Abstract
Sepsis is a systemic inflammatory response to infection and includes severe sepsis, septic shock and death. Platelet endothelial cell adhesion molecule-1 (PECAM-1) is one cell adhesion molecule expressed on platelets and leukocytes. It regulates platelet activation and mediates transendothelial migration of leukocytes, thus maintaining the integrity of the vasculature. There are some animal experiments associated with the protective role of PECAM-1 against septic shock. However few host genetic risk factors have been identified for sepsis severity and susceptibility to septic shock. A case-control study was conducted, which included 217 patients with sepsis and 90 control subjects recruited from our hospital. One single nucleotide polymorphisms (SNP) of PECAM-1 gene Leu125Val (C373G) was analyzed by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) analysis. Serum soluble PECAM-1 (sPECAM-1) levels were determined by enzyme-linked immunosorbent assay (ELISA). Our results showed that the CG and GG genotypes of SNP in Leu125Val of PECAM-1 (rs668: C>G) was significantly associated with increased susceptibility to septic shock compared with CC genotype in sepsis patients (CG genotype, OR: 2.493, 95% CI: 1.175~5.287, P = 0.016; GG genotype: OR: 3.328, 95% CI: 1.445~7.666, P = 0.004). The serum levels of sPECAM-1 in the sepsis patients (47.1 ± 17.5 ng/ml) were significantly higher than those in the healthy controls (61.3 ± 20.9 ng/ml, P<0.01). Among sepsis patients, the serum levels of sPECAM-1 were significantly higher in CG and GG genotype than in CC genotype. In septic shock patients, nonsurvivors (83.7 ± 12.6 ng/ml, n = 69) had a significantly higher serum sPECAM-1 level than the survivors (76.9 ± 12.7 ng/ml, n = 53) (P<0.01). In conclusion, PECAM-1 Leu125Val polymorphism and its sPECAM-1 levels are associated with sepsis severity and susceptibility to septic shock.
Keywords: Sepsis, septic shock, platelet endothelial cell adhesion molecule-1 (PECAM-1), single nucleotide polymorphisms (SNPs)
Introduction
Sepsis is a systemic inflammatory response to infection and constitutes the leading cause of death in intensive care units (ICUs). Septic shock is severe form of sepsis with refractory hypotension and demonstrates high mortality rate caused by multiple organ dysfunction syndrome (MODS) [1]. A variety of studies have confirmed that individuals demonstrate different susceptibility to sepsis or septic shock. Genetic polymorphisms, such as those in E-selectin, apolipoprotein E, C-reactive protein, interleukin-10 and CD14 alleles, have been reported to determine the risk and outcome of sepsis [2-4]. In sepsis endothelium is activated and subsequent endothelial dysfunction lead to inflammation response, impaired microvascular barrier integrity, tissue edema, shock and MODS, thereby contributing to the morbidity and mortality of sepsis [5]. In septic shock the endothelial barrier function is mostly impaired, which might contribute to its adverse outcomes [6]. Therefore, endothelial barrier repair is a promising treatment strategy for sepsis. Furthermore, molecules that could regulate endothelial integrity may participate in the pathophysiology of sepsis and their genetic polymorphisms may influence the risk and outcome of sepsis.
Platelet endothelial cell adhesion molecule-1 (PECAM-1, CD31) is a 130-kDa cell adhesion molecule that is expressed on platelets and leukocytes and is highly enriched in endothelial cell intercellular junctions [7]. As a signaling adhesion molecule, PECAM-1 shows diverse roles in vascular biology, including platelet activation, thrombosis, angiogenesis, endothelial cell response to shear stress, and transendothelial migration of leukocytes [8]. Recently, there is growing evidence that PECAM-1 functions as one regulator of endothelial junctional integrity [9]. PECAM-1-overexpressing endothelial cells exhibit increased steady-state barrier function and more rapid restoration of barrier integrity induced by thrombin perturbation [10]. Furthermore, PECAM-1 deficient mice showed reduced survival in endotoxic shock induced by LPS, which was associated with enhanced vascular permeability [11]. The increased LPS-induced mortality in PECAM-1 deficient mice was caused by reduced expression of PECAM-1 at endothelial cell-cell junctions [12]. The SNP of PECAM-1 gene have been reported to be correlated with atherosclerosis and myocardial infarction [13,14]. However, currently it remains unclear about the association between PECAM-1 gene SNP and sepsis.
In this study, we investigated the relationship between SNP in PECAM-1 gene and the serum concentration of sPECAM-1 in sepsis patients. We evaluated whether the SNP and serum sPECAM-1 influence the severity and outcome of sepsis.
