Abstract
Purpose: Many scientific evidences suggested that the methylation of p16INK4a (p16) was associated with bladder cancer, but some existing studies have yielded inconclusive results about the relationship between p16 promoter methylation and pathological features or the tumor grade of bladder cancer. This meta-analysis of studies aims to evaluate the clinical and prognostic significance of p16 methylation in bladder carcinogenesis. Methods: Studies were systemically searched via PubMed and Google Scholar in English up to Sept 2015 and a total of ten appropriate studies (693 cases and 290 controls) with an average NOS score of 6.8 were included. The quality of the appropriate studies was measured by the Newcastle-Ottawa Scale (NOS) assessment. Results: The meta-analysis results revealed that the methylation state of p16 was statistically significantly associated with an increased risk of bladder cancer (OR=6.71, 95% CI=3.79-11.87) compared to control, and there is no statistically significantly association between the p16 methylation and the tumor pTNM staging (OR=0.59, 95% CI=0.22-1.60) or the tumor grade (OR=1.01, 95% CI=0.52-1.94) in p16 methylated patients compared to unmethylated patients. Conclusions: our meta-analysis indicates that p16 promoter methylation may be a promising biomarker for the diagnosis of bladder cancer and the inactivation of p16 may be an early event in bladder carcinogenesis. More studies with larger numbers of participants worldwide are needed to further identify the obvious association above.
Keywords: p16, methylation, bladder cancer, meta-analysis
Introduction
As the most common malignancy of the urinary tract, bladder cancer is the 11th most common cancer diagnosis worldwide and is responsible for 386,300 new cases and more than 150,200 deaths per year [1,2]. Though up to 80% of the cases possess a favorable prognosis for the superficial lesions, many of them will recur and some will eventually develop a more aggressive phenotype [3]. However, up till the present moment, the clinical method for detecting bladder cancer is still mainly rely on cystoscopy, an invasive procedure which will bring pain to patients and has a low sensitivity mostly for detection of low-grade and low-stage disease [4]. So, exploring the reliable biological markers for detecting bladder cancer is of significance and urgency.
DNA methylation, which is one of the important research content of epigenetics, is related to various biological processes including cancer [5-8]. It is a procedure of chemical modification which will specifically methylate the cytosines located 5’ to guanosines in CpG dinucleotides and give rise to 5-methylcytosine (m5C) via the DNA methyltransferases (DNMTs) [9]. This modification play a significant role in regulating gene expression, especially when involving CpG-rich areas known as CpG islands, located in the promoter regions of many genes [10]. CpG islands are usually not methylated in the germline and, with some exceptions, in the normal somatic cells [11]. In contrast, the aberrant methylation of CpG islands always occurs on autosomal genes during carcinogenesis, suggesting that DNA methylation provide a promising method for monitoring the occurrence and progression of cancer [5]. The p16INK4a protein (referred to as p16 throughout) located in the INK4a/ARF locus (9p21) belongs to a family of regulators of the cell cycle, called cyclin dependent kinase inhibitors (CDKI), which bind themselves to cyclin-CDK complexes. As a result, the cell cycle in the G1 phase would be arrested, making p16 stop the proliferation of neoplastic cells [12]. The association of inactivated p16 with hypermethylation has expanded p16 as a tumor suppressor, suggesting that this gene is one of the most frequently inactivated in human neoplasms [13,14]. Studies have indicated that the methylation of the CpG islands within p16 promoter correlates with the loss of expression and in general inactivation of p16 is associating with a more aggressive phenotype and worse prognosis [15,16]. On the other hand, reexpression of p16 caused by demethylation agents also further confirming the role of DNA methylation in the inactivation of p16 [17]. What’s more, p16 methylation was reported as a prognostic indicator in many tumors, including gastric, breast, multiple myeloma, Leukemia, melanoma, and prostate cancer [18-23]. All of the above suggesting that p16 might play a significant role in the development of human cancer.
