Abstract
Background: Several single nucleotide polymorphisms (SNPs), rs16892766 in the 8q23.3 region and rs6983267, rs10505477, rs7014346 and rs7837328 in the 8q24.21 region, have been identified by genome-wide association studies (GWAS) and a number of case-control studies to be closely associated with risk of colorectal cancer (CRC). In the present study, a meta-analysis was performed to confirm if these loci are risk factors for susceptibility to CRC, taking heterogeneity of population into consideration. Methods: The whole literature search was conducted via database of MEDLINE and Embase, through which 33 articles with 49 studies (141,899 cases and 157,536 controls) were finally included in this meta-analysis to evaluate the association between the 5 polymorphisms and risk of CRC under allelic model. Results: A meta-analysis of the pooled data showed that the G allele of rs6983267, the A allele of rs7014346, the T allele of rs10505477, the C allele of rs16892766 and the A allele of rs7837328 were associated with significantly increased risk of CRC under allelic model. Additionally, subgroup analyses of four SNPs (rs7837328 excluded) by ethnicity witnessed a notable association between the G allele of rs6983267 and increased risk of CRC among Caucasians, Asians and Africans. Furthermore, the C allele of rs16892766 was strongly linked with elevated risk of CRC among Caucasians and Africans. However, the A allele of rs7014346 and T allele of rs10505477 only heightened risk for CRC among Caucasians and showed no effects among Asians. Conclusion: In summary, rs6983267 is a risk factor for CRC among Caucasians, Asians and Africans; rs7014346 and rs10505477 are risky genetic polymorphisms only among Caucasians; rs16892766 is a hazardous element among populations with Caucasian and African ancestry; and rs7837328 could elevate the susceptibility to CRC in a multinational group. However, more potential factors related with CRC risk should be investigated in further studies.
Keywords: Colorectal cancer (CRC), 8q23.3, 8q24.21, single nucleotide polymorphism (SNP), genome-wide association studies (GWAS), meta-analysis
Introduction
Colorectal cancer (CRC), a malignant cancer developing between dentate line and rectosigmoid junction within the digestive tract, has been regarded as the fourth culprit for cancer-related mortality worldwide [1], injuring 1.23 million people and generating 0.6 million deaths globally [2]. While incremental risk of CRC could be explained partially by lifestyle factors (smoking, high protein and fat-rich diet, shortage of exercise etc.), genetic disorders also contribute to 35% of CRC cases as demonstrated by twin- and family- based studies [3]. High penetrance genes (DNA mismatch repair genes, APC, SMAD4, BMPR1A, MUTYH and STK11) are estimated to explain < 5% of total CRC, while much of the remaining genetic variation may be owing to multiple common alleles with low penetrance [2]. Until now, however, the molecular basis of CRC is still obscure, even more than 90% of the genetic pathogenesis for CRC remains unclear [4]. In fact, a significant aspect of the hereditary predisposition to CRC lies in the presence of single nucleotide polymorphisms (SNPs) [3].
Previously published genome-wide association studies (GWAS) have identified 5 SNPs located in the 8q24.21 (rs6983267, rs10505477, rs7837328, rs10505477) or 8q23.3 (rs16892766) chromosome region to show strong associations with the development of CRC [5-9]. Moreover, strong linkage disequilibrium (LD) were observed between rs6983267 and rs10505477 [10], rs6983267 and rs7837328 (r2 = 0.71 among Caucasian population) [11], rs6983267 and rs7014346 (r2 = 0.55 among European-American population) [12]; rs16892766 was also found to be in a high LD region [13]. Therefore, the 5 SNPs were selected as representative polymorphisms on the association studies of 8q23-24 region with risk of CRC.