Materials and methods
Subjects
A total of 217 patients with sepsis were included in this study between January 2013 and December 2014 in our hospital. All patients were admitted to intensive care unit (ICU) and treated following sepsis management protocol. The average age of all patients were 58.2 ± 14.5 (male: female (M: F) = 131:86). All 217 patients were divided into two groups: the severe sepsis group (mean age 59.6 ± 14.5 years; M: F = 56:39) and the septic shock group (mean age 57.1 ± 14.5 years; M: F = 75:47). The severe sepsis or septic shock were diagnosed based on the criteria proposed at the American College of Chest Physicians/Society of Critical Care Medicine Consensus Conference in 1992 [15]. Patients were followed until death or hospital discharge. A total of 90 healthy blood donors were recruited as control subjects with matched age and sex ratios. Informed written consent was obtained from all subjects or patients’ surrogates. This study was approved by the Research Ethics Committee of our hospital.
SIRS (systemic inflammatory response syndrome) was defined as the presence of at least two symptoms in the following: ① fever or hypothermia (core temperature >38°C or <36°C); ② tachycardia (>90 beats/min); ③ tachypnea or hyperventilation (breaths/min >20 or PaCO2<32 mmHg); ④ leukocytosis (WBC >12.000 mm3) or leucopenia (WBC <4.000 mm3). Severe sepsis was defined as SIRS and organ dysfunction secondary to infection, and septic shock was defined as severe sepsis complicated with refractory arterial hypotension which need fluid replacement and vasopressors.
APACHE-II (Acute Physiology and Chronic Health Evaluation II) score was used to evaluation illness severity [16]. SOFA (Sequential Organ Failure Assessment) score was used to evaluate organ dysfunction [17]. The clinical data, including demographic details and age were collected from each subject. The APACHE II score and SOFA score were obtained on ICU admission day of severe sepsis or septic shock patients. Length of ICU stay was recorded for severe sepsis or septic shock patients. Mortality was defined as death of patients during total hospital stay, and correspondingly divided patients into survivors and nonsurvivors. Within 24 hours of the onset of severe sepsis or septic shock, blood samples were collected for further evaluations on PECAM-1 polymorphism and serum sPECAM-1 levels.
Determination of PECAM-1 genotype
Peripheral venous blood was obtained from severe sepsis patients, septic shock patients and healthy controls. Genomic DNA was extracted from leucocytes by a DNA extraction kit (Qiagen, Crawley, UK), according to the manufacturer’s protocols, and was stored at -70°C until use. The genotypes of Leu125Val were determined by polymerase chain reaction-restriction fragment length polymorphism method (PCR-RFLP). The PCR primers were designed based on the GenBank reference sequence (accession no. NC-000017) and primer sequences were as follows. Leu125Val: sense: 5’-GCTCCATCTGCTTGCCTGT-3’; antisense: 5’-TGTCAGCACCACCTCTCACG-3’. The cycling conditions for PCR were as follows: after initiation at 95°C for 5 min, 30 cycles were performed by denaturation at 95°C for 30 sec, annealing at 60°C for 5 sec and extension at 72°C for 10 sec, with a final extension step at 72°C for 7 min. After overnight incubation with restriction endonuclease PvuII, the amplified PCR products were separated on 8% polyacrylamide gel electrophoresis. PvuII digestion produced 245 bp fragment in Leu125Leu (C373C) homozygous genotype, 52+193+245 bp fragment in Leu125Val (C373G) heterozygous genotype, and 52+193 bp fragment in Val125Val (G373G) homozygous genotype.
Determination of sPECAM-1 level
Venous blood collected from severe sepsis and septic shock patients within 24 hours after disease onset, or from healthy controls. Serum was acquired by centrifugation was performed at 1000 g for 10 min, and was stored in aliquots at -70°C until use. The serum sPECAM-1 were measured by Human sPECAM-1 enzyme-linked immunosorbant assay kit (ELISA) (Bender MedSystems, Vienna, Austria), according to the manufacturer’s protocol. The developed color reaction was determined at OD450 wavelength by an ELISA plate reader (Ricso RK201, Shenzhen Ricso Technology Co., Ltd, Shenzhen, Guangdong, China). The serum concentrations of sPECAM-1 was determined by standard curve which was constructed with the kit’s standards (Range: 0~1000 ng/ml).
Statistical analysis
All quantitative data were expressed in mean ± standard deviation (SD). The statistical analysis was performed by the commercially available software SPSS 19.0 (SPSS Inc., Chicago, IL, USA). The comparisons between two groups were determined by an independent t-test. Chi-squared test or a Fisher’s exact test was applied to compare categorical data. The consistency of genotype frequencies among patients and controls were checked by Hardy-Weinberg equilibrium separately. The differences in the genotype frequencies were compared using Chi-squared test between controls and sepsis, and between severe sepsis and septic shock patients. The odds ratios (ORs) and 95% confidence intervals (95% CIs) were determined by unconditional logistic regression models. A probability value of P<0.05 was considered as statistically significant difference.
Results
Demographics of the subjects
The clinical characteristics of the patients at the time of ICU admission are shown in Table 1. There are no statistically significant differences in age and gender between severe sepsis group and septic shock group (P>0.05), which indicates the two groups were matched in age and gender. The septic shock group showed higher APACHE-II score, higher SOFA score and increased length of ICU stay compared with the severe sepsis group (P<0.001). The overall mortality rate at 28 days was 40.1% among all 217 sepsis patients. The septic shock group had higher mortality rate compared with the severe sepsis group (56.6% vs. 32.6%; P<0.01).