For all that subsequent individual studies conducted in bladder cancer patients, the value in prognosis of p16 methylation status in bladder cancer patient’s diagnosis remains controversial, especially when involving the relationship between p16 methylation and common clinical and pathologic features of bladder cancer. In order to evaluate this question, we conducted a meta-analysis in the present study to assess precisely of its prognostic value in bladder cancer.
Materials and methods
Publication selection
The appropriate literature in English prior to Sept 2015 were systemically searched via the online databases of PubMed and Google Scholar with the following key words and MeSH terms: (“p16” or “p16INK4a” or “CDKN2A”) and “methylation” and (“bladder cancer” or “bladder neoplasm” or “bladder tumor” or “bladder carcinoma” or “bladder carcinogenesis”). A manual search was also performed in order to retrieve other potential study in the references of original articles.
Inclusion and exclusion criteria
Studies included in this meta-analysis should meet the following criteria: 1) The article focus on the role of p16 promoter methylation in prognosis of bladder cancer patients and the correlation of p16 hypermethylation with clinicopathological markers such as pathological features or the tumor grade of bladder cancer; 2) Only the case-control studies were taken into account; 3) When authors published several studies using the same subjects, only the most recently published or with the largest sample size was included. 4) The minimum number of cases in the included studies should be more than 5 respectively. 5) The candidates were limited to human studies which were published as full-text articles. The study failed to meet the inclusion criteria should be excluded.
Data collection
Relevant data were carefully extracted from all eligible studies which were independently reviewed by two investigators according to the inclusion criteria listed above. The researchers collected the following data: the first author’s surname, publication year of article, country of origin, inclusion criteria, exclusion criteria, pathological features, tumor grade, the frequency of p16 methylation in case and control and the testing material. The non-cancer patients, defined as normal healthy persons or people with disease of urinary system but no prior history of genitourinary malignancy, were used as control in eligible studies. And we united non-cancer patients in this meta-analysis based on their original group in each individual study since redefining non-cancer-patients on a unified standard is impossible. In this meta-analysis, tumor grade ≤1 was defined as low-grade and tumor grade ≥2 was defined as high-grade, which were defined by cellular differentiation. In case of conflicting evaluation, disagreements were resolved by discussion and consensus. To obtain the missing data and additional information, authors were contacted by phone or e-mail. Since there were several kinds of testing material, tumor tissues was classified into a group while the serum and urine were classified into the group of body fluids, which were classified by morphology differentiation.
Quality assessment of study
The quality of studies was evaluated independently by two authors according to the Newcastle-Ottawa Scale (NOS) assessment [24]. Any disagreements were dealt by discussion. The NOS assessment consists of eight items of methodology which were grouped into three major classifications: selection of cases and controls, comparability of case and control groups, and ascertainment of exposure. Each item was graded for a maximum score of 1, except that related to comparability, which allowed for 2. The scores ranged from 0 (lowest) to 9 (highest), and studies with more than 5 points were evaluated as qualified.
Statistical analysis
A meta-analysis was performed to estimate the differences in the frequency of p16 methylation between bladder cancer and control in previously published studies by pooled odds ratio (OR) with the corresponding 95% confidence interval (CI) in a fixed- or random-effect model. What’s more, the power of the association of the p16 methylation and patients’ pTNM (p, pathologic stage; T, tumor size; N, node status; M, metastatic status) and tumor grade were also evaluated by OR with the corresponding 95% CI. In additional, stratified analyses were performed by material. The Cochran’s Q-statistic and I2 test were used to evaluate potential heterogeneity between studies [25,26]. If the Q-test showed a P<0.05 or I2 test exhibits >50%, indicating evidence of heterogeneity, then the random-effect model was conducted; otherwise the fixed-effect model was performed [27]. Sensitivity analysis was performed by deleting each study in turn to determine the influence of the individual data, confirming the stability and reliability of the result. Begger’s funnel plots was conducted as a visual aid to investigate the potential bias or systematic heterogeneity, and publication bias was further measured by Egger’s test (P<0.05 was considered statistically significant) [28]. STATA version 12.0 (Stata Corp, College Station, TX, USA) was used for this meta-analysis. All the P values were two-sided.
