Abstract
This study is to estimate the association between polymorphisms in the tumor necrosis factor alpha (TNF-α) gene and pulmonary tuberculosis susceptibility (pTB). Studies were identified by searching PubMed and ISI web of Knowledge. The strength of association between the TNF-α gene and pTB susceptibility was assessed by odds ratios. Totals of 18 studies including 2, 735 cases and 3, 177 controls were identified referring to four single-nucleotide polymorphisms: -308G>A, -863C>A, -857C>T and -238G>A. The significantly associations were found between -308G>A (Dominant model: OR 0.53, 95% CI 0.35-0.81, P=0.004; Homozygote model: OR 0.51, 95% CI 0.33-0.78, P=0.002), -238G>A (Dominant model: OR 0.33, 95% CI 0.18-0.57, P<0.001) and pTB susceptibility. The results showed that the variant genotype of TNF-α -308G>A was protective in pooled groups of patients with pTB in the dominant genetic model (OR 0.16, 95% CI 0.06-0.39, P<0.001), the homozygote comparison (OR 0.14, 95% CI 0.06-0.36, P<0.001) in African, while that was with -238G>A in the dominant genetic model (OR 0.31, 95% CI 0.18-0.56, P<0.001) in Asian. Our meta-analysis suggest TNF-α -308G>A and -238G>A polymorphisms increases the risk of pTB susceptibility regardless of ethnicity and HIV statue. In Asian population, the significantly association with pTB is TNF-α -238G>A, while TNF-α -308G>A is in African population.
Keywords: TNF-α, pulmonary tuberculosis, meta-analysis
Introduction
Tuberculosis (TB) which caused by the intracellular pathogen Mycobacterium tuberculosis (MTB) still remains a major public health. It is estimated that about 8.6 million new TB cases and 1.3 million deaths globally according to the World Health Organization (WHO) in 2012 [1]. TB is developed by the complex interactions of genetic and environment. Thus, apart from environmental factors, genetic variability is regarded to be responsible for the infection [2]. Some gene polymorphisms have been demonstrated to be associated with TB susceptibility [3-5]. It is expected that the identification of TB susceptibility host genetic factors could contribute greatly to global TB control.
The prominent role played by tumor necrosis factor (TNF) in inflammation and its relevance to both infectious and autoimmune diseases [6] has led to great interest in both the regulation of the TNF gene, and the possibility that variants of the gene or deregulation of its production may be associated with pathology. Also, the serum level of TNF-α was higher in the hyperplasia group of spinal TB than that in the necrosis group, which suggested that it played an essential role in the formation and maintenance of granulomas [7]. On the other hand, patients using anti-TNF agents significantly increase the risk of TB infection [8,9]. Based on those, tumor necrosis factor alpha (TNF-α) gene was focused in TB susceptibility. Allelic polymorphisms of TNF-α have been identified at positions -238, -308, -857, -863 [10,11]. Several meta-analysis studies have reported that the association between TNF-α and TB risk [12-14]. However, they are focused on the all kinds of TB other than pTB and needed to be updated. Also, different results were discovered in our meta-analysis.
Materials and methods
Publication search strategy
The literature search was performed among two English databases (NCBI PubMed and Web of Knowledge) until to 1st April, 2015. The strategies were on the following keywords: ‘tuberculosis’ or ‘TB’ in combination with ‘tumor necrosis factor’ or ‘TNF’, and in combination with ‘polymorphism’ or ‘variant’ or ‘genotype’ or ‘allele’ or ‘SNP’. Moreover, the reference lists of all retrieved articles were also searched to identify more studies. Only English studies were applied.
Selection criteria
To be included, studies had to meet the following criteria: i) evaluated the association between TNF-α polymorphism and pTB susceptibility, ii) was case-control study in design, iii) provided genotype frequencies or numbers for cases and controls. In additional, when the same study was included by more than one article, the study with the largest sample size was selected. The studies were excluded if i) the studies were reviews, abstracts, letters, or communications, ii) Its targets were animals rather than human beings, iii) genotype frequencies or numbers were not retrieved, iv) studies were conducted among the patients with some potential confounding diseases, such as extrapulmonary TB, or pneumoconiosis.
Data extraction and quality score assessment
Two reviewers (Wei Lin and Lin-xiang Jin) independently searched and selected studies, and then extracted following items: author, publication year, original country, ethnicity, sample size, diagnostic methods, genotype and allele number in cases and controls, and HIV status. Disagreements were resolved in consultation with a third investigator (Pei-pei Fang).
The Newcastle Ottawa Scale (NOS) was used to assess the quality of studies included in this meta-analysis, basing on three aspects: selection, comparability and exposure, with scores ranging from 0 to 9 [15]. Studies with a NOS score ≥7 were identified to be of high quality.
Statistical analysis
First, we assessed Hardy-Weinberg Equilibrium (HWE) for each included study using Chi-square test in the controls groups, and P<0.05 was considered as deviation from HWE. The strength of association between TNF-α polymorphisms and pTB susceptibility was estimated by odds ratio (OR) with the corresponding 95% confidence interval (CI). Heterogeneity among included studies was checked by chi-square based Q test and I2 test. If no heterogeneity (P>0.05, I2<50%) was shown in data, the fix effect model [16] was used, otherwise the random effect model [17] was used. Sensitivity analyses were also performed to assess the stability of the meta-analysis by excluding studies deviating from HWE. Publication bias was tested by Begg’s funnel plot and Egger’s weighted regression test. All statistical analyses were done with STATA version 11.0 (Stata Corp., College Station, TX, USA), using two-sided p values.