Table 1.
Baseline characteristics of the patients at day one of severe sepsis or septic shock
| Characteristics | Severe sepsis (n = 95) | Septic shock (n = 122) | P value |
|---|---|---|---|
| Age | 59.6 ± 14.5 | 57.1 ± 14.5 | 0.218 |
| Gender (Male %) | 58.9 | 61.5 | 0.588 |
| APACHE II score | 19.3 ± 1.9 | 26.6 ± 2.5 | <0.001 |
| SOFA score | 8.9 ± 1.1 | 12.5 ± 1.2 | <0.001 |
| Length of ICU stay | 4.2 ± 1.2 | 9.9 ± 2.8 | <0.001 |
| Mortality | 32.6 | 56.6 | 0.002 |
Note: Data are expressed as the mean ± standard deviation (SD). APACHE II = Acute Physiology, Age, and Chronic Health Evaluation II; NS = not significant; SOFA = Sequential Organ; ICU = intensive care unit.
Association of the PECAM-1 gene polymorphisms with susceptibility to septic shock
We performed PCR-RFLP assay to determine the genotype at Leu125Val of PECAM-1 gene. The restriction endonuclease PvuII cleaved sequence in 373G allele but not in 373C allele, therefore produced one DNA fragment (245 bp) in 373CC homozygous subjects, three DNA fragments (52, 193 and 245 bp) in 373CG heterozygous subjects and two DNA fragments (52 and 193 bp) in 373GG homozygous subjects (Figure 1). The 52 bp fragment was too small to be visible in the electrophoresis gel. In Leu125Val of PECAM-1, the frequencies of the 373CC, 373CG and 373GG genotypes were 23.3%, 55.6%, and 21.1% in healthy controls, and were 18.0%, 53.0%, and 29.0% in all sepsis patients, respectively. There was no significant difference in the genotype frequencies between controls and sepsis patients (Table 2). The frequencies of the 373CC, 373CG and 373GG genotypes were 26.3%, 50.5%, 23.2% in severe sepsis group, and were 11.5%, 54.9%, 33.6% in septic shock group, respectively (Table 3). Compared with the 373CC genotype, patients who were heterozygous (373CG) or homozygous (373GG) for the Leu125Val polymorphism were more likely to have septic shock in the all sepsis patients (373CG, OR: 2.493, 95% CI: 1.175~5.287, P = 0.016; 373GG, OR: 3.328, 95% CI: 1.445~7.666, P = 0.004). The general genotype and allele frequencies did not differ from the values expected by the Hardy-Weinberg model among the controls, severe sepsis and septic shock groups (Table 4).
Figure 1.
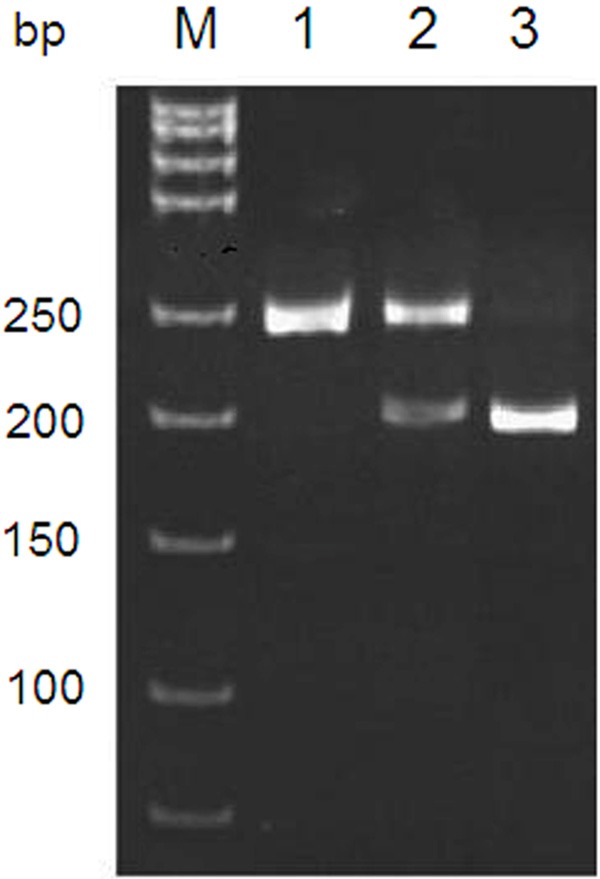
PCR-RFLP assay for PECAM-1 gene Leu125Val (C373G) polymorphism. Representative electrophoresis of PCR products cleaved with restriction endonuclease PvuII was shown. M: DNA molecular weight marker. Lane 1: 373CC (125 Leu/Leu) homozygous genotype, 245 bp; lanes 2: 373CG (125 Leu/Val) heterozygous genotype, 245 bp and 193 bp; lane 3: 373GG (125 Val/Val) homozygous genotype, 193 bp. The 52 bp in 373CG and 373GG genotypes was not visible in electrophoresis gel due to too small size. PECAM-1: platelet endothelial cell adhesion molecule-1.