Several replication studies targeting diverse ethnicities (British, American, Japanese, Chinese etc.) have also confirmed the association of 5 loci mentioned above with susceptibility to CRC [9,14-16]. Nonetheless, heterogeneity of populations in certain studies makes it tough to deem 5 loci as risk factors for susceptibility to CRC confidently (Table 1). A series of meta-analyses have already been conducted to confirm the association of rs6983267 and rs10505477 polymorphisms with CRC risk [10,17]. In the present study, a meta-analysis incorporating more related case-control studies (GWAS included) about a specific chromosome region (8q23-24) was carried out to confirm the association of rs16892766, rs7014346, rs7837328, rs6983267 and rs10505477 with susceptibility to CRC, through which the combined effects of 8q23-24 on CRC might be estimated.
Table 1.
Main characteristics of studies selected in the meta-analysis
| ID | First author | Year | Genotyping method | Country | Ethnicity | SNP | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| rs6983267 | rs10505477 | rs7014346 | rs7837328 | rs16892766 | ||||||
|
| ||||||||||
| Case/Cont | Case/Cont | Case/Cont | Case/Cont | Case/Cont | ||||||
| 1 | Gruber | 2007 | GeneChip | Northern Israel | Asian | -- | 1861/1937 | -- | -- | -- |
| 2 | Poynter | 2007 | PCR | Europe | Caucasian | 1339/2191 | 1341/2193 | -- | -- | -- |
| 3 | Tomlinson | 2007 | Illumina | United Kingdom | Caucasian | 7954/6202 | -- | -- | -- | -- |
| 4 | Zanke | 2007 | TaqMan | Newfoundland, Canada | Caucasian | -- | 445/366 | -- | -- | -- |
| 5 | Zanke | 2007 | TaqMan | America | Caucasian | -- | 1859/1882 | -- | -- | -- |
| 6 | Zanke | 2007 | TaqMan | Scotland | Caucasian | -- | 2809/2912 | -- | -- | -- |
| 7 | Zanke | 2007 | TaqMan | France | Caucasian | -- | 1415/1656 | -- | -- | -- |
| 8 | Zanke | 2007 | TaqMan | Europe | Caucasian | -- | 761/749 | -- | -- | -- |
| 9 | Li | 2008 | TaqMan | America | Caucasian | 527/679 | -- | -- | -- | -- |
| 10 | Pittman | 2008 | PCR | United Kingdom | Caucasian | 3583/2579 | -- | -- | -- | -- |
| 11 | Schafmayer | 2008 | SNPlex | German | Caucasian | 2712/2713 | 2713/2718 | 2713/2718 | -- | -- |
| 12 | Tenesa | 2008 | Illumina | Scotland | Caucasian | -- | -- | 2986/3059 | -- | -- |
| 13 | Tenesa | 2008 | Illumina | Japan | Caucasian | -- | -- | 4395/3179 | -- | -- |
| 14 | Tenesa | 2008 | Illumina | Canada | Caucasian | -- | -- | 1175/1183 | -- | -- |
| 15 | Tenesa | 2008 | Illumina | England | Caucasian | -- | -- | 2233/2248 | -- | -- |
| 16 | Tenesa | 2008 | Illumina | Spain | Caucasian | -- | -- | 349/292 | -- | -- |
| 17 | Tenesa | 2008 | Illumina | Germany | Caucasian | -- | -- | 3455/3563 | -- | -- |
| 18 | Tenesa | 2008 | Illumina | Scotland | Caucasian | -- | -- | 826/892 | -- | -- |
| 19 | Tenesa | 2008 | Illumina | Israel | Caucasian | -- | -- | 1517/1466 | -- | -- |
| 20 | Tomlinson | 2008 | Illumina | United Kingdom | Caucasian | -- | -- | -- | -- | 18831/18540 |
| 21 | Tuupanen | 2008 | PCR | Finland | Caucasian | 996/1012 | -- | -- | -- | -- |
| 22 | Wokołorczyk | 2008 | RFLP-PCR | Poland | Caucasian | 779/1910 | -- | -- | -- | -- |
