling AI, Wan R, Gentle A, Bui BV, McBrien NA. Retinal and choroidal TGF-beta in the tree shrew model of myopia: isoform expression, activation and effects on function. Exp Eye Res. 2009;88:458–66. doi: 10.1016/j.exer.2008.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–60. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- 31.Lin HJ, Wan L, Tsai Y, Tsai YY, Fan SS, Tsai CH, Tsai FJ. The TGFbeta1 gene codon 10 polymorphism contributes to the genetic predisposition to high myopia. Mol Vis. 2006;12:698–703. [PubMed] [Google Scholar]
- 32.Zha Y, Leung KH, Lo KK, Fung WY, Ng PW, Shi MG, Yap MK, Yip SP. TGFB1 as a susceptibility gene for high myopia: a replication study with new findings. Arch Ophthalmol. 2009;127:541–8. doi: 10.1001/archophthalmol.2008.623. [DOI] [PubMed] [Google Scholar]
- 33.Khor CC, Fan Q, Goh L, Tan D, Young TL, Li YJ, Seielstad M, Goh DL, Saw SM. Support for TGFB1 as a susceptibility gene for high myopia in individuals of Chinese descent. Arch Ophthalmol. 2010;128:1081–4. doi: 10.1001/archophthalmol.2010.149. [DOI] [PubMed] [Google Scholar]
- 34.Zhou G, Liu B, Yang YF, MB Y. Single nucleotide polymorphism analysis of multi-loci and -genes in primary open-angle glaucoma with pathological myopia. International Journal of Ophthalmology. 2007;7:1619–23. [Google Scholar]
- 35.Little J, Bradley L, Bray MS, Clyne M, Dorman J, Ellsworth DL, Hanson J, Khoury M, Lau J, O’Brien TR, Rothman N, Stroup D, Taioli E, Thomas D, Vainio H, Wacholder S, Weinberg C. Reporting, appraising, and integrating data on genotype prevalence and gene-disease associations. Am J Epidemiol. 2002;156:300–10. doi: 10.1093/oxfordjournals.aje.a000179. [DOI] [PubMed] [Google Scholar]
- 36.Wang P, Li S, Xiao X, Jia X, Jiao X, Guo X, Zhang Q. High myopia is not associated with the SNPs in the TGIF, lumican, TGFB1, and HGF genes. Invest Ophthalmol Vis Sci. 2009;50:1546–51. doi: 10.1167/iovs.08-2537. [DOI] [PubMed] [Google Scholar]
- 37.Ahmed I, Rasool S, Jan T, Qureshi T, Naykoo NA, Andrabi KI. TGIF1 is a potential candidate gene for high myopia in ethnic Kashmiri population. Curr Eye Res. 2014;39:282–90. doi: 10.3109/02713683.2013.841950. [DOI] [PubMed] [Google Scholar]
- 38.Hayashi T, Inoko H, Nishizaki R, Ohno S, Mizuki N. Exclusion of transforming growth factor-beta1 as a candidate gene for myopia in the Japanese. Jpn J Ophthalmol. 2007;51:96–9. doi: 10.1007/s10384-006-0417-y. [DOI] [PubMed] [Google Scholar]
- 39.Rasool S, Ahmed I, Dar R, Ayub SG, Rashid S, Jan T, Ahmed T, Naikoo NA, Andrabi KI. Contribution of TGFbeta1 codon 10 polymorphism to high myopia in an ethnic Kashmiri population from India. Biochem Genet. 2013;51:323–33. doi: 10.1007/s10528-012-9565-6. [DOI] [PubMed] [Google Scholar]
- 40.Shen L, Ahuja N, Shen Y, Habib NA, Toyota M, Rashid A, Issa JP. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–61. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 41.Arikoglu H, Ozdemir H, Kaya DE, Ipekci SH, Arslan A, Kayis SA, Gonen MS. The Adiponectin Variants Contribute to the Genetic Background of Type 2 Diabetes in Turkish Population. Gene. 2013 [PubMed] [Google Scholar]
- 42.Lee S, Kwon MS, Park T. Network graph analysis of gene-gene interactions in genome-wide association study data. Genomics Inform. 2012;10:256–62. doi: 10.5808/GI.2012.10.4.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.PLOS ONE Staff. Correction: Rapid, Accurate, and Non-Invasive Measurement of Zebrafish Axial Length and Other Eye Dimensions Using SD-OCT Allows Longitudinal Analysis of Myopia and Emmetropization. PLoS One. 2015;10:e0119779. doi: 10.1371/journal.pone.0119779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo L, Frost MR, Siegwart JT Jr, Norton TT. Scleral gene expression during recovery from myopia compared with expression during myopia development in tree shrew. Mol Vis. 2014;20:1643–59. [PMC free article] [PubMed] [Google Scholar]
- 45.Zou L, Liu R, Zhang X, Chu R, Dai J, Zhou H, Liu H. Upregulation of regulator of G-protein signaling 2 in the sclera of a form deprivation myopic animal model. Mol Vis. 2014;20:977–87. [PMC free article] [PubMed] [Google Scholar]
- 46.Shehata M, Schwarzmeier JD, Hilgarth M, Hubmann R, Duechler M, Gisslinger H. TGF-beta1 induces bone marrow reticulin fibrosis in hairy cell leukemia. J Clin Invest. 2004;113:676–85. doi: 10.1172/JCI19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–61. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 48.Awad MR, El-Gamel A, Hasleton P, Turner DM, Sinnott PJ, Hutchinson IV. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factorbeta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation. 1998;66:1014–20. doi: 10.1097/00007890-199810270-00009. [DOI] [PubMed] [Google Scholar]