Abstract
Background: MiRNAs might function as oncogenes or tumor suppressor genes in the tumorigenesis process. Dysregulation of miR-345 is a frequent event in many types of human cancers. However, the tissue miR-345 expression level in non-small cell lung cancer (NSCLC) and its potential clinical significance remains unknown. Materials and methods: Real-time PCR was conducted to evaluate the expression level of miR-345 in NSCLC tissues as well as cell lines. Then the association between tissue miR-345 expression level and clinical outcome was investigated. Results: The expression level of miR-345 was significantly decreased in NSCLC tissues and cell lines compared with the controls (P<0.05; P<0.01). Tissue miR-345 expression level was associated with various clinicopathological parameters including LN metastasis (P=0.012), distant metastasis (P=0.007), TNM stage (P=0.008) and grade (P=0.030). In addition, the NSCLC patients in thelow tissue miR-345 expression group had significantly shorter 5-year overall survival time than those in the high tissue miR-345expression group (P=0.016). Multivariate analysis showed that tissue miR-345 was an independent risk factor for NSCLC (HR=3.921, 95% CI: 2.285-10.540; P=0.008). Conclusions: The expression level of miR-345 was reduced in NSCLC tissues and cell lines. Low tissue miR-345 expression was associated with progression and poor prognosis of NSCLC, indicating that tissue miR-345 may serve as a novel prognostic marker in NSCLC.
Keywords: Biomarker, MiR-345, non-small cell lung cancer, prognosis
Introduction
Non-small cell lung cancer (NSCLC), accounting for approximately 80% of all lung cancer, is the leading cause of cancer-related mortality worldwide [1]. Despite progress in surgical techniques, chemoradio therapy as well as target therapy has significantly improved the clinical outcome of patients with lung cancer; the five-year survival rate of NSCLC patients remains poor [2]. Therefore, it is important to screen biomarkers with high specificity and sensitivity for early detecting NSCLC and predicting the prognosis of this malignant disease.
MicroRNAs (miRNAs) are a class of small non-coding RNAs and considered asmaster regulators of gene expression [3]. Altered expression of miRNAs contributes to the initiation and development of many human diseases including cancer; and some miRNAs have shown great promising for lung cancer detection, diagnosis and treatment [4,5]. Nadal et al. compared the serum miRNAs expression profile between patients with NSCLC and healthy volunteers. A large number of differently expressed serum miRNAs was identified and combination of four selected miRNAs (miR-193b, miR-301, miR-141 and miR-200b) could discriminate NSCLC patients from healthy controls with high accuracy [6]. Recently Mavridiset al. reported that the expression level of miR-197 was upregulated in NSCLC tissues and it was associated with tumor size as well as histotype. In addition, miR-197 overexpression was an independent predictor of unfavorable prognosis for NSCLC; indicating miR-197 might function as an oncogene in NSCLC [7].
Dysregulation of miR-345 is a common feature in many types of cancers such as prostate cancer, colorectal cancer andacute lymphocytic leukemia [8-10]. However, the expression profile of tissue miR-345 in patients with NSCLC is unknown. Thus the aim of the present study was to elucidate the miR-345 expression level in NSCLC and its potential clinical significance.
Materialsand methods
Study population and clinical samples
Clinical tissue samples were obtained fromDepartment of Oncology, The First Affiliated Hospital of Shantou University Medical College. Written informed consent was obtained from all participants and the study was approved by the Ethics Committee of The First Affiliated Hospital of Shantou University Medical College. All the patients with NSCLC werepathological confirmed and diagnosed. NSCLC was staged according to the sixth edition of the American Joint Committee on Cancer (AJCC) TNM staging system for lung cancer. Overall survival was defined as the time interval from the date of diagnosis at our department to the date of death or the last follow-up. The clinical data of the NSCLC patients were summarized in Table 1.
Table 1.