| 23 | Curtin | 2009 | SNPlex | United Kingdom | Caucasian | 1069/1040 | 1071/1040 | -- | -- | -- |
| 24 | Kupfer | 2009 | Sequenom MassARRAY | America (European) | Caucasian | 288/202 | -- | -- | -- | -- |
| 25 | Matsuo | 2009 | TaqMan | Japan | Asian | 476/961 | -- | -- | -- | -- |
| 26 | Middeldorp | 2009 | PCR | Dutch | Caucasian | 995/1340 | -- | -- | -- | -- |
| 27 | Cui | 2010 | Illumina | Japan | Asian | 6161/4494 | -- | -- | 6163/4494 | -- |
| 28 | Ghazi | 2010 | TaqMan | Sweden | Caucasian | 511/1017 | -- | -- | -- | -- |
| 29 | Holst | 2010 | TaqMan | Sweden | Caucasian | 1737/1738 | -- | -- | -- | 1755/1691 |
| 30 | Hutter | 2010 | TaqMan | Iran | Asian | 1962/2418 | 2089/2443 | -- | -- | -- |
| 31 | Kupfer | 2010 | Sequenom MassARRAY | America (European) | Caucasian | 399/367 | -- | 399/367 | 399/367 | 399/367 |
| 32 | Kupfer | 2010 | Sequenom MassARRAY | America (African) | African | 795/985 | -- | 795/985 | 795/985 | 795/985 |
| 33 | Xiong | 2010 | RFLP-PCR | China | Asian | 2124/2124 | -- | -- | -- | -- |
| 34 | He | 2011 | TaqMan | America (European) | Caucasian | 1171/1543 | -- | -- | -- | 1171/1543 |
| 35 | He | 2011 | TaqMan | America (African) | African | 382/510 | -- | -- | -- | 382/510 |
| 36 | He | 2011 | TaqMan | America (Native Hawaiians) | Caucasian | 323/472 | -- | -- | -- | 323/472 |
| 37 | He | 2011 | TaqMan | America (Japanese) | Asian | 1042/1426 | -- | -- | -- | 1042/1426 |
| 38 | He | 2011 | TaqMan | America (Latino) | Caucasian | 393/524 | -- | -- | -- | 393/524 |
| 39 | Ho | 2011 | Sequenom MassARRAY | China | Asian | 716/714 | -- | 716/714 | -- | -- |
| 40 | Li | 2011 | TaqMan | China | Asian | 430/786 | -- | -- | -- | -- |
| 41 | Lubbe | 2011 | PCR | United Kingdom | Caucasian | 8878/6051 | -- | -- | -- | -- |
| 42 | Daraei | 2012 | PCR–RFLP | Iran | Asian | 110/120 | -- | -- | -- | -- |
| 43 | Peters | 2012 | Illumina | -- | Caucasian | 4166/4990 | -- | -- | -- | 7686/8977 |
| 44 | Thean | 2012 | Affymetrix GeneChip | Singapore | Asian | 1000/1000 | -- | -- | -- | 991/993 |
| 45 | Hong | 2013 | TaqMan | Korea | Asian | 198/328 | -- | -- | -- | -- |
| 46 | Nan | 2013 | TaqMan | America | Caucasian | 807/1623 | -- | -- | -- | -- |
| 47 | Wang | 2013 | Illumina | America (African) | African | 1894/4703 | 1894/4703 | 1894/4703 | 1894/4703 | |
| 48 | Yang | 2014 | TaqMan | America | Caucasian | 90/132 | -- | -- | 90/132 | -- |
| 49 | Yang | 2014 | Sequenom MassARRAY | Taiwan | Asian | 705/1802 | 705/1802 | 705/1802 | -- | -- |
Moreover, the incidence rate of CRC varies between populations partly because the variation of SNPs differs among distinct ethnicities [18]. However, there have been few reports about how to categorize 8q23-24 related SNPs identified by GWAS as common disease-common variants (CD-CV) and common disease-rare variants (CD-RV) among populations of different ethnicities. Therefore, subgroup analyses based on ethnicity are conducted to distinguish the susceptibility alleles that are frequent among population on a larger scale from those that are limited to specific ethnicities, in which way CRC risk could be predicted in different populations with identification of particular risk variants.