Results
Study characteristics
Base on the inclusion criteria, a total of ten eligible studies [29-38] involving 693 cases and 290 controls were included in this meta analysis (Figure 1). The features of these studies are summarized in Table 1. The studies originated from 9 countries or regions (including the Indian, Hong Kong, Poland, Finland, Taiwan, Japan, Germany, Israel and the Spain) and were published between 2002 and 2013. To the testing material, six of the studies evaluated the methylation status of p16 promoter in body fluids [33-38] and the other in tissues [29-32]. Bladder cancers were confirmed histologically or pathologically in all the studies.
Figure 1.
Flow diagram of study selection.
Table 1.
Characteristics of studies included in this meta-analysis
| First author | Year | Location | Study design | p16 (M/U)b | pTNMa (M/U)b | Grade (M/U)b | |||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| case | control | ≤T1 | ≥T2 | Low grade | High grade | ||||
| Seyed Ali Hosseini | 2010 | India | Case-control | 12/68 | 0/80 | 4/59 | 8/9 | - | - |
| Sonata Jarmalaite | 2008 | Finland | Case-control | 36/22 | 0/5 | 3/35 | 5/15 | 0/10 | 8/44 |
| Hui-Hui Lin | 2010 | Taiwan | Case-control | 20/37 | 0/20 | 19/13 | 13/8 | 9/10 | 23/15 |
| M.T. Valenzuela | 2002 | Spain | Case-control | 19/67 | 1/48 | 14/43 | 4/17 | 7/17 | 11/49 |
| Michael W.Y. Chan | 2002 | Hong Kong | Case-control | 26/72 | 0/11 | - | - | 2/7 | 1/12 |
| Zbigniew Jabłonowski | 2010 | Poland | Case-control | 17/25 | 0/36 | - | - | - | - |
| Tohru Nakagawa | 2005 | Japan | Case-control | 16/64 | 0/9 | 14/57 | 4/21 | - | - |
| Yasuhiro Tada | 2002 | Japan | Case-control | 6/49 | 0/5 | - | - | 1/4 | 5/45 |
| Michael B. Scher | 2012 | Israel | Case-control | 3/39 | 1/21 | 3/28 | 0/9 | 0/12 | 1/17 |
| Stefan Hauser | 2013 | Germany | Case-control | 33/62 | 9/44 | - | - | - | - |
pTNM (p, pathologic stage; T, tumor size; N, node status; M, metastatic status). Tumor grade ≤1 was defined as low-grade, and tumor grade ≥2 was defined as high-grade.
P, pathologic stage; T, tumor size; N, node status; M, metastatic status;
p16 methylated/p16 unmethylated.
Meta-analysis
Figure 2 shows the meta-analysis of the selected studies. The role of the of p16 methylation in bladder carcinogenesis were tested in ten case-control studies. The main results were summarized in Table 2. Since there was no obviously heterogeneity existed in all included studies (P=0.213, I2=25.0%), the fixed effects model was conducted. The meta-analysis results revealed that the methylation state of p16 was statistically significantly associated with an increased risk of bladder cancer (adjusted OR=6.71, 95% CI=3.79-11.87) (Figure 2A) compared to control. When it came to determine the association between p16 methylation and pTNM/grade in bladder cancer, each was carried out in six studies. The main results were also summarized in Table 2. Under the random-effects model, the pooled OR of pTNM in p16 methylated patients, compared to unmethylated patients was 0.59 (95% CI=0.22-1.60) (Figure 2B). Under the fixed-effects model, the pooled OR of tumor grade in p16 methylated patients, compared to unmethylated patients was 1.01 (95% CI=0.52-1.94) (Figure 2C). In the stratified analysis by material, significantly increased risks were found in tissues in detection p16 methylation in bladder cancer (OR=6.49, 95% CI=1.53-27.50) (Figure 2D) and in body fluids (OR=8.15, 95% CI=2.36-28.20) (Figure 2E).