Tissue miR-345 expression and clinicopathological characteristics in NSCLC
| Parameters | Group | Total | Tissue miR-345 | P | |
|---|---|---|---|---|---|
|
|
|||||
| Low | High | ||||
| Age | ≤60 | 45 | 22 | 23 | 0.441 |
| >60 | 42 | 24 | 18 | ||
| Gender | Female | 38 | 21 | 17 | 0.694 |
| Male | 49 | 25 | 24 | ||
| LN metastasis | No | 56 | 24 | 32 | 0.012 |
| Yes | 31 | 22 | 9 | ||
| Distant metastasis | No | 70 | 32 | 38 | 0.007 |
| Yes | 17 | 14 | 3 | ||
| Surgery margins | free | 68 | 33 | 35 | 0.125 |
| Not free | 19 | 13 | 6 | ||
| TNM stage | Ι-II | 53 | 22 | 31 | 0.008 |
| III-IV | 34 | 24 | 10 | ||
| Grade | Well/Moderate | 51 | 22 | 29 | 0.030 |
| Poor | 36 | 24 | 12 | ||
| Family history | No | 81 | 44 | 37 | 0.320 |
| Yes | 6 | 2 | 4 | ||
Cell culture
Three NSCLC cell lines (A549, H157, and H460) and one normal control lung cell line (MRC-9) wereobtained from AmericanType Culture Collection (ATCC). The basic culture medium for NSCLC cell lines was RPMI 1640 (Invitrogen, Carlsbad, CA) and MRC-9 was grown in α-MEM. The two basic culture media were supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. All cell lines were stored in incubators at 37°C with 5% CO2.
Real-time PCR
Total RNA was extracted from tissues and cell lines using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Briefly, 20 μL of total RNA samplewas transcribed to cDNA using miScript-II-RT-Kit (Qiagen, Germany). Then real-time PCR was performed with the Applied Biosystems prism 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Reactions were performed in triplicateand the miRNA relative expression was calculated using a 2-ΔΔCt method. RNU6 was used as an endogenous control.
Statistical analysis
Mann-Whitney U-test was performed to compare the expression level of miR-345 between NSCLC tissues and adjacent normal tissues. The expression level of miR-345 among different cell lines was evaluated using one-way ANOVA. The association between tissue miR-345 expression level and clinicopathological parameters of NSCLC was evaluated by Chi-square test. The overall survival was analyzed by log-rank test, and survival curve was plotted based on Kaplan-Meier method. Univariate and multivariate analyses were conducted to explore the independent risk factors for NSCLC using the Cox proportional hazard model. All statistical analyses were performed using SPSS 21.0 software (SPSS Inc., Chicago, IL, USA), and P values of <0.05 were considered significant.
Results
The expression level of miR-345 in NSCLC tissues and cell lines
Real-time PCR was performed to compare the miR-345 expression level in NSCLC tissues and cell lines. The results showed that the expression level of miR-345 was significantly reduced in NSCLC tissues compared with the adjacent normal tissues (P<0.01) (Figure 1). In addition, miR-345 was downregulated in all three NSCLC cell lines relative to normal human fibroblast cells (P<0.05; P<0.01) (Figure 2).
Figure 1.

The expression level of miR-345 in NSCLC tissues.
Figure 2.
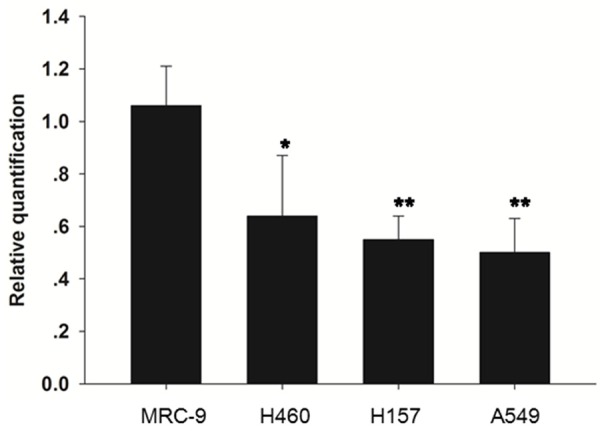
Theexpression level of miR-345 in NSCLC cell lines.
The association between tissue miR-345 and clinical parameters of NSCLC
The median expression level of tissue miR-345 (0.58 fold) was used as the cutoff value to divide the 87 NSCLC patients into high tissue miR-345 expression group and low tissue miR-345 expression group. Our results showed thattissue miR-345 expression level was associated with various clinicopathological parameters including LN metastasis (P=0.012), distant metastasis (P=0.007), TNM stage (P=0.008) and grade (P=0.030). However, it was not correlated with age (P=0.441), gender (P=0.694), surgery margins (P=0.125) and family history (P=0.320) (Table 1).
Overall survival analysis
The mean overall survival time of NSCLC patients in thehigh tissue miR-345 expression group was 45.78±3.20 months, which was significantly longer than that of the patients in the lowtissue miR-345 expression group (37.87±3.09 months) (P=0.016) (Figure 3).