Methods
Search strategy and selection
A meta-analysis was conducted in accordance with PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement [19]. Articles were searched by usage of the MeSH terms “colorectal cancer”, “SNP”, “rs6983267”, “rs10505477”, “rs7014346”, “7837328”, “16892766”, “8q24.21”, “8q23.3”, “case-control” and “meta-analysis” in MEDLINE and Embase without language limitations, with the latest search being updated in January 2015. The reference lists were sought for by hand for other pertinent publications.
Data extraction
Studies were included in the meta-analysis if they met the following criteria: (i) study patients were diagnosed with CRC at any tumorigenesis stage; (ii) availability of genotype or allele of both case and control groups or minor allele frequency (MAF) of patients and control groups or related odds ratio (OR) and confidence interval (95% CI) of the allelic model for CRC; and (iii) genotype frequencies were congruent with Hardy-Weinberg equilibrium (HWE) in the control group. Major exclusion criteria were as follows: (i) combined data of CRC with other cancers; (ii) no available genotype or allele frequencies or MAF or related OR; (iii) family-based studies; and (iv) abstracts and reviews.
Studies were screened by two investigators and information was extracted from all candidate publications independently. Disagreements were recorded and settled via discussion with a third author. The following characteristics were collected from each study: first author’s surname, year of publication, ethnicity of patients, genotyping method, total number of CRC patients and controls, methods of genotyping, loci, frequencies of genotype and allele, OR and 95% CI of allelic model.
Statistical analysis
A chi-squared (χ 2) test was performed when the frequencies of genotypes in controls satisfied HWE. Crude OR with 95% confidence interval (CI) and P-values were calculated to assess the stability of the results of the meta-analyses. Pooled ORs were calculated for the allelic model of rs6983267 (G vs. T), rs10505477 (T vs. C), rs7014346 (A vs. G), rs7837328 (A vs. G), rs16892766 (C vs. A). The statistical significance of pooled ORs was assessed by Z test. To measure the strength of genetic association, Cochran’s Q test and Higgins’s (I2) test were used to assess between-study heterogeneity. In case of I2 < 50% and P > 0.1, the fixed-effects model was employed to evaluate the pooled ORs; otherwise, the random-effects model was applied. Begg’s funnel plots test and Egger’s regression test were carried out to estimate publication bias. A value of P < 0.1 was regarded as statistically-significant publication bias. STATA software (version 12.0) was utilized to conduct statistical analyses, and a two-tailed P value less than 0.05 was considered to be statistically significant.
Results
Study characteristics
As shown in Figure 1, 121 reports were initially searched on account of the subject terms mentioned above. After excluding articles that described reviews, uncorrelated SNPs and diseases, 29 articles were finally selected for full assessment and another 4 studies were added through manual searching from references. On the whole, 33 articles containing 49 case-control studies were eligible for this meta-analysis study, among which the researches by Kupfer et al., Tenesa et al., Zanke et al. and He et al. contained 2, 9, 8 and 5 case-control studies, respectively [5,8,9,11,12,14-18,20-42]. The characteristics of the included studies were presented in Table 1. Moreover, the full-text reports of susceptibility to CRC were classified into Cauca-sian, Asian or African subgroups: of 34 studies on rs6983267 (56712 cases and 60691 controls), 20 studies were from Caucasian subjects (38317 cases and 38320 controls), 11 studies were from Asian subjects (14924 cases and 16173 controls) and 3 studies were from African subjects (3071 cases and 6198 controls); of 12 studies on rs10505477 (18962 cases and 24400 controls), 8 studies were from Caucasian subjects (12414 cases and 13516 controls), 3 studies were from Asian subjects (4654 cases and 6181 controls); of 14 studies on rs7014346 (24158 cases and 27171 controls), 8 studies were from Caucasian subjects (14136 cases and 14322 controls), 4 studies were from Asian subjects (7333 cases and 7161 controls); of 11 studies on rs16892766 (34620 cases and 39296 controls), 7 studies were from Caucasian subjects (30558 cases and 32105 controls) and 3 studies were from African subjects (3071 cases and 6198 controls).