Figure 2.

Forest plot of comparison. A: The frequency of p16 methylation between bladder cancer patients and non-cancer people; B and C: The association of the p16 methylation and patients’ pTNM and tumor grade respectively; D and E: The analyses of p16 methylation in tissues and body fluids respectively.
Table 2.
Stratified analyses of p16 methylation and bladder cancer risk
| Variable | pa | OR | 95% CI | Heterogeneity | |
|---|---|---|---|---|---|
|
| |||||
| P | I2 | ||||
| p16 | |||||
| Total | 10 | 6.71 | 3.79-11.87 | 0.213 | 25.0% |
| pTNM | 6 | 0.59 | 0.22-1.60 | 0.015 | 64.6% |
| Grade | 6 | 1.01 | 0.52-1.94 | 0.487 | 0% |
| Material | |||||
| Body fluids | 6 | 8.15 | 2.36-28.20 | 0.061 | 52.6% |
| Tissues | 4 | 6.49 | 1.53-27.50 | 0.686 | 0% |
Number of comparisons.
Quality assessment results
The details of the quality evaluation for each study are shown in Table 3. The average NOS score was 6.8. The most common biases were selection of controls and control for important factor.
Table 3.
Results of quality assessment by NOS for case-control studies
| Study | Selection | Comparability | Exposure | Scores | |||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | ||
| Chan et al. | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | ★ | 8 |
| Jarmalaite et al. | ★ | ★ | - | ★ | ★ | ★ | ★ | ★ | 6 |
| Tada et al. | ★ | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 7 |
| Nakagawa et al. | ★ | ★ | ★ | ★ | - | ★ | ★ | ★ | 6 |
| Lin et al. | ★ | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 7 |
| Jablonowski et al. | ★ | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 7 |
| Valenzuela et al. | ★ | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 7 |
| Ali et al. | ★ | ★ | ★ | ★ | ★★ | ★ | ★ | ★ | 8 |
| Hauser et al. | ★ | ★ | ★ | ★ | ★ | ★ | ★ | ★ | 7 |
| Scher et al. | ★ | ★ | - | ★ | - | ★ | ★ | ★ | 5 |
1. Adequate definition of cases; 2. Representativeness of cases; 3. Selection of controls; 4. Definition of controls; 5. Control for important factor; 6. Ascertainment of Exposure; 7. Same method to ascertain for cases and controls; 8. Non-response rate.
Sensitivity analyses
The influence of each individual study on the pooled OR was evaluated by sensitivity analysis which omitting each individual studies in turn to assess the quality and consistency of the results (Table 4). The results revealed that no single study exhibited excessive influence and the results of this meta-analysis are stable.
Table 4.
Sensitivity analysis after each study was excluded by turns
| Study omitted | OR (95% CI) for remainders | Heterogeneity | |
|---|---|---|---|
|
| |||
| I2 | P | ||
| Chan et al. (2002) | 6.56 (3.67, 11.74) | 32.7% | 0.156 |
| Jarmalaite et al. (2008) | 6.42 (3.58, 11.49) | 28.7% | 0.189 |
| Tada et al. (2002) | 7.04 (3.93, 12.60) | 29.3% | 0.185 |
| Nakagawa et al. (2005) | 6.81 (3.81, 12.19) | 33.7% | 0.149 |
| Lin et al. (2010) | 6.14 (3.42, 11.01) | 24.8% | 0.223 |
| Jablonowski et al. (2010) | 5.66 (3.13, 10.23) | 9.2% | 0.358 |
| Valenzuela et al. (2002) | 6.16 (3.40, 11.18) | 26.4% | 0.209 |
| Ali et al. (2010) | 5.97 (3.32, 10.74) | 20.7% | 0.259 |
| Hauser et al. (2013) | 11.93 (5.07, 28.04) | 0.0% | 0.559 |
| Scher et al. (2012) | 7.22 (3.99, 13.05) | 27.2% | 0.202 |
Publication bias
To the methylation comparison between bladder cancer patients and controls, the shape of the funnel plots seemed symmetry as shown in Figure 3, suggesting that there was no obvious asymmetry. Then, Egger’s test further confirm it by providing statistical evidence (P=0.05). What’s more, funnel plot were also conducted to evaluate the publication bias in studies of association between p16 methylation and pTNM/grade. There was no any evidence of obvious asymmetry was demonstrated through the shape of the funnel plot (figure not show) and Egger’s test suggested the absence of publication bias (pTNM: P=0.925; grade: P=0.91).