Figure 3.
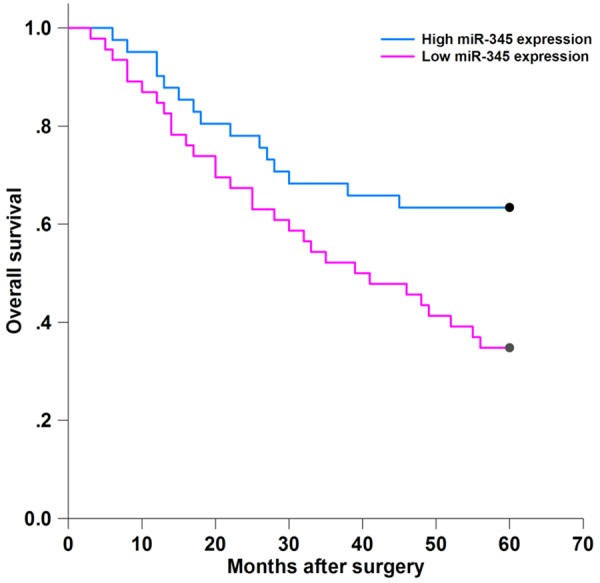
The association between overall survival and tissue miR-345 expression.
Tissue miR-345 was an independent risk factor for NSCLC
Univariate analysis showed that LN metastasis (HR=2.823, 95% CI: 1.393-5.709; P=0.026), distant metastasis (HR=3.562, 95% CI: 1.830-7.814; P=0.009), TNM stage (HR=4.067, 95% CI: 2.535-9.952; P=0.005) and tissue miR-345 expression level (HR=3.398, 95% CI: 1.763-7.260; P=0.011) was significantly correlated with worse overall survival (Table 2).
Table 2.
Univariate and multivariate analyses for overall survival by Cox regression model
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Age | 1.014 | 0.281-1.635 | 0.810 | |||
| Gender | 1.473 | 0.544-2.162 | 0.502 | |||
| LN metastasis | 2.823 | 1.393-5.709 | 0.026 | 3.027 | 1.721-7.039 | 0.034 |
| Distant metastasis | 3.562 | 1.830-7.814 | 0.009 | 3.653 | 2.350-8.931 | 0.013 |
| Surgery margins | 1.215 | 0.687-1.891 | 0.628 | |||
| TNM stage | 4.067 | 2.535-9.952 | 0.005 | 4.815 | 2.960-12.851 | 0.002 |
| Grade | 2.058 | 0.975-3.857 | 0.054 | |||
| Family history | 1.793 | 0.611-2.925 | 0.124 | |||
| Tissue miR-345 | 3.398 | 1.763-7.260 | 0.011 | 3.921 | 2.285-10.540 | 0.008 |
Multivariate analysis revealed thatLN metastasis (HR=3.027, 95% CI: 1.721-7.039; P=0.034), distant metastasis (HR=3.653, 95% CI: 2.350-8.931; P=0.013), TNM stage (HR=4.815, 95% CI: 2.960-12.851; P=0.002) and tissue miR-345 expression level (HR=3.921, 95% CI: 2.285-10.540; P=0.008) were independent risk prognostic factors for NSCLC (Table 2).
Discussion
MicroRNAs have increasinglybeen recognized as major players in the development of cancer [11,12]. Mutation or aberrantexpression of microRNAs is a common feature in patients with lung cancer, thus miRNA-based therapy has great potential to improve the clinical outcome of this malignant disease [13,14]. Alterations of miR-345 might be associated with cancer, systemic lupus erythematosus and status epilepticus [15,16]. Our study showed that the expression level of miR-345 was decreased in NSCLC tissues and cell lines. In addition, tissue miR-345 expression was correlated with various important NSCLC clinical parameters including LN metastasis, distant metastasis, TNM stage and grade. Moreover, the NSCLC patients with lower tissue miR-345 expression had shorter OS. Downregulation of tissue miR-345 was an independent predictor for poor survival in NSCLC. The findings from the current study suggested that miR-345 might play a tumor suppressive role in the development of NSCLC. As tissue miR-345 expression was closely associated with the clinical outcome of NSCLC, it may therefore be a promising diagnostic and prognostic biomarker for this malignance. Similar to our observations, Chen et al. showed that ectopic expression of miR-345 could suppress the proliferation, migration and invasion capacity of prostate cancer cells both in vivo and in vitro. In addition, Smad1 was identified as a direct target of miR-345; indicating miR-345 might act as a tumor suppressor gene in prostate cancer [17]. Srivastava et al. reported that the expression level of miR-345 was significantly reduced in pancreatic cancer (PC) tissues and cell lines in comparison with the controls. Forced expression of miR-345 in vitro suppressed the proliferation of PC cells byinducing apoptosis and BCL-2 was validated as a target of miR-345, suggesting restoration of miR-345 might be a practical and effective method for treating PC [18]. Similarly, miR-345 was demonstrated to be a methylation-sensitive miRNA and it was downregulated in colorectal cancer (CRC) tissues. In addition, reduced expression of miR-345 was correlated with associated with lymph node metastasis and worse histological type. Overexpression of miR-345 could inhibit the proliferation and invasion capacity of CRC cell lines by targeting BCL2-associated athanogene 3. Thus miR-345 may be a negative regulator of CRC tumorigenesis [19].