Results
Study characteristics
A total of 419 relevant articles were identified from the two databases. After excluding those overlapped studies between the databases, 162 titles and abstracts were retrieved for further evaluation. Subsequently, 18 studies with full texts that met the inclusion criteria were analyzed in this meta-analysis [11,18-34]. The flow chart of study selection was shown in Figure 1.
Figure 1.
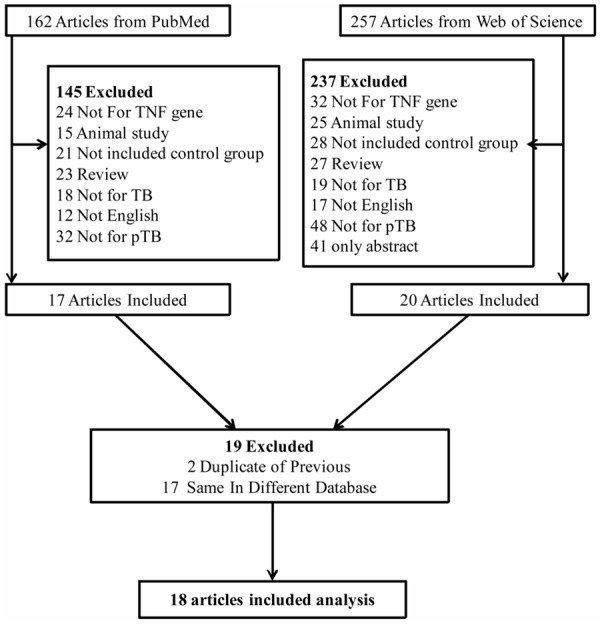
Flow charts of included studies.
The characteristics of included studies were listed in Table 1. These 18 case-control studies were published from 2001 to 2014, including 2, 735 pTB cases and 3, 177 controls. Among them, twelve were performed in Asian population, four in Caucasian population and two in African population. In addition, the HIV (Human Immunodeficiency Virus) status was not available in ten (55.56%) articles. The methodological quality scores of all included studies range from 4 to 7, with 38.89% of the studies (7 of 18) identified to be high quality.
Table 1.
Characteristics of the 18 included studies
| Author | Year | Country | Ethnicity | Diagnosis of case | HIV status | Polymorphisms | Cases/Controls | HWE | NOS scores |
|---|---|---|---|---|---|---|---|---|---|
| Mabunda N, et al | 2014 | Mozambique | Africa | Smear, biopsy, radiological | Negative | -308 | 98/367 | 0.39 | 5 |
| Varahram M, et al | 2014 | Iran | Asian | Culture | NA | -308, -857, -238 | 151/83 | 0.48 | 6 |
| Amirzargar AA, et al | 2006 | Iran | Asian | Smear, radiological | NA | -308, -857, -238 | 40/123 | 0.27 | 4 |
| Anoosheh S, et al | 2011 | Iran | Asian | Smear, cultures | NA | -308, -863, -857, -238, -244 | 93/103 | 0.39 | 6 |
| Delgado JC, et al | 2002 | Cambodia | Asian | Smear | Negative | -308, -863, -857, -238, -1030 | 358/106 | <0.001 | 5 |
| Fan HM, et al | 2010 | China | Asian | Culture, smear, bronchial aspirate/washing | NA | -308, -238 | 113/113 | 0.77 | 6 |
| Merza M, et al | 2009 | Iran | Asian | Smear, radiological | NA | -308, -863, -857, -238, -244 | 117/60 | 0.79 | 7 |
| Oh JH, et al | 2007 | Korea | Asian | Smear, radiological and culture | Negative | -308 | 145/117 | 0.18 | 7 |
| Qu Y, et al | 2007 | China | Asian | Smear, radiological and culture | Negative | -308 | 184/111 | 0.11 | 7 |
| Selvaraj P, et al | 2001 | India | Asian | Smear, radiological and culture | NA | -308, -238 | 210/120 | 0.4 | 7 |
| Tang MQ, et al | 2008 | China | Asian | Smear, radiological | NA | -308 | 44/108 | 0.54 | 6 |
| Vejbaesya S, et al | 2007 | Thailand | Asian | Smear, radiological and culture | Negative | -308, -238, +488 | 149/147 | 0.51 | 5 |
| Correa PA, et al | 2005 | Colombia | Caucasian | Smears and cultures | Negative | -308, -238 | 135/430 | 0.82 | 7 |
| Scola L, et al | 2003 | Italy | Caucasian | Smear, radiological and culture | NA | -308 | 45/114 | 0.81 | 6 |
| Ben-Selma W, et al | 2011 | Tunisia | African | Smear, radiological and culture | Negative | -308 | 95/95 | 0.95 | 7 |
| Henao MI, et al | 2006 | Colombia | Caucasian | Smears, culture, biopsy, radiological | Negative | -308 | 140/135 | 0.39 | 5 |
| Trajkov D, et al | 2009 | Macedonia | Caucasian | Smear, culture | NA | -308 | 75/301 | - | 6 |
| Ma MJ, et al | 2010 | China | Asian | Smear, radiological and culture | NA | -863, -857 | 543/524 | 0.04 | 7 |
Note. NA: Not association, HWE: Hardy-Weinberg Equilibrium, -: No statistics.