Figure 3.
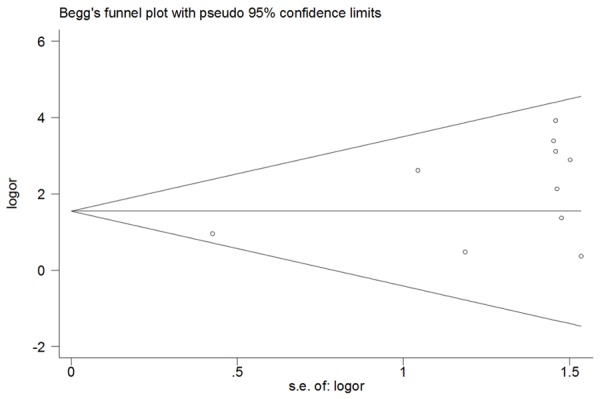
Begg’s funnel plots of the meta-analysis for publication bias in selection of studies on association between p16 methylation and bladder cancer.
Discussion
Ten studies were used in the present study to estimate overall results. The results of our meta-analysis indicated that p16 methylation in bladder cancer was associated with tumor risk as either detected in body fluids or tissues. But, the p16 methylation was not associated with increased risk for pathological features or the tumor grade of bladder cancer in comparison between p16 methylated bladder cancer patients and unmethylated patients.
Though the development of bladder cancer biomarker has advanced rapidly over the last decades, it has not yet been able to produce a significant influence on the diagnosis and management of the disease. Many researchers agree that hypermethylation is an excellent marker for cancer diagnosis and treatment target since it is among the earliest and most common phenomenon known to appear in human cancers [39]. Methylation of CpG islands, for example, happens in the promoter regions, which was observed in various types of human tumors. As a consequence, hypermethylation plays an important role in the induction of the cancer genesis process because it can decrease the transcription activity of specific genes. Esteller et al. indicated that even up to 65% of cases of neoplastic disease may be linked to these epigenetic changes [40]. Chromosome region 9p21 harbors genes p16INK4a, which have growth inhibition activities. Herman et al. have found that the hypermethylation of promoter regions mainly involves genes responsible for control of the cell cycle and apoptosis [41]. Previous reports also demonstrated that bladder cancer patients always show p16 methylation [42]. To further confirm the status of p16 promoter methylation in bladder cancer patient’s diagnosis, we carried out a meta-analysis to conduct a more accurate estimation of the association. The frequency of p16 methylation in bladder cancer patients was 7.13 times higher than that in non-cancer people, suggesting that p16 methylation may be a potential risk factor for bladder cancer.
The traditional material used by researchers to estimate the degree of DNA hypermethylation is neoplastic tissues. The noninvasive detection of DNA hypermethylation in serum/plasma and urine samples can improves the effectiveness of therapies for reducing morbidity and mortality in urological and other tumor diseases, especially in early stage cancer [43]. Michael et al. and Lin et al. reported that the methylation of p16 promoter can be detected in the bladder cancer patients’ urine samples. What’s more, their results indicated that detection of bladder cancer in urine using methylation markers appeared to be more sensitive than conventional urine cytology, especially in low grade cases [29,33]. When it comes to serum, Valenzuela et al. reported that there is a statistically close relationship between methylation of p16 in tumor and in serum matched samples and its identification in serum is strongly related with the aberrant methylation of p16 in tumor and with cancer diagnosis [35]. In this study, the results also confirmed that p16 methylation is a potential risk factor for bladder cancer as detected both in body fluids, suggesting that serum/plasma could be potential material for detecting bladder cancer.