Figure 1.
Selection of the related studies.
Meta-analysis
The association results of 5 polymorphisms on the 8q23-24 genetic stripe with CRC in allelic model were shown in Table 2, and detailed analysis about the relationship between every specific SNP and susceptibility to CRC was demonstrated in Supplementary Figures 1, 2, 3, 4 and 5. A notable association between chromosome 8q23-24 (5 SNPs involved) and CRC was also achieved on the foundation of meta-analysis of all related studies, including a total of 141899 cases and 157536 controls. As shown in Figure 2, the G allele of rs6983267 was associated with significantly increased risk of CRC under allelic model [OR = 1.16 (95% CI = 1.12-1.20), P < 0.001]. Furthermore, it was found that the A allele of rs7014346 remarkably increased CRC risk with OR of 1.12 (95% CI = 1.03-1.21, P < 0.001) and the T allele of rs10505477 could also elevate risk for CRC intensively [OR = 1.11 (95% CI = 1.06-1.16), P < 0.001]. Similarly, either the C allele of rs16892766 or the A allele of rs7837328 could promote CRC occurrence with OR of 1.23 (95% CI = 1.19-1.28, P < 0.001) and OR of 1.17 (95% CI = 1.11-1.23, P < 0.001), respectively.
Table 2.
Main results of meta-analysis of 5 polymorphisms on chromosome 8q23-24 (8q24.21 and 8q23.3) region and susceptibility to CRC
| ID | SNP | Location (Hapmap) | Ethnicity | No. of studies | Case/Control | OR (95%CI) | Z | P-value | Model | I2 | P for Q-test | P* |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs6983267 | 8: 127,401,060 | All | 34 | 56712/60691 | 1.16 (1.12, 1.20) | 56.43 | < 0.001 | Random | 0.687 | 0.000 | 0.848 |
| Caucasian | 20 | 38717/38320 | 1.15 (1.10, 1.21) | 42.43 | < 0.001 | Random | 0.750 | 0.000 | 0.719 | |||
| Asian | 11 | 14924/16173 | 1.17 (1.09, 1.24) | 32.14 | < 0.001 | Random | 0.624 | 0.003 | 0.793 | |||
| African | 3 | 3071/6198 | 1.20 (1.06, 1.34) | 17.03 | < 0.001 | Fixed | 0.063 | 0.344 | 0.180 | |||
| 2 | rs10505477 | 8: 127,395,198 | All | 12 | 18962/24400 | 1.11 (1.06, 1.16) | 43.84 | < 0.001 | Random | 0.575 | 0.007 | 0.067 |
| Caucasian | 8 | 12414/13516 | 1.15 (1.11, 1.19) | 56.52 | < 0.001 | Fixed | 0.402 | 0.110 | 0.122 | |||
| Asian | 3 | 4654/6181 | 1.05 (0.94, 1.15) | 19.28 | < 0.001 | Random | 0.697 | 0.037 | 0.160 | |||
| 3 | rs7014346 | 8: 127,412,547 | All | 14 | 24158/27171 | 1.12 (1.03, 1.21) | 25.25 | < 0.001 | Random | 0.880 | 0.000 | 0.628 |
| Caucasian | 8 | 14136/14322 | 1.20 (1.16, 1.24) | 57.61 | < 0.001 | Fixed | 0.000 | 0.524 | 0.941 | |||
| Asian | 4 | 7333/7161 | 1.01 (0.85, 1.17) | 12.45 | < 0.001 | Random | 0.874 | 0.000 | 0.209 | |||
| 4 | rs7837328 | 8: 127,410,882 | All | 4 | 7447/5978 | 1.17 (1.11, 1.23) | 38.45 | < 0.001 | Fixed | 0.076 | 0.355 | 0.433 |
| 5 | rs16892766 | 8: 116,618,444 | All | 11 | 34620/39296 | 1.23 (1.19, 1.28) | 52.44 | < 0.001 | Fixed | 0.000 | 0.961 | 0.720 |
| Caucasian | 7 | 30558/32105 | 1.25 (1.20, 1.30) | 46.81 | < 0.001 | Fixed | 0.000 | 0.995 | 0.955 | |||
| African | 3 | 3071/6198 | 1.16 (1.05, 1.26) | 20.88 | < 0.001 | Fixed | 0.000 | 0.758 | 0.987 |
P: publication bias.