The genotype and allele distributions of TNF-α polymorphisms were shown in Table 1. Seven polymorphisms were identified in this meta-analysis. They were TNF-α -308G>A (17 articles), -863C>A (4 articles), -857C>T (5 articles), -238G>A (8 articles), -244G>A (2 articles), -1030T>C (1 article) and +488G>A (1 article). The genotype distributions among the controls of all studies for TNF-α -308G>A, -863C>A, -857C>T and -238G>A were consistent with HWE except for one [26], one [33], one [26] and three studies [20,23,25], respectively. As the polymorphism of TNF-α -244G>A with only GG genotype, it cannot be analyzed. Also, the polymorphisms of TNF-α -1030T>C and +488G>A were reported in one study, they were not included. There was one study of -308G>A with additive genetic model (G vs. A) [24]. The results of this meta-analysis were shoun in Tables 2, 3; Figures 2, 3.
Table 2.
Summary of different comparative results
| No. of studies | OR (95% CI) | P | I2 | ph | Model | Publication bias | ||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Egger | Begg | |||||||
| -308G>A | ||||||||
| Dominant model (GG+GA vs. AA) | 14 | 0.53 (0.35-0.81) | 0.004 | 26.2% | 0.17 | Fix | 0.83 | 0.66 |
| Recessive model (GG vs. GA+AA) | 17 | 0.83 (0.62-1.10) | 0.19 | 65.4% | <0.001 | Random | 0.90 | 0.82 |
| Homozygote model (GG vs. AA) | 14 | 0.51 (0.33-0.78) | 0.002 | 35.6% | 0.09 | Fix | 0.84 | 0.83 |
| Additive model (G vs. A) | 17 | 0.81 (0.63-1.04) | 0.11 | 68.0% | <0.001 | Random | 0.43 | 0.77 |
| -863C>A | ||||||||
| Dominant model (CC+CA vs. AA) | 4 | 0.78 (0.25-2.47) | 0.67 | 72.8% | 0.01 | Random | 0.03 | 0.73 |
| Recessive model (CC vs. CA+AA) | 4 | 1.01 (0.83-1.22) | 0.96 | 0 | 0.72 | Fix | 0.06 | 0.31 |
| Homozygote model (CC vs. AA) | 4 | 0.80 (0.25-2.53) | 0.70 | 71.7% | 0.01 | Random | 0.10 | 0.73 |
| Additive model (C vs. A) | 4 | 1.08 (0.80-1.47) | 0.61 | 61.0% | 0.05 | Random | 0.88 | 0.73 |
| -857C>T | ||||||||
| Dominant model (CC+CT vs. TT) | 5 | 2.10 (0.77-5.75) | 0.15 | 54.0% | 0.07 | Random | 0.42 | 0.81 |
| Recessive model (CC vs. CT+TT) | 5 | 0.84 (0.45-1.57) | 0.59 | 85.0% | <0.001 | Random | 0.24 | 0.46 |
| Homozygote model (CC vs. TT) | 5 | 2.11 (0.72-6.12) | 0.17 | 58.6% | 0.047 | Random | 0.61 | 1.00 |
| Homozygote model (C vs. T) | 5 | 0.92 (0.55-1.53) | 0.73 | 84.5% | <0.001 | Random | 0.13 | 0.46 |
| -238G>A | ||||||||
| Dominant model (GG+GA vs. AA) | 4 | 0.33 (0.18-0.57) | <0.001 | 19.9% | 0.29 | Fix | 0.33 | 0.09 |
| Recessive model (GG vs. GA+AA) | 8 | 0.62 (0.30-1.30) | 0.21 | 85.4% | <0.001 | Random | 0.59 | 0.71 |
| Homozygote model (GG vs. AA) | 4 | 0.34 (0.06-2.12) | 0.25 | 56.1% | 0.078 | Random | 0.17 | 0.31 |
| Additive model (G vs. A) | 8 | 0.67 (0.35-1.26) | 0.21 | 85.4% | <0.001 | Random | 0.32 | 0.39 |
Note. OR: Odds ratio; CI: Confidence interval; ph: P value of heterogeneity.
Table 3.