Sequential studies have demonstrated that the methylation of p16 may be related with advanced stage and tumor metastasis [36,44]. However, some reports indicated that there was no association between the methylation status of p16 promoter and grading or muscle invasiveness [29,33,35]. In order to deal with the contradictory results, we also carried out a meta-analysis which indicated that the frequency of p16 methylation did not correlate with the pTNM or tumor grade of bladder cancer patients, suggesting the inactivation of p16 may be an early event in bladder cancer.
Some limitations of the present meta-analysis should be mentioned. First, only a small number of publications met the criteria for the present meta-analysis, so the number of included studies was relatively small and may not provide sufficient statistical power to explore the real association of methylation and pTNM/grade, and to estimate the diagnostic value of body fluids and tumor tissues in detecting p16 methylation in bladder cancer. Hence, more studies with larger sample size are still needed to accurately provide a more representative statistical analysis. Then, similar to other hospital-based case-control studies, the control subjects in our study may not be representative of the general population. Finally, since our results were based on unadjusted estimates, which may cause serious confounding bias to the data analysis, a more precise analysis needs to be performed if individual data such as age and sex are available.
In conclusion, our meta-analysis indicated that detection of p16 methylation in body fluids, such as serum and urine, is a potential non-invasive diagnostic tool in bladder cancer and the inactivation of p16 may be an early event in bladder cancer. It is necessary to conduct large sample size studies of the association between p16 methylation and bladder cancer risk, eventually leading to our better understanding.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (No. 81472999, No. 81272350), The Key Natural Science Foundation of Guangdong (N0. 2015A030311038; W Ji) the Program of Health Department of Guangdong province (No. C2013025).
Disclosure of conflict of interest
None.
References
- 1.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Jerónimo C, Henrique R. Epigenetic biomarkers in urological tumors: A systematic review. Cancer Lett. 2014;342:264–274. doi: 10.1016/j.canlet.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 4.Brown AJ, Zietman AL, Shipley WU, Kaufman DS. An organ-preserving approach to muscle-invading transitional cell cancer of the bladder. Hematol Oncol Clin North Am. 2001;15:345–358. doi: 10.1016/s0889-8588(05)70216-1. [DOI] [PubMed] [Google Scholar]
- 5.Laird PW, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 6.Jones PA. DNA methylation errors and cancer. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 7.Liu L, Wylie RC, Andrews LG, Tollefsbol TO. Aging, cancer and nutrition: the DNA methylation connection. Mech Ageing Dev. 2003;124:989–998. doi: 10.1016/j.mad.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Davis CD, Uthus EO. DNA methylation, cancer susceptibility, and nutrient interactions. Exp Biol Med (Maywood) 2004;229:988–995. doi: 10.1177/153537020422901002. [DOI] [PubMed] [Google Scholar]
- 9.Scarano MI, Strazzullo M, Matarazzo MR, D’Esposito M. DNA methylation 40 years later: Its role in human health and disease. J Cell Physiol. 2005;204:21–35. doi: 10.1002/jcp.20280. [DOI] [PubMed] [Google Scholar]
- 10.Maruyama R, Toyooka S, Toyooka KO, Harada K, Virmani AK, Zochbauer-Muller S, Farinas AJ, Vakar-Lopez F, Minna JD, Sagalowsky A, Czerniak B, Gazdar AF. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 2001;61:8659–8663. [PubMed] [Google Scholar]