Table 2.
The genotype frequencies for Leu125Val in PECAM-1 between controls and sepsis patients
| Genotype | Controls (n = 90) | Sepsis (n = 217) | OR (95% CI) | P value |
|---|---|---|---|---|
|
| ||||
| n (%) | n (%) | |||
| Leu/Leu | 21 (23.3) | 39 (18.0) | 1 | |
| Leu/Val | 50 (55.6) | 115 (53.0) | 1.238 (0.662, 2.316) | 0.503 |
| Val/Val | 19 (21.1) | 63 (29.0) | 1.785 (0.854, 3.735) | 0.122 |
Note: OR = odds ratio; CI = confidence interval.
Table 3.
The genotype frequencies for Leu125Val in PECAM-1 between severe sepsis and septic shock patients
| Genotype | Severe sepsis (n = 95) | Septic shock (n = 122) | OR (95% CI) | P value |
|---|---|---|---|---|
|
| ||||
| n (%) | n (%) | |||
| Leu/Leu | 25 (26.3) | 14 (11.5) | 1 | |
| Leu/Val | 48 (50.5) | 67 (54.9) | 2.493 (1.175, 5.287) | 0.016 |
| Val/Val | 22 (23.2) | 41 (33.6) | 3.328 (1.445, 7.666) | 0.004 |
Note: Compared with the Leu/Leu genotype, the Leu/Val and Val/Val genotypes were significantly correlated with an increased risk of septic shock. OR = odds ratio; CI = confidence interval.
Table 4.
Hardy-Weinberg equilibrium of PECAM-1 genotype for the study population (healthy controls and septic patients)
| Groups | Genotype | Observed value | Predicted value | χ2 | P value |
|---|---|---|---|---|---|
|
|
|||||
| n (%) | n (%) | ||||
| Controls | Leu/Leu | 21 (23.3) | 23.5 (26.1) | 1.12 | 0.289 |
| Leu/Val | 50 (55.6) | 45.0 (50.0) | |||
| Val/Val | 19 (21.1) | 21.5 (23.9) | |||
| Sepsis patients | Leu/Leu | 39 (18.0) | 42.9 (19.8) | 1.16 | 0.282 |
| Leu/Val | 115 (53.0) | 107.2 (49.4) | |||
| Val/Val | 63 (29.0) | 66.9 (30.8) | |||
| Severe sepsis | Leu/Leu | 25 (26.3) | 25.3 (26.6) | 0.01 | 0.910 |
| Leu/Val | 48 (50.5) | 47.4 (49.9) | |||
| Val/Val | 22 (23.2) | 22.3 (23.5) | |||
| Septic shock | Leu/Leu | 14 (11.5) | 18.5 (15.2) | 2.93 | 0.087 |
| Leu/Val | 67 (54.9) | 58.0 (47.5) | |||
| Val/Val | 41 (33.6) | 45.5 (37.3) | |||
Note: To test the Hardy-Weinberg equilibrium for consistency, the genotype frequencies of Leu125Val were compared with those of expected values among healthy controls cases (n = 90), sepsis patients (n = 27), severe sepsis (n = 95) and septic shock (n = 122) separately.
Association of serum sPECAM-1 levels with PECAM-1 gene Leu125Val genotypes
To investigate the association between serum sPECAM-1 levels and Leu125Val genotype, we performed ELISA assay to measure serum sPECAM-1 levels in three genotypes of controls, severe sepsis group and septic shock group. The serum sPECAM-1 levels were closely associated with various PECAM-1 genotypes. In controls, severe sepsis group and septic shock group, the serum sPECAM-1 levels was significantly higher in subjects with heterozygous 373CG genotype (Controls: 69.5 ± 12.0 ng/ml; severe sepsis: 70.2 ± 11.0 ng/ml, 81.1 ± 12.2 ng/ml) or homozygous 373GG genotype (Controls: 72.3 ± 11.3 ng/ml; severe sepsis: 74.5 ± 11.2 ng/ml, 85.4 ± 12.1 ng/ml) than subjects with homozygous 373CC genotype (Controls: 59.1 ± 8.6 ng/ml; severe sepsis: 63.3 ± 9.4 ng/ml, 65.4 ± 6.9 ng/ml. P< 0.01, respectively). However, there were no significant differences in serum sPECAM-1 levels between 373CG subjects and 373GG subjects in controls, severe sepsis group and septic shock group, respectively (Figure 2).
Figure 2.