Results of subgroup analysis
| Polymorphisms | No. of studies | Dominant model | Recessive model | Homozygote model | Additive model | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| OR (95% CI) | p | OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |||
| -308G>A | Ethnicity | |||||||||
| Asian | 12 | 0.76 (0.44-1.32) | 0.33 | 0.82 (0.61-1.10) | 0.18 | 0.71 (0.42-1.20) | 0.20 | 0.86 (0.69-1.08) | 0.20 | |
| African | 2 | 0.16 (0.06-0.39) | <0.001 | 0.60 (0.24-1.54) | 0.29 | 0.14 (0.06-0.36) | <0.001 | 0.50 (0.24-1.08) | 0.08 | |
| Caucasian | 3 | 0.83 (0.35-0.81) | 0.75 | 1.10 (0.48-2.51) | 0.83 | 1.17 (0.24-5.71) | 0.84 | 0.81 (0.63-1.05) | 0.97 | |
| HIV Status | ||||||||||
| HIV negative | 7 | 0.55 (0.25-1.21) | 0.14 | 0.78 (0.51-1.18) | 0.23 | 0.49 (0.22-1.08) | 0.08 | 0.79 (0.55-1.14) | 0.21 | |
| HIV NA | 7 | 0.52 (0.32-0.87) | 0.012 | 0.88 (0.57-1.35) | 0.55 | 0.52 (0.31-0.85) | 0.01 | 0.83 (0.56-1.22) | 0.33 | |
| -863C>A | HIV Status | |||||||||
| HIV negative | 1 | - | - | - | - | - | - | - | - | |
| HIV NA | 3 | 0.43 (0.24-0.78) | 0.005 | 0.98 (0.79-1.21) | 0.83 | 0.44 (0.24-0.79) | 0.006 | 1.12 (0.93-1.34) | 0.24 | |
| -857C>T | HIV Status | |||||||||
| HIV negative | 1 | - | - | - | - | - | - | - | - | |
| HIV NA | 4 | 2.61 (0.73-9.31) | 0.14 | 0.85 (0.41-1.78) | 0.66 | 2.65 (0.70-10.05) | 0.85 | 0.91 (0.49-1.68) | 0.76 | |
| -238G>A | Ethnicity | |||||||||
| Asian | 7 | 0.31 (0.18-0.56) | <0.001 | 0.69 (0.28-1.71) | 0.42 | 0.33 (0.03-3.27) | 0.34 | 0.72 (0.31-1.68) | 0.45 | |
| Caucasian | 1 | - | - | - | - | - | - | - | - | |
| HIV Status | ||||||||||
| HIV negative | 2 | - | - | 0.68 (0.23-2.02) | 0.48 | 0.68 (0.06-2.12) | 0.81 | 0.51 (0.33-0.81) | 0.04 | |
| HIV NA | 6 | 0.32 (0.17-0.57) | <0.001 | 0.48 (0.24-0.96) | 0.04 | 0.33 (0.03-3.27) | 0.34 | 0.72 (0. 27-1.92) | 0.51 |
Figure 2.

Forest plots of a meta-analysis of the association between the -308G>A and -238G>A polymorphisms of the TNF-α gene and pTB susceptibility. A. Dominant model (GG+GA vs. AA) of -308G>A polymorphism. B. Homozygote model (GG vs. AA) of -308G>A polymorphism. C. Dominant model (GG+GA vs. AA) of -238G>A polymorphism.
Figure 3.
Funnel plots of -308G>A, -863C>A, -857C>T and -238G>A. A1-D1. Funnel plots of Dominant, Recessive model, Homozygote and Additive model of -308G>A. A2-D2. Funnel plots of Dominant, Recessive model, Homozygote and Additive model of -863C>A. A3-D3. Funnel plots of Dominant, Recessive model, Homozygote and Additive model of -857C>T. A4-D4. Funnel plots of Dominant, Recessive model, Homozygote and Additive model of -238G>A.
TNF-α -308G>A polymorphism
Seventeen case-control studies on relationship between TNF-α -308G>A polymorphism and pTB risk were identified, including 2, 192 cases and 2, 633 controls. The results of four different genetic models testing this polymorphism and pTB susceptibility were shown in Table 2. For the dominant model (GG+GA vs. AA) and homozygote model (GG vs. AA), fourteen studies were analyzed as to two studies [18,32] with the no people for AA genotype. The p value for heterogeneity was 0.17 and 0.09, and I2 was 26.2% and 35.6%, showing no heterogeneity. Thus, the fix-effect model was used. The overall OR for the dominant model and homozygote model was 0.53 (95% CI: 0.35-0.81, P=0.032, Figure 2A) and 0.51 (95% CI: 0.33-0.78, P=0.002, Figure 2B), suggesting that significantly association was in the dominant model and homozygote model. The Egger’s test and Begg’s test indicated no publication bias in this model (Egger P=0.21, 0.21; Begg, P=0.67, 0.83) (Table 2; Figure 3A, 3C). We also performed comparisons for other two genetic models (Table 2), but no associations were found (GG vs. GA+AA and G vs. A). Also, after removing the study deviating from HWE, the results had not been changed.
TNF-α -238G>A polymorphism
Eight case-control studies on relationship between TNF-α -238G>A polymorphism and pTB risk were identified, including 1, 008 cases and 1, 178 controls. For the dominant model (GG+GA vs. AA) and homozygote model (GG vs. AA), four studies were analyzed as the other four studies [18,25,30,35] with no people for AA genotype. In the dominant model, the p value for heterogeneity was 0.29, and I2 was 19.9%, showing no heterogeneity. Thus, the fix-effect model was used. The overall OR for the dominant model was 0.33 (95% CI: 0.18-0.57, P<0.001), suggesting that significantly association was in the dominant model (Figure 2C). The Egger’s test and Begg’s test indicated no publication bias in this model (Egger P=0.33; Begg, P=0.09) (Table 2; Figure 3A4). We also performed comparisons for other three genetic models (Table 2), but no associations were found (GG vs. GA+AA, GG vs. AA and G vs. A). Also, after removing the study deviating from HWE, the results had not been changed (Data not shown).