- 11.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 12.Brown I, Milner BJ, Rooney PH, Haites NE. Inactivation of the p16INK4A gene by methylation is not a frequent event in sporadic ovarian carcinoma. Oncol Rep. 2001;8:1359–1362. doi: 10.3892/or.8.6.1359. [DOI] [PubMed] [Google Scholar]
- 13.Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- 14.Serrano M. The tumor suppressor protein p16INK4a. Exp Cell Res. 1997;237:7–13. doi: 10.1006/excr.1997.3824. [DOI] [PubMed] [Google Scholar]
- 15.Nishikawa R, Furnari FB, Lin H, Arap W, Berger MS, Cavenee WK, Su HH. Loss of P16INK4 expression is frequent in high grade gliomas. Cancer Res. 1995;55:1941–1945. [PubMed] [Google Scholar]
- 16.Kees UR, Burton PR, Lu C, Baker DL. Homozygous deletion of the p16/MTS1 gene in pediatric acute lymphoblastic leukemia is associated with unfavorable clinical outcome. Blood. 1997;89:4161–4166. [PubMed] [Google Scholar]
- 17.Yoo CB, Jeong S, Egger G, Liang G, Phiasivongsa P, Tang C, Redkar S, Jones PA. Delivery of 5-aza-2’-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res. 2007;67:6400–6408. doi: 10.1158/0008-5472.CAN-07-0251. [DOI] [PubMed] [Google Scholar]
- 18.Bernal C, Aguayo F, Villarroel C, Vargas M, Diaz I, Ossandon FJ, Santibanez E, Palma M, Aravena E, Barrientos C, Corvalan AH. Reprimo as a potential biomarker for early detection in gastric cancer. Clin Cancer Res. 2008;14:6264–6269. doi: 10.1158/1078-0432.CCR-07-4522. [DOI] [PubMed] [Google Scholar]
- 19.Sharma G, Mirza S, Prasad CP, Srivastava A, Gupta SD, Ralhan R. Promoter hypermethylation of p16INK4A, p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast cancer patients. Life Sci. 2007;80:1873–1881. doi: 10.1016/j.lfs.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Chim CS, Kwong YL, Liang R. Gene hypermethylation in multiple myeloma: lessons from a cancer pathway approach. Clin Lymphoma Myeloma. 2008;8:331–339. doi: 10.3816/CLM.2008.n.048. [DOI] [PubMed] [Google Scholar]
- 21.Williams RT, Sherr CJ. The INK4-ARF (CDKN2A/B) locus in hematopoiesis and BCRABL-induced leukemias. Cold Spring Harb Symp Quant Biol. 2008;73:461–467. doi: 10.1101/sqb.2008.73.039. [DOI] [PubMed] [Google Scholar]
- 22.Gallagher SJ, Thompson JF, Indsto J, Scurr LL, Lett M, Gao BF, Dunleavey R, Mann GJ, Kefford RF, Rizos H. p16INK4a expression and absence of activated B-RAF are independent predictors of chemosensitivity in melanoma tumors. Neoplasia. 2008;10:1231–1239. doi: 10.1593/neo.08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan C, He L, Kapoor A, Gillis A, Rybak AP, Cutz JC, Tang D. Bmi1 promotes prostate tumorigenesis via inhibiting p16(INK4A) and p14(ARF) expression. Biochim Biophys Acta. 2008;1782:642–648. doi: 10.1016/j.bbadis.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–605. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 25.Zintzaras E, Ioannidis JP. HEGESMA: genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–3673. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 26.Jackson D, White IR, Riley RD. Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med. 2012;31:3805–3820. doi: 10.1002/sim.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 28.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan MW, Chan LW, Tang NL, Tong JH, Lo KW, Lee TL, Cheung HY, Wong WS, Chan PS, Lai FM, To KF. Hypermethylation of multiple genes in tumor tissues and voided urine in urinary bladder cancer patients. Clin Cancer Res. 2002;8:464–470. [PubMed] [Google Scholar]
- 30.Jarmalaite S, Jankevicius F, Kurgonaite K, Suziedelis K, Mutanen P, Husgafvel-Pursiainen K. Promoter hypermethylation in tumour suppressor genes shows association with stage, grade and invasiveness of bladder cancer. Oncology. 2008;75:145–151. doi: 10.1159/000158665. [DOI] [PubMed] [Google Scholar]