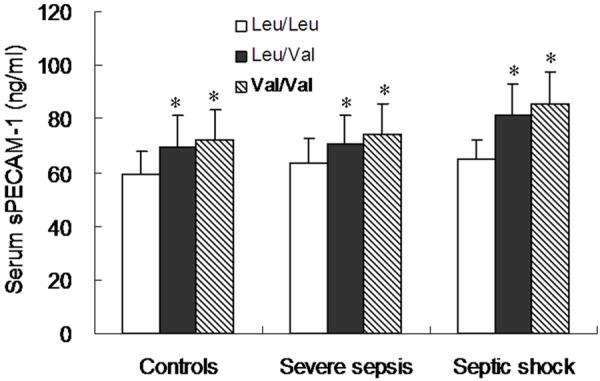
Comparison of serum sPECAM-1 levels in various genotypes of PECAM-1 Leu125Val polymorphism in controls, severe sepsis and septic shock patients. The serum sPECAM-1 levels in the 373CG heterozygous subjects or the 373GG homozygous subjects were significantly higher than those in the 373CC homozygous subjects among controls, severe sepsis group and septic shock group, respectively (All P<0.001). There were no significant difference in serum sPECAM-1 levels between 373CG heterozygous subjects and 373GG homozygous subjects among three groups. Data are expressed as the mean and standard deviation (SD). Significant differences from the 373CC genotype are denoted by “*” (P<0.001).
Associations of serum sPECAM-1 levels with outcome
The serum sPECAM-1 level was significantly higher in the septic shock group (80.8 ± 13.0 ng/ml, n = 122) than the severe sepsis (69.4 ± 11.3 ng/ml, n = 95) or control groups (67.3 ± 12.1 ng/ml, n = 90) (P<0.001) (Figure 3). However, there was no significant difference in the serum sPECAM-1 level between the severe sepsis and control groups. We further divided severe sepsis and septic shock patients into survivors and nonsurvivors respectively. Our results showed that nonsurvivors in septic shock patients (87.4 ± 10.4 ng/ml, n = 69) had a significantly higher serum sPECAM-1 level than the survivors (72.1 ± 10.9 ng/ml, n = 53) (P<0.01) (Figure 4). There was no significant difference in serum sPECAM-1 level between survivors and nonsurvivors in severe sepsis patients (P>0.05).
Figure 3.
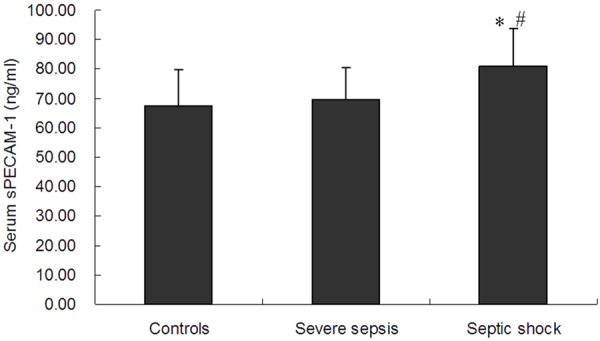
The serum sPECAM-1 levels in controls, severe sepsis and septic shock patients. The serum sPECAM-1 level in the septic shock group was higher than that in the severe sepsis or control group. Data are expressed as the mean and standard deviation (SD). Significant difference from the control group is denoted by “*” (P<0.001). Significant difference from the severe sepsis group is denoted by “#” (P<0.001).
Figure 4.
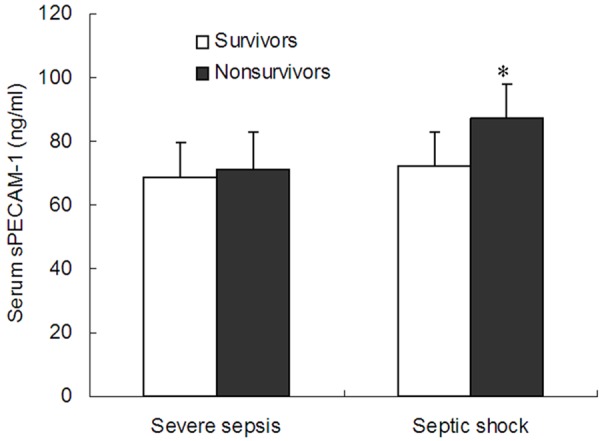
Comparison of serum sPECAM-1 levels between survivors and nonsurvivors in severe sepsis and septic shock groups. The nonsurvivors had a higher serum sPECAM-1 level than the survivors in the septic shock group. Data are expressed as the mean and standard deviation (SD). Significant difference from the survivors group is denoted by “*” (P<0.001).
Discussion
In this study, we show that the SNP in Leu125Val of PECAM-1 may be associated with increased susceptibility to septic shock in sepsis patients. The serum levels of sPECAM-1 in sepsis patients were significantly higher in CG and GG genotype than in CC genotype. In septic shock patients, nonsurvivors had a significantly higher serum sPECAM-1 level than the survivors. However, we did not observe associations of SNP in Leu125Val of PECAM-1 with the risk of sepsis. Our data suggest that PECAM-1 may play protective roles in the development of septic shock, and Leu125Val polymorphism of PEC-AM-1 gene may serve as a novel genetic marker for susceptibility to septic shock in sepsis patients.