TNF-α -863C>A and -857C>T polymorphisms
There were four and five researches for TNF-α -863C>A (1, 109 cases and 813 controls) and -857C>T (1, 261 cases and 896 controls) polymorphisms, and all the population were Asian. There was no significantly association between TNF-α -863C>A and -857C>T polymorphisms and pTB risks. Also, after removing the study deviating from HWE, the results had not been changed.
Sensitivity analysis
Sensitivity analyses were carried out by omitting certain studies each time, such as studies carry out in ethnicity (Asian, African and Caucasian), or studies carry out in certain population (HIV status). The results showed in Table 3. We performed subgroup analyses in TNF-α -308G>A polymorphism by ethnicity. Significantly reduced risk of pTB was found among African population under two genetic models (Dominant model: OR 0.16, 95% CI 0.06-0.39, P<0.001 and Homozygote model: OR 0.14, 95% CI 0.06-0.36, P<0.001), but not for Asian population and Caucasian population. However, in TNF-α -238G>A polymorphism, significantly reduced risk of pTB was found among Asian population under two genetic models (Dominant model: OR 0.31, 95% CI 0.18-0.56, P<0.001). Also, significantly reduced risk of pTB was found among patients without HIV test. Subgroup analyses in the other TNF-α polymorphisms (-863C>A, -857C>T and -238G>A), significantly reduced risk of pTB was found among patients without HIV test.
Discussion
Though only a minority of individuals will develop clinical disease, even if infected with Mycobacterium tuberculosis (MT), TB infections still considered to be substantial threat to public health. As we known, the complex interactions of genetic and environment can lead to multifactorial disease of TB. Thus, besides environmental factors, the variability in the human genes is also contributed to inter-individual variability of disease susceptibility and clinical outcome. Moreover, it was recently reported success in host genotype-specific therapies upon zebrafish with TB infection, which also promote our interest of exploring the host genetic characteristics in TB [36]. Though there were few Genome-wide association studies (GWAS) on the relationship between TNF-α gene polymorphisms and pTB risk. These polymorphisms were still widely focused to experts. TNF-α, being highly involved in the tuberculosis related immunity, has attracted considerable concerns. Also, in the spine TB patients, expression imbalance of TNF-α was existed [7], and anti-TNF agents significantly increase the risk of TB infection in Korean patients with inflammatory bowel disease [8]. Although evidence gradually accumulates regarding the potential association between TNF-α and TB risk, the result remains complex. Several meta-analysis had been show that TNF-α polymorphism was no association with TB risk [12,13], but few studies had analyzed the association with pTB risk. Therefore, we performed this meta-analysis, adding more recently published studies, to evaluate the association between TNF-α polymorphisms and pTB risk more comprehensively and rigorously.
In the present study, 17 case-control studies for analyzing TNF-α -308G>A were analyzed with the main results in pTB infection in two genetic models (GG+GA vs. AA and GG vs. AA), which is not showed in previous meta-analysis [13,14]. Although one of the studies included was deviating from HWE, the sensitivity analysis by excluding this study yielded the similar results, indicating that the results were reliable. Though no obvious heterogeneity when pooling all the studies, we did further stratified analysis by ethnicity (Asian, African and Caucasian). The results appeared that significantly association was only found in African population, while not found in Asian and Caucasian population. The reason for these discrepant results was unclear and differences in sample size, methodologies, ethnicities and dominance of different etiologic factors in different populations might contribute to this heterogeneity results. Also, for analyzing TNF-α -238G>A, the significantly association was shown in Asian population with one genetic model (GG+GA vs. AA). The presence of variation among different ethnicities might be explained partly by the difference in gene allele frequencies, as studies have suggested that the frequency of genetic markers often showed high variation among various racial and ethnic groups [37,38]. Meanwhile, interactions between genes along with environmental factors may also lead to the complexity of effect.
The overall analysis of TNF-α -308G>A and-238G>A showed significantly relationship with pTB risk, while no association in TNF-α -863C>A and -857C>T. In the subgroup analysis stratified of HIV status, this association was existed in HIV undone subgroups (TNF-α -308G>A: dominant model and homozygote model; TNF-α -238G>A: dominant model, recessive model and additive model; TNF-α -863C>A: dominant model and homozygote model). These results suggest that HIV infection is probably a confounding factor which could affect the disease susceptibility and development. So far, we have known that MT is associated with lower HIV specific T cell function in co-infection patients [39]. Thus, the interaction of HIV and MT may lead to disease development.
The mechanism by which TNF-α polymorphism confers association with pTB is unknown. But, TNF as a pro-inflammatory cytokine, plays an important role in immune response against TB activating microbicidal response in macrophages and also during granuloma organization, and patients with anti-TNF agent shows higher risk for TB infection and reaction [9,40,41]. These suggest that the higher level of TNF can control TB infection and reaction. However, not all the TNF-α polymorphisms were association with TB risk. It is that the location of the TNF genes was within the MHC region and HLA gene polymorphism also contributed the TB infection [42]. The possibility that some haplotypes of HLA may be associated due to linkage disequilibrium gains significance.
We should also pay attention to the several limitations in our study, which may affect the result. Firstly, we only included published English studies. Relevant articles published in other language may have been missed. Secondly, the small sample sizes in some subgroup analyses may not comprehensively represent the population. Thirdly, the subgroup analyses were only implemented among some explicitly described population due to the lack of original studies. Studies conduct among African and Caucasian population were needed in the future. Fourthly, the included studies were most middle quality of NOS scores, which might weaker the results.