Figure 2.
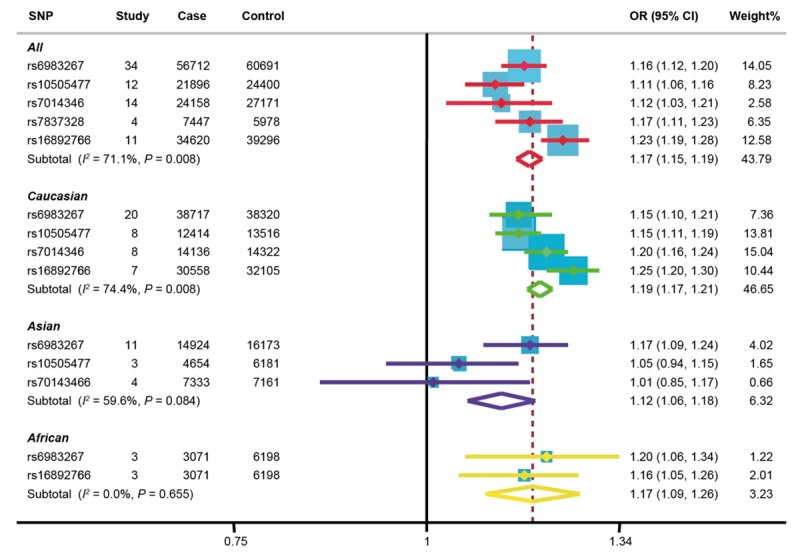
Forest plot presenting the meta-analysis of the association between 5 polymorphisms (rs6983267, rs10505477, rs7014346, rs7837328 and rs16892766) in 8q23.3 or 8q24.21 region and risk of CRC as well as subgroup analysis of 4 polymorphisms (rs7837328 excluded) by ethnicity in allelic model.
Moreover, subgroup analyses of four SNPs (rs7837328 excluded) by ethnicity were further performed. Figure 2 witnessed a remarkable association between the G allele of rs6983267 and increased risk of CRC among Caucasians [OR = 1.15 (95% CI = 1.10-1.21), P < 0.001], Asians [OR = 1.17 (95% CI = 1.09-1.24), P < 0.001] and Africans [OR = 1.20 (95% CI = 1.06-1.34), P < 0.001], which were consistent with the overall analysis. However, strong associations between the T allele of rs10505477 polymorphism and enhancive susceptibility to CRC were found only within Caucasians [OR = 1.15 (95% CI = 1.11-1.19), P < 0.001], and the results among Asians were not positive [OR = 1.05 (95% CI = 0.94-1.15), P < 0.001]. Likewise, the A allele of rs7014346 only elevated risk for CRC among Caucasians [OR = 1.20 (95% CI = 1.16-1.24), P < 0.001] and showed no effect among Asians. Regarding rs16892766, only meta-analysis of Caucasians and Africans were carried out due to shortage of Asian studies, revealing that the C allele of rs16892766 was closely related with increased risk of CRC among Caucasians [OR = 1.25 (95% CI = 1.20-1.30), P < 0.001] and Africans [OR = 1.16 (95% CI = 1.05-1.26), P < 0.001].