However, miR-345 might also function as an oncogene in the carcinogenesis process. Schou et al. showed that high miR-345 expression in whole blood wasa prognostic biomarker for poor overall survival and progression-free survival of patients with CRC. Moreover, upregulated expression of miR-345 was associated with lack of response to treatment with cetuximab and irinotecan [20]. MiR-345 may be an oncogene in CRC, which was inconsistent with the results reported by Tang and his colleagues [19]. Higher expression of miR-345 was observed in oral leukoplakia tissues with an increased number and size of nucleoli or increased nuclear/cytoplasmic ratio compared with the normal oral mucosa, suggesting that miR-345 might play an important role in promoting the oral precancerous lesions progression into oral squamous cell carcinoma [21]. Guled et al. compared the differentially expressed miRNAs between malignant mesothelioma tissues and normal samples. The expression level of tissue miR-345 was found to be significantly upregulated in malignant mesothelioma, indicating miR-345 might promote the progression of this malignance [22]. Two major reasons might be accounting for the contradictory role of miR-345 in different types of cancers or even in the same type of cancer. Firstly,one miRNA may regulate many genes as its targets and the concrete function of miRNA depends on its target genes, thus a specific miRNA might inhibit tumor progression in one type of cancer, while promote tumorigenesis in another. In addition, the biological function of miR-345 might be associated with tumor microenvironment and each type of cancer has its own specific microenvironment.
There are some limitations in our study. Firstly, the sample size is relatively small, large clinical trials are needed to conduct. Secondly, we fail to evaluate the expression profile of miR-345 in the serum/plasma samples in patients with NSCLC. Examining the biomarkers in the serum/plasma samples might help monitor the therapy response in real-time. Thirdly, the role of miR-345 in the regulation of NSCLC tumorigenesis at the cellular and molecular level is poorly known.
Conclusion
The expression level of tissue miR-345 was significantly decreased in patients with NSCLC. Reduced tissue miR-345 expression was correlated with development and poor prognosis of NSCLC, indicating miR-345 acted as a tumor suppressor in this deadly disease and might be a promising biomarker for predicting NSCLC progression.