- 31.Tada Y, Wada M, Taguchi K, Mochida Y, Kinugawa N, Tsuneyoshi M, Naito S, Kuwano M. The association of death-associated protein kinase hypermethylation with early recurrence in superficial bladder cancers. Cancer Res. 2002;62:4048–4053. [PubMed] [Google Scholar]
- 32.Nakagawa T, Kanai Y, Ushijima S, Kitamura T, Kakizoe T, Hirohashi S. DNA hypermethylation on multiple CpG islands associated with increased DNA methyltransferase DNMT1 protein expression during multistage urothelial carcinogenesis. J Urol. 2005;173:1767–1771. doi: 10.1097/01.ju.0000154632.11824.4d. [DOI] [PubMed] [Google Scholar]
- 33.Lin HH, Ke HL, Huang SP, Wu WJ, Chen YK, Chang LL. Increase sensitivity in detecting superficial, low grade bladder cancer by combination analysis of hypermethylation of E-cadherin, p16, p14, RASSF1A genes in urine. Urol Oncol. 2010;28:597–602. doi: 10.1016/j.urolonc.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Jablonowski Z, Reszka E, Gromadzinska J, Wasowicz W, Sosnowski M. Hypermethylation of p16 and DAPK promoter gene regions in patients with non-invasive urinary bladder cancer. Arch Med Sci. 2011;7:512–516. doi: 10.5114/aoms.2011.23421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valenzuela MT, Galisteo R, Zuluaga A, Villalobos M, Núñez MI, Oliver FJ, Ruiz de Almodóvar JM. Assessing the use of p16(INK4a) promoter gene methylation in serum for detection of bladder cancer. Eur Urol. 2002;42:622–628. doi: 10.1016/s0302-2838(02)00468-2. [DOI] [PubMed] [Google Scholar]
- 36.Ali HS, Sobti RC, Malekzadeh K, Singh SK, Joshi K. Frequency of P16INK4a and P14ARF genes methylation and its impact on bladder cancer cases in north Indian population. Dis Markers. 2010;28:361–368. doi: 10.3233/DMA-2010-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauser S, Kogej M, Fechner G, VON Pezold J, Vorreuther R, Lummen G, Muller SC, Ellinger J. Serum DNA hypermethylation in patients with bladder cancer: results of a prospective multicenter study. Anticancer Res. 2013;33:779–784. [PubMed] [Google Scholar]
- 38.Scher MB, Elbaum MB, Mogilevkin Y, Hilbert DW, Mydlo JH, Sidi AA, Adelson ME, Mordechai E, Trama JP. Detecting DNA methylation of the BCL2, CDKN2A and NID2 genes in urine using a nested methylation specific polymerase chain reaction assay to predict bladder cancer. J Urol. 2012;188:2101–2107. doi: 10.1016/j.juro.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 39.Vucic EA, Brown CJ, Lam WL. Epigenetics of cancer progression. Pharmacogenomics. 2008;9:215–234. doi: 10.2217/14622416.9.2.215. [DOI] [PubMed] [Google Scholar]
- 40.Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. Cancer Res. 1999;59:67–70. [PubMed] [Google Scholar]
- 41.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 42.Dominguez G, Silva J, Garcia JM, Silva JM, Rodriguez R, Munoz C, Chacon I, Sanchez R, Carballido J, Colas A, Espana P, Bonilla F. Prevalence of aberrant methylation of p14ARF over p16INK4a in some human primary tumors. Mutat Res. 2003;530:9–17. doi: 10.1016/s0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 43.Anker P, Mulcahy H, Chen XQ, Stroun M. Detection of circulating tumour DNA in the blood (plasma/serum) of cancer patients. Cancer Metastasis Rev. 1999;18:65–73. doi: 10.1023/a:1006260319913. [DOI] [PubMed] [Google Scholar]
- 44.Kawamoto K, Enokida H, Gotanda T, Kubo H, Nishiyama K, Kawahara M, Nakagawa M. p16INK4a and p14ARF methylation as a potential biomarker for human bladder cancer. Biochem Biophys Res Commun. 2006;339:790–796. doi: 10.1016/j.bbrc.2005.11.072. [DOI] [PubMed] [Google Scholar]