In conclusion, our meta-analysis suggest that the TNF-α -308G>A and -238G>A polymorphisms are significantly associated with pTB susceptibility regardless of ethnicity and HIV statue. TNF-α -308G>A, -863C>A and -238G>A polymorphisms are significantly associated with pTB susceptibility of HIV-undone patients. In Asian population, the significantly association with pTB is TNF-α -238G>A, while TNF-α -308G>A is in African population. For future studies, well-designed, larger scale epidemiological studies among different ethnicities are needed. More detailed information concerning the potential confounding factors are wanted and multiple SNPs should also be considered in the future studies.
Acknowledgements
This work was supported by Wenzhou Public Welfare Foundation of Science and Technology Project, Science and Technology Bureau of Wenzhou (Y20150023), Nanjing Medical Science and Technique Development Foundation, Nanjing Department of Health (Grant: QRX11235 and Grant: ZDX12008), Jiangsu Science and Technology Project of Clinical Medicine Foundation, Science and Technology Department of Jiangsu Province (BL2014005).
Disclosure of conflict of interest
None.
References
- 1.World HO. Global tuberculosis report 2013. World Health Organization. 2013 [Google Scholar]
- 2.Schurr E. Is susceptibility to tuberculosis acquired or inherited? J Intern Med. 2007;261:106–11. doi: 10.1111/j.1365-2796.2006.01741.x. [DOI] [PubMed] [Google Scholar]
- 3.Grant AV, El BJ, Sabri A, El Azbaoui S, Alaoui-Tahiri K, Abderrahmani Rhorfi I, Gharbaoui Y, Abid A, Benkirane M, Raharimanga V, Richard V, Orlova M, Boland A, Migaud M, Okada S, Nolan DK, Bustamante J, Barreiro LB, Schurr E, Boisson-Dupuis S, Rasolofo V, Casanova JL, Abel L. Age-dependent association between pulmonary tuberculosis and common TOX variants in the 8q12-13 linkage region. Am J Hum Genet. 2013;92:407–14. doi: 10.1016/j.ajhg.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park BL, Kim LH, Namgoong S, Kim JO, Kim JY, Chang HS, Park JS, Jang AS, Park SW, Kim do J, Kim KU, Kim YG, Uh ST, Seo KH, Kim YH, Park CS, Shin HD. Association analysis of melanocortin 3 receptor polymorphisms with the risk of pulmonary tuberculosis. Lung. 2014;192:857–62. doi: 10.1007/s00408-014-9625-2. [DOI] [PubMed] [Google Scholar]
- 5.Qi H, Sun L, Wu X, Jin Y, Xiao J, Wang S, Shen C, Chu P, Qi Z, Xu F, Guo Y, Jiao W, Tian J, Shen A. Toll-like receptor 1(TLR1) Gene SNP rs5743618 is associated with increased risk for tuberculosis in Han Chinese children. Tuberculosis (Edinb) 2015;95:197–203. doi: 10.1016/j.tube.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Beutler BA. The role of tumor necrosis factor in health and disease. J Rheumatol Suppl. 1999;57:16–21. [PubMed] [Google Scholar]
- 7.Chen H, Cheng C, Li M, Gao S, Li S, Sun H. Expression of TNF-alpha, IFN-gamma, TGF-beta, and IL-4 in the spinal tuberculous focus and its impact on the disease. Cell Biochem Biophys. 2014;70:1759–64. doi: 10.1007/s12013-014-0125-z. [DOI] [PubMed] [Google Scholar]
- 8.Kim ES, Song GA, Cho KB, Park KS, Kim KO, Jang BI, Kim EY, Jeon SW, Lee HS, Yang CH, Lee YK, Lee DW, Kim SK, Kim TO, Lee J, Kim HW, Jee SR, Park SJ, Kim HJ. Significant risk and associated factors of active tuberculosis infection in Korean patients with inflammatory bowel disease using anti-TNF agents. World J Gastroenterol. 2015;21:3308–16. doi: 10.3748/wjg.v21.i11.3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ergun T, Seckin D, Baskan BE, Onsun N, Ozgen Z, Unalan P, Alpsoy E, Karakurt S. The risk of tuberculosis in patients with psoriasis treated with anti-tumor necrosis factor agents. Int J Dermatol. 2015;54:594–9. doi: 10.1111/ijd.12628. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S, Rathored J, Ghosh B, Sharma SK. Genetic polymorphisms in TNF genes and tuberculosis in North Indians. BMC Infect Dis. 2010;10:165. doi: 10.1186/1471-2334-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merza M, Farnia P, Anoosheh S, Varahram M, Kazampour M, Pajand O, Saeif S, Mirsaeidi M, Masjedi MR, Velayati AA, Hoffner S. The NRAMPI, VDR and TNF-alpha gene polymorphisms in Iranian tuberculosis patients: The study on host susceptibility. Braz J Infect Dis. 2009;13:252–6. doi: 10.1590/s1413-86702009000400002. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Zhu H, Pu X, Meng S, Zhang F, Xun L, Liu Q, Wang Y. Association between tumor necrosis factor alpha-238G/a polymorphism and tuberculosis susceptibility: a meta-analysis study. BMC Infect Dis. 2012;12:328. doi: 10.1186/1471-2334-12-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q, Zhan P, Qiu LX, Qian Q, Yu LK. TNF-308 gene polymorphism and tuberculosis susceptibility: a meta-analysis involving 18 studies. Mol Biol Rep. 2012;39:3393–400. doi: 10.1007/s11033-011-1110-x. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Zhang Z, Lei X, Feng J, Zhang F, Wang Y. Tumor necrosis factor alpha -308G>A, -863C>A, -857C>T gene polymorphisms and tuberculosis susceptibility: a meta-analysis. Gene. 2012;509:206–14. doi: 10.1016/j.gene.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Zhou YP, Peng HJ, Zhang XH, Zhou FY, Liu ZH, Chen XG. Predictive symptoms and signs of severe dengue disease for patients with dengue fever: a meta-analysis. Biomed Res Int. 2014;2014:359308. doi: 10.1155/2014/359308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies. J Natl Cancer Inst. 1959;22:719–48. [PubMed] [Google Scholar]
- 17.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 18.Anoosheh S, Farnia P, Kargar M. Association between TNF-Alpha (-857) Gene Polymorphism and Susceptibility to Tuberculosis. Iran Red Crescent Med J. 2011;13:243–8. [PMC free article] [PubMed] [Google Scholar]
- 19.Trajkov D, Trajchevska M, Arsov T, Petlichkovski A, Strezova A, Efinska-Mladenovska O, Sandevski A, Spiroski M. Association of 22 cytokine gene polymorphisms with tuberculosis in Macedonians. Indian J Tuberc. 2009;56:117–31. [PubMed] [Google Scholar]
- 20.Varahram M, Farnia P, Nasiri MJ, Karahrudi MA, Dizagie MK, Velayati AA. Association of Mycobacterium Tuberculosis Lineages with IFN-gamma and TNF-alpha Gene Polymorphisms among Pulmonary Tuberculosis Patient. Mediterr J Hematol Infect Dis. 2014;6:e2014015. doi: 10.4084/MJHID.2014.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Selma W, Harizi H, Boukadida J. Association of TNF-alpha and IL-10 polymorphisms with tuberculosis in Tunisian populations. Microbes Infect. 2011;13:837–43. doi: 10.1016/j.micinf.2011.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Fan HM, Wang Z, Feng FM, Zhang KL, Yuan JX, Sui H, Qiu HY, Liu LH, Deng XJ, Ren JX. Association of TNF-alpha-238G/A and 308 G/A gene polymorphisms with pulmonary tuberculosis among patients with coal worker's pneumoconiosis. Biomed Environ Sci. 2010;23:137–45. doi: 10.1016/S0895-3988(10)60043-8. [DOI] [PubMed] [Google Scholar]
- 23.Correa PA, Gomez LM, Cadena J, Anaya JM. Autoimmunity and tuberculosis. Opposite association with TNF polymorphism. J Rheumatol. 2005;32:219–24. [PubMed] [Google Scholar]
- 24.Henao MI, Montes C, Paris SC, Garcia LF. Cytokine gene polymorphisms in Colombian patients with different clinical presentations of tuberculosis. Tuberculosis (Edinb) 2006;86:11–9. doi: 10.1016/j.tube.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Amirzargar AA, Rezaei N, Jabbari H, Danesh AA, Khosravi F, Hajabdolbaghi M, Yalda A, Nikbin B. Cytokine single nucleotide polymorphisms in Iranian patients with pulmonary tuberculosis. Eur Cytokine Netw. 2006;17:84–9. [PubMed] [Google Scholar]
- 26.Delgado JC, Baena A, Thim S, Goldfeld AE. Ethnic-specific genetic associations with pulmonary tuberculosis. J Infect Dis. 2002;186:1463–8. doi: 10.1086/344891. [DOI] [PubMed] [Google Scholar]
- 27.Mabunda N, Alvarado-Arnez LE, Vubil A, Mariamo A, Pacheco AG, Jani IV, Moraes MO. Gene polymorphisms in patients with pulmonary tuberculosis from Mozambique. Mol Biol Rep. 2015;42:71–6. doi: 10.1007/s11033-014-3741-1. [DOI] [PubMed] [Google Scholar]
- 28.Qu Y, Tang Y, Cao D, Wu F, Liu J, Lu G, Zhang Z, Xia Z. Genetic polymorphisms in alveolar macrophage response-related genes, and risk of silicosis and pulmonary tuberculosis in Chinese iron miners. Int J Hyg Environ Health. 2007;210:679–89. doi: 10.1016/j.ijheh.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Scola L, Crivello A, Marino V, Gioia V, Serauto A, Candore G, Colonna-Romano G, Caruso C, Lio D. IL-10 and TNF-alpha polymorphisms in a sample of Sicilian patients affected by tuberculosis: implication for ageing and life span expectancy. Mech Ageing Dev. 2003;124:569–72. doi: 10.1016/s0047-6374(03)00038-1. [DOI] [PubMed] [Google Scholar]