The PECAM-1 gene is located on human chromosome 17q23, including 16 exons. Currently, a number of SNP have been identified in PECAM-1 gene [18], among which 3 SNP, including Leu125Val (C373G), Asn563Ser (T1688C) and Gly670Arg (C2008T), have been reported to be associated with some diseases [19]. Leu125Val polymorphism is located in the coding region (exon 3), causing a mutation of leucine to valine. The Leu125Val polymorphism in PECAM-1 gene has been reported to be associated with coronary artery disease [20,21], ischemic stroke [22], atherosclerotic cerebral infarction [23], bronchial asthma [24] and Deep vein thrombosis [25]. However, though PECAM-1 deficient mice showed higher susceptibility to septic shock and higher mortality [11,12], no studies has investigated the association between PECAM-1 gene polymorphisms and septic shock in sepsis patients.
In the present study, the genotypes of Leu125Val were not associated with the risk of sepsis. However, compared with 373GG (125 Leu/Leu) homozygous genotype, 373CG (125 Leu/Val) heterozygous genotype and 373GG (125 Val/Val) homozygous genotype for the PECAM-1 were significantly associated with the progression from severe sepsis to septic shock. To our knowledge, this is the first report on the association about PECAM-1 gene polymorphism with septic shock in sepsis patients. Our results are also in accordance with animal experiments that PECAM-1 deletion increases the risk from sepsis to septic shock [11,12]. This indicates that the C is normal allele that may confer preventive effect on the progression of septic shock by intact PECAM-1 function, while G is mutational allele that impairs the PECAM-1 function as the effect of PECAM-1 deletion in mice.
PECAM-1 functions to maintain endothelial barrier integrity in various conditions [9,10], and promotes recover of the vascular permeability barrier after histamine challenge [26]. Breakdown of endothelial barrier functions is a prominent feather in severe sepsis, and is caused by systemic inflammation, subsequently contributing to septic shock [6,27]. Therefore, PECAM-1 is a protective molecule for septic shock in sepsis patients and G allele in Leu125Val may serve as a prognostic factor for high risk subjects in individualized treatment of sepsis patients. Furthermore, G allele in Leu125Val increases the susceptibility to septic shock other than sepsis. This result is consistent with one hypothesis that PECAM-1 plays roles in modulating and maintaining endothelial junctional integrity under inflammatory conditions but in the normal quiescent state [28,29]. Indeed, though PECAM-1 deficient mice showed enhanced susceptibility to endotoxic shock, they have no structural or functional abnormalities in the vasculature [30] and showed normal blood pressure and heart rate under basal conditions [12].
We then investigated the relationship between serum sPECAM-1 level and SNP in Leu125Val. Serum sPECAM-1 levels in 373CG subjects and 373GG subjects were significantly higher than those in 373CC subjects among healthy controls, severe sepsis and septic shock, respectively. This indicates that serum sPECAM-1 is influenced by genotypes in Leu125Val and G allele can increase the serum sPECAM-1 levels universally in healthy and sepsis subjects. The association between higher circulating sPECAM-1 levels and G allele in Leu125Val has been reported in coronary artery disease [20], ischemic stroke [22], atherosclerotic cerebral infarction [23] and Deep vein thrombosis [25]. sPECAM-1 can be cleaved at the cell surface of PECAM-1 protein [31] and show competitive inhibition on the function of membrane-bound PECAM-1 [32]. Leu125Val is located in the first loop of the extracellular domain of PECAM-1 protein, and leucine to valine mutation may facilitate the cleavage of sPECAM-1 from cell surface, thereby increasing the serum sPECAM-1 level and decreasing the endothelium barrier function of PECAM-1. It deserves further study whether increased susceptibility to septic shock in subjects with G allele is caused by competitive inhibition of the membrane-bound PECAM-1 by serum sPECAM-1, or caused by reduced function of PECAM-1 protein with mutated extracellular domain. Our results also showed that there were no significant differences in serum sPECAM-1 levels between 373CG subjects and 373GG subjects. This means one mutated Val is enough to decrease the function of PECAM-1. Indeed, homophilic interactions of PECAM-1 proteins are essential for maintaining integrity of endothelial cell junctions [10]. Whether dominant negative inhibition effect is involved in 373CG heterozygous subjects in septic shock remains further investigation.
Our results showed a higher serum sPECAM-1 level in the septic shock group than in the severe sepsis group and control group, with no significant difference between severe sepsis group and control group. This further confirmed sPECAM-1 is a serum marker for sepsis severity and susceptibility to septic shock, rather than sepsis risk. Furthermore, we also found that nonsurvivors had a significantly higher serum sPECAM-1 level than the survivors in septic shock patients, but not in severe sepsis patients. Therefore, serum sPECAM-1 may act as one prognostic factor on ICU admission day of sepsis patients that elevated serum sPECAM-1 can predict poor survival for septic shock patients.
In conclusion, we found the subjects with 373G allele of PECAM-1 gene have an increased susceptibility to septic shock and increased serum sPECAM-1 levels. Septic shock patients show higher serum sPECAM-1 levels than severe sepsis patients and healthy controls. In septic shock patients, higher serum sPECAM-1 levels are associated with poor survival. Our study will provide Leu125Val SNP of PECAM-1 gene and serum sPECAM-1as a prognostic marker to predict septic shock susceptibility and mortality.