Publication bias
As revealed in Table 2 and Supplementary Figure 6, no obvious asymmetry could be observed in the shape of funnel plots (P = 0.848 for rs6983267, P = 0.067 for rs10505477, P = 0.628 for rs7014346, P = 0.433 for rs7837328 and P = 0.720 for rs16892766).
Discussion
There seems to exist potent associations between genetic markers at human chromosome 8q23.3 (rs16892766) or 8q24.21 (rs6983267, rs10505477, rs7014346 and rs7837328) and increased susceptibility to CRC [5-7,9,34]. This hypothesis was confirmed in the present study by conducting an allelic meta-analysis of involved association studies of the 5 polymorphisms worldwide, deriving consequences that the rs7014346 and rs10505477 polymorphisms were CRC-associated locus in both the Caucasians and the Asians while the rs6983267, rs16892766 and rs7837328 variants served as risk locus, respectively, among triple (Caucasian, Asian and African), double (Caucasian and African) and multi- ethnicities. The diverse magnitudes of increased risk of CRC in different populations (also shown as different values of I2 in Table 2) conferred by the above genetic polymorphisms could be attributed to the discrepancy in allelic frequencies. Categorizing nearly all the case-control studies for each polymorphism on the basis of ethnicity, interesting results about allelic frequencies (case and control, respectively) are drawn as follows: Asian (0.54 and 0.56, respectively) > Caucasian (0.44 and 0.48, respectively) for T allele of rs6983267; Caucasian (0.46 and 0.50, respectively) ≈ Asian (0.46 and 0.47, respectively) for G allele of rs10505477; Caucasian (0.59 and 0.63, respectively) > Asian (0.40 and 0.46, respectively) for C allele of rs7014346; Caucasian (0.1171 and 0.1003, respectively) > Asian (0.0040 and 0.0005, respectively) for C allele of rs16892766. The possible reason might be that external environment would render one allele more frequent in one population than another, indicating that the allele could be associated with susceptibility to CRC [43]. Another probable explanation for the distinct proportion of different populations suffering from CRC could be clarified by diversified linkage disequilibrium (LD) structure [12,44-47]. Various genotyping methods could also account for why the research results on a particular polymorphism are different from one another; for instance, SNPlex chemistry (Applied Biosystems, Foster City) [33], Sequenom MassARRAY platform [18,41], Illumina [8,34], PCR [37] were employed for the rs6983267 polymorphism in different published studies and distinct levels of risk for CRC were observed.
In fact, the characteristic function of 5 polymorphisms for CRC could be partly expatiated by the genomic organization, where the 5 SNPs reside. For instance, the 8q24.21 genomic region is featured by gene desert with 14807 bp away from pseudogene POU5F1P1, which is followed by oncogene MYC and the nearest proximal gene FAM84B. The above two genes, MYC and FAM84B, are respectively 335 kb and 849 kb from the rs6983267 polymorphism (the tag SNP in the 8q24.21 region) [37]. SNP rs6983267 is reported to either exert direct differential effects on MYC expression [48] or indirectly regulate expression of MYC through binding to some spicing forms of the transcription factor 7-like 2 (TCF7L2) [49]. According to Tuupanen et al., TCF7L2 is a main transcriptional effector of the Wnt signaling pathway, co-activating β-catenin in CRC [50]. Since the other three SNPs (rs7014346, rs7837328 and rs10505477) are in strong LD with rs6983267 [10-12], they might indirectly influence CRC through affiliation with cancer risk-associated rs6983267 polymorphism [10]. Hence, the four SNPs on the 8q24.21 region might carry an integrated and greater risk effect [11]. Still, clarification of the plausible regulatory role of these four loci and their LD in risk of CRC is required to be expatiated in further studies.
Besides, errors in meiotic cross-over events might also lead to chromosome abnormalities or non-disjunction, further causing loss of heterozygotes (LOH), which is closely associated with neoplastic progression [41,51]. The meiotic cross-over events often take place in the recombination hotspots, where the locus are characteristic of higher exchange frequencies than others in the chromosome [51]. Certain variants harboring within the hotspots could thereby regulate hotspot