- 30.Vejbaesya S, Chierakul N, Luangtrakool P, Sermduangprateep C. NRAMP1 and TNF-alpha polymorphisms and susceptibility to tuberculosis in Thais. Respirology. 2007;12:202–6. doi: 10.1111/j.1440-1843.2006.01037.x. [DOI] [PubMed] [Google Scholar]
- 31.Oh JH, Yang CS, Noh YK, Kweon YM, Jung SS, Son JW, Kong SJ, Yoon JU, Lee JS, Kim HJ, Park JK, Jo EK, Song CH. Polymorphisms of interleukin-10 and tumour necrosis factor-alpha genes are associated with newly diagnosed and recurrent pulmonary tuberculosis. Respirology. 2007;12:594–8. doi: 10.1111/j.1440-1843.2007.01108.x. [DOI] [PubMed] [Google Scholar]
- 32.Tang MQ, Mo HW, Cheng YQ. The relationship between the gene polymorphisms of TNF alpha and the genetic susceptibility to chronic obstructive pulmonary disease (COPD) with tuberculosis. J Qiqihar Med Coll. 2008;29:2949–51. [Google Scholar]
- 33.Ma MJ, Xie LP, Wu SC, Tang F, Li H, Zhang ZS, Yang H, Chen SL, Liu N, Liu W, Cao WC. Toll-like receptors, tumor necrosis factor-alpha, and interleukin-10 gene polymorphisms in risk of pulmonary tuberculosis and disease severity. Hum Immunol. 2010;71:1005–10. doi: 10.1016/j.humimm.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Selvaraj P, Sriram U, Mathan KS, Reetha AM, Narayanan PR. Tumour necrosis factor alpha (-238 and -308) and beta gene polymorphisms in pulmonary tuberculosis: haplotype analysis with HLA-A, B and DR genes. Tuberculosis (Edinb) 2001;81:335–41. doi: 10.1054/tube.2001.0307. [DOI] [PubMed] [Google Scholar]
- 35.Merza M, Farnia P, Anoosheh S, Varahram M, Kazampour M, Pajand O, Saeif S, Mirsaeidi M, Masjedi MR, Velayati AA, Hoffner S. The NRAMPI, VDR and TNF-alpha gene polymorphisms in Iranian tuberculosis patients: the study on host susceptibility. Braz J Infect Dis. 2009;13:252–6. doi: 10.1590/s1413-86702009000400002. [DOI] [PubMed] [Google Scholar]
- 36.Tobin DM, Roca FJ, Oh SF, McFarland R, Vickery TW, Ray JP, Ko DC, Zou Y, Bang ND, Chau TT, Vary JC, Hawn TR, Dunstan SJ, Farrar JJ, Thwaites GE, King MC, Serhan CN, Ramakrishnan L. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148:434–46. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–8. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 38.Garte S, Gaspari L, Alexandrie AK, Ambrosone C, Autrup H, Autrup JL, Baranova H, Bathum L, Benhamou S, Boffetta P, Bouchardy C, Breskvar K, Brockmoller J, Cascorbi I, Clapper ML, Coutelle C, Daly A, Dell’Omo M, Dolzan V, Dresler CM, Fryer A, Haugen A, Hein DW, Hildesheim A, Hirvonen A, Hsieh LL, Ingelman-Sundberg M, Kalina I, Kang D, Kihara M, Kiyohara C, Kremers P, Lazarus P, Le Marchand L, Lechner MC, van Lieshout EM, London S, Manni JJ, Maugard CM, Morita S, Nazar-Stewart V, Noda K, Oda Y, Parl FF, Pastorelli R, Persson I, Peters WH, Rannug A, Rebbeck T, Risch A, Roelandt L, Romkes M, Ryberg D, Salagovic J, Schoket B, Seidegard J, Shields PG, Sim E, Sinnet D, Strange RC, Stücker I, Sugimura H, To-Figueras J, Vineis P, Yu MC, Taioli E. Metabolic gene polymorphism frequencies in control populations. Cancer Epidemiol Biomarkers Prev. 2001;10:1239–48. [PubMed] [Google Scholar]
- 39.Chetty S, Govender P, Zupkosky J, Pillay M, Ghebremichael M, Moosa MY, Ndung’u T, Porichis F, Kasprowicz VO. Co-Infection with Mycobacterium tuberculosis Impairs HIV-Specific CD8+ and CD4+ T Cell Functionality. PLoS One. 2015;10:e0118654. doi: 10.1371/journal.pone.0118654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harris J, Keane J. How tumour necrosis factor blockers interfere with tuberculosis immunity. Clin Exp Immunol. 2010;161:1–9. doi: 10.1111/j.1365-2249.2010.04146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solovic I, Sester M, Gomez-Reino JJ, Rieder HL, Ehlers S, Milburn HJ, Kampmann B, Hellmich B, Groves R, Schreiber S, Wallis RS, Sotgiu G, Schölvinck EH, Goletti D, Zellweger JP, Diel R, Carmona L, Bartalesi F, Ravn P, Bossink A, Duarte R, Erkens C, Clark J, Migliori GB, Lange C. The risk of tuberculosis related to tumour necrosis factor antagonist therapies: A TBNET consensus statement. Eur Respir J. 2010;36:1185–206. doi: 10.1183/09031936.00028510. [DOI] [PubMed] [Google Scholar]
- 42.Tong X, Chen L, Liu S, Yan Z, Peng S, Zhang Y, Fan H. Polymorphisms in HLA-DRB1 Gene and the Risk of Tuberculosis: A Meta-analysis of 31 Studies. Lung. 2015;193:309–18. doi: 10.1007/s00408-015-9692-z. [DOI] [PubMed] [Google Scholar]