Disclosure of conflict of interest
None.
References
- 1.Esper AM, Martin GS. Extending international sepsis epidemiology: the impact of organ dysfunction. Crit Care. 2009;13:120. doi: 10.1186/cc7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Namath A, Patterson AJ. Genetic polymorphisms in sepsis. Crit Care Clin. 2009;25:835–56. doi: 10.1016/j.ccc.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Ouyang L, Lv YD, Hou C, Wu GB, He ZH. Quantitative analysis of the association between interleukin-10 1082 A/G polymorphism and susceptibility to sepsis. Mol Biol Rep. 2013;40:4327–4332. doi: 10.1007/s11033-013-2520-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang AQ, Yue CL, Gu W, Du J, Wang HY, Jiang J. Association between CD14 promoter -159C/T polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One. 2013;8:e71237. doi: 10.1371/journal.pone.0071237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee WL, Liles WC. Endothelial activation, dysfunction and permeability during severe infections. Curr Opin Hematol. 2011;18:191–196. doi: 10.1097/MOH.0b013e328345a3d1. [DOI] [PubMed] [Google Scholar]
- 6.Opal SM, van der Poll T. Endothelial barrier dysfunction in septic shock. J Intern Med. 2015;277:277–293. doi: 10.1111/joim.12331. [DOI] [PubMed] [Google Scholar]
- 7.Berman ME, Muller WA. Ligation of platelet/endothelial cell adhesion molecule 1 (PECAM-1/CD31) on monocytes and neutrophils increases binding capacity of leukocyte CR3 (CD11b/CD18) J Immunol. 1995;154:299–307. [PubMed] [Google Scholar]
- 8.Woodfin A, Voisin MB, Nourshargh S. PECAM-1: a multi-functional molecule in inflammation and vascular biology. Arterioscler Thromb Vasc Biol. 2007;27:2514–2523. doi: 10.1161/ATVBAHA.107.151456. [DOI] [PubMed] [Google Scholar]
- 9.Privratsky JR, Newman PJ. PECAM-1: regulator of endothelial junctional integrity. Cell Tissue Res. 2014;355:607–19. doi: 10.1007/s00441-013-1779-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Privratsky JR, Paddock CM, Florey O, Newman DK, Muller WA, Newman PJ. Relative contribution of PECAM-1 adhesion and signaling to the maintenance of vascular integrity. J Cell Sci. 2011;124:1477–1485. doi: 10.1242/jcs.082271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrithers M, Tandon S, Canosa S, Michaud M, Graesser D, Madri JA. Enhanced susceptibility to endotoxic shock and impaired STAT3 signaling in CD31-deficient mice. Am J Pathol. 2005;166:185–196. doi: 10.1016/S0002-9440(10)62243-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maas M, Stapleton M, Bergom C, Mattson DL, Newman DK, Newman PJ. Endothelial cell PECAM-1 confers protection against endotoxic shock. Am J Physiol Heart Circ Physiol. 2005;288:H159–H164. doi: 10.1152/ajpheart.00500.2004. [DOI] [PubMed] [Google Scholar]
- 13.Listì F, Caruso C, Di Carlo D, Falcone C, Boiocchi C, Cuccia M, Candore G. Association between platelet endothelial cellular adhesion molecule-1 polymorphisms and atherosclerosis: results of a study on patients from northern Italy. Rejuvenation Res. 2010;13:237–241. doi: 10.1089/rej.2009.0940. [DOI] [PubMed] [Google Scholar]
- 14.Sahebkar A, Morris DR, Biros E, Golledge J. Association of single nucleotide polymorphisms in the gene encoding platelet endothelial cell adhesion molecule-1 with the risk of myocardial infarction: a systematic review and meta-analysis. Thromb Res. 2013;132:227–233. doi: 10.1016/j.thromres.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, Knaus WA, Schein RM, Sibbald WJ. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–1655. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 16.Knaus WA, Draper EA, Wagner DP, Zimmerman JE. APACHE II: a severity of disease classification system. Crit Care Med. 1985;13:818–829. [PubMed] [Google Scholar]
- 17.Vincent JL, de Mendonça A, Cantraine F, Moreno R, Takala J, Suter PM, Sprung CL, Colardyn F, Blecher S. Use of the SOFA score to assess the incidence of organ dysfunction/failure in intensive care units: results of a multicenter, prospective study. Working group on “sepsis-related problems” of the European Society of Intensive Care Medicine. Crit Care Med. 1998;26:1793–1800. doi: 10.1097/00003246-199811000-00016. [DOI] [PubMed] [Google Scholar]
- 18.Robbins FM, Hartzman RJ. CD31/PECAM-1 genotyping and haplotype analyses show population diversity. Tissue Antigens. 2007;69:28–37. doi: 10.1111/j.1399-0039.2006.00722.x. [DOI] [PubMed] [Google Scholar]
- 19.Novinska MS, Pietz BC, Ellis TM, Newman DK, Newman PJ. The alleles of PECAM-1. Gene. 2006;376:95–101. doi: 10.1016/j.gene.2006.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei H, Fang L, Chowdhury SH, Gong N, Xiong Z, Song J, Mak KH, Wu S, Koay E, Sethi S, Lim YL, Chatterjee S. Platelet-endothelial cell adhesion molecule-1 gene polymorphism and its soluble level are associated with severe coronary artery stenosis in Chinese Singaporean. Clin Biochem. 2004;37:1091–1097. doi: 10.1016/j.clinbiochem.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 21.Fang L, Wei H, Chowdhury SH, Gong N, Song J, Heng CK, Sethi S, Koh TH, Chatterjee S. Association of Leu125Val polymorphism of platelet endothelial cell adhesion molecule-1 (PECAM-1) gene & soluble level of PECAM-1 with coronary artery disease in Asian Indians. Indian J Med Res. 2005;121:92–99. [PubMed] [Google Scholar]
- 22.Wei YS, Lan Y, Liu YG, Meng LQ, Xu QQ, Xie HY. Platelet-endothelial cell adhesion molecule-1 gene polymorphism and its soluble level are associated with ischemic stroke. DNA Cell Biol. 2009;28:151–158. doi: 10.1089/dna.2008.0817. [DOI] [PubMed] [Google Scholar]
- 23.Song Y, Zhao R, Long L, Zhang N, Liu Y. Leu125Val polymorphism of platelet endothelial cell adhesion molecule-1 is associated with atherosclerotic cerebral infarction in Chinese Han population. Int J Clin Exp Med. 2014;7:5808–5813. [PMC free article] [PubMed] [Google Scholar]
- 24.Nadi E, Hajilooi M, Babakhani D, Rafiei A. Platelet endothelial cell adhesion molecule-1 polymorphism in patients with bronchial asthma. Iran J Allergy Asthma Immunol. 2012;11:276–281. [PubMed] [Google Scholar]
- 25.Li G, Han ZL, Dong HG, Zhang X, Kong XQ, Jin X. Platelet endothelial cell adhesion molecule 1 gene 125C/G polymorphism is associated with deep vein thrombosis. Mol Med Rep. 2015;12:2203–2210. doi: 10.3892/mmr.2015.3586. [DOI] [PubMed] [Google Scholar]
- 26.Graesser D, Solowiej A, Bruckner M, Osterweil E, Juedes A, Davis S, Ruddle NH, Engelhardt B, Madri JA. Altered vascular permeability and early onset of experimental autoimmune encephalomyelitis in PECAM-1-deficient mice. J Clin Invest. 2002;109:383–392. doi: 10.1172/JCI13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flemming S, Burkard N, Renschler M, Vielmuth F, Meir M, Schick MA, Wunder C, Germer CT, Spindler V, Waschke J, Schlegel N. Soluble VE-cadherin is involved in endothelial barrier breakdown in systemic inflammation and sepsis. Cardiovasc Res. 2015;107:32–44. doi: 10.1093/cvr/cvv144. [DOI] [PubMed] [Google Scholar]
- 28.Ilan N, Madri JA. PECAM-1: old friend, new partners. Curr Opin Cell Biol. 2003;15:515–524. doi: 10.1016/s0955-0674(03)00100-5. [DOI] [PubMed] [Google Scholar]
- 29.Payne GW, Madri JA, Sessa WC, Segal SS. Histamine inhibits conducted vasodilation through endothelium-derived NO production in arterioles of mouse skeletal muscle. FASEB J. 2004;18:280–286. doi: 10.1096/fj.03-0752com. [DOI] [PubMed] [Google Scholar]
- 30.Duncan GS, Andrew DP, Takimoto H, Kaufman SA, Yoshida H, Spellberg J, de la Pompa JL, Elia A, Wakeham A, Karan-Tamir B, Muller WA, Senaldi G, Zukowski MM, Mak TW. Genetic evidence for functional redundancy of Platelet/Endothelial cell adhesion molecule-1 (PECAM-1): CD31-deficient mice reveal PECAM-1-dependent and PECAM-1-independent functions. J Immunol. 1999;162:3022–3030. [PubMed] [Google Scholar]
- 31.Eugenin EA, Gamss R, Buckner C, Buono D, Klein RS, Schoenbaum EE, Calderon TM, Berman JW. Shedding of PECAM-1 during HIV infection: a potential role for soluble PECAM-1 in the pathogenesis of NeuroAIDS. J Leukoc Biol. 2006;79:444–452. doi: 10.1189/jlb.0405215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao F, Ali J, Greene T, Muller WA. Soluble domain 1 of platelet-endothelial cell adhesion molecule (PECAM) is sufficient to block transendothelial migration in vitro and in vivo. J Exp Med. 1997;185:1349–1357. doi: 10.1084/jem.185.7.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]