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Wink KC, Roelofs E, Solberg T, Lin L, Simone CB 2nd, Jakobi A, Richter C, Lambin P, Troost EG. Particle therapy for non-small cell lung tumors: where do we stand? A systematic review of the literature. Front Oncol. 2014;4:292. doi: 10.3389/fonc.2014.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 4.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, Su TJ, Chiang CC, Li HN, Hong QS, Su HY, Chen CC, Chen WJ, Liu CC, Chan WK, Chen WJ, Li KC, Chen JJ, Yang PC. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13:48–57. doi: 10.1016/j.ccr.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Nadal E, Truini A, Nakata A, Lin J, Reddy RM, Chang AC, Ramnath N, Gotoh N, Beer DG, Chen G. A novel serum 4-microRNA signature for lung cancer detection. Sci Rep. 2015;5:12464. doi: 10.1038/srep12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mavridis K, Gueugnon F, Petit-Courty A, Courty Y, Barascu A, Guyetant S, Scorilas A. The oncomiR miR-197 is a novel prognostic indicator for non-small cell lung cancer patients. Br J Cancer. 2015;112:1527–1535. doi: 10.1038/bjc.2015.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang SY, Shiboski S, Belair CD, Cooperberg MR, Simko JP, Stoppler H, Cowan J, Carroll PR, Blelloch R. miR-19, miR-345, miR-519c-5p serum levels predict adverse pathology in prostate cancer patients eligible for active surveillance. PLoS One. 2014;9:e98597. doi: 10.1371/journal.pone.0098597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaur S, Lotsari JE, Al-Sohaily S, Warusavitarne J, Kohonen-Corish MR, Peltomäki P. Identification of subgroup-specific miRNA patterns by epigenetic profiling of sporadic and Lynch syndrome-associated colorectal and endometrial carcinoma. Clin Epigenetics. 2015;7:20. doi: 10.1186/s13148-015-0059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Li D, Zhuang Y, Shi Q, Wei W, Ju X. Overexpression of miR-708 and its targets in the childhood common precursor B-cell ALL. Pediatr Blood Cancer. 2013;60:2060–2067. doi: 10.1002/pbc.24583. [DOI] [PubMed] [Google Scholar]
- 11.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett. 2009;285:116–126. doi: 10.1016/j.canlet.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 13.Pichler M, Calin GA. MicroRNAs in cancer: from developmental genes in worms to their clinical application in patients. Br J Cancer. 2015;113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue W, Dahlman JE, Tammela T, Khan OF, Sood S, Dave A, Cai W, Chirino LM, Yang GR, Bronson R, Crowley DG, Sahay G, Schroeder A, Langer R, Anderson DG, Jacks T. Small RNA combination therapy for lung cancer. Proc Natl Acad Sci U S A. 2014;111:E3553–3561. doi: 10.1073/pnas.1412686111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun Z, Yu JT, Jiang T, Li MM, Tan L, Zhang Q, Tan L. Genome-wide microRNA profiling of rat hippocampus after status epilepticus induced by amygdala stimulation identifies modulators of neuronal apoptosis. PLoS One. 2013;8:e78375. doi: 10.1371/journal.pone.0078375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Ramos R, García-Lozano JR, Lucena JM, Castillo-Palma MJ, García-Hernández F, Rodríguez MC, Núñez-Roldán A, González-Escribano MF. Differential expression pattern of microRNAs in CD4+ and CD19+ cells from asymptomatic patients with systemic lupus erythematosus. Lupus. 2014;23:353–359. doi: 10.1177/0961203314522335. [DOI] [PubMed] [Google Scholar]
- 17.Chen QG, Zhou W, Han T, Du SQ, Li ZH, Zhang Z, Shan GY, Kong CZ. MiR-345 suppresses proliferation, migration and invasion by targeting Smad1 in human prostate cancer. J Cancer Res Clin Oncol. 2015 doi: 10.1007/s00432-015-2016-0. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srivastava SK, Bhardwaj A, Arora S, Tyagi N, Singh S, Andrews J, McClellan S, Wang B, Singh AP. MicroRNA-345 induces apoptosis in pancreatic cancer cells through potentiation of caspase-dependent and -independent pathways. Br J Cancer. 2015;113:660–668. doi: 10.1038/bjc.2015.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang JT, Wang JL, Du W, Hong J, Zhao SL, Wang YC, Xiong H, Chen HM, Fang JY. MicroRNA 345, a methylation-sensitive microRNA is involved in cell proliferation and invasion in human colorectal cancer. Carcinogenesis. 2011;32:1207–1215. doi: 10.1093/carcin/bgr114. [DOI] [PubMed] [Google Scholar]
- 20.Schou JV, Rossi S, Jensen BV, Nielsen DL, Pfeiffer P, Høgdall E, Yilmaz M, Tejpar S, Delorenzi M, Kruhøffer M, Johansen JS. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS One. 2014;9:e99886. doi: 10.1371/journal.pone.0099886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brito JA, Gomes CC, Guimarães AL, Campos K, Gomez RS. Relationship between microRNA expression levels and histopathological features of dysplasia in oral leukoplakia. J Oral Pathol Med. 2014;43:211–216. doi: 10.1111/jop.12112. [DOI] [PubMed] [Google Scholar]
- 22.Guled M, Lahti L, Lindholm PM, Salmenkivi K, Bagwan I, Nicholson AG, Knuutila S. CDKN2A, NF2, and JUN are dysregulated among other genes by miRNAs in malignant mesothelioma-A miRNA microarray analysis. Genes Chromosomes Cancer. 2009;48:615–623. doi: 10.1002/gcc.20669. [DOI] [PubMed] [Google Scholar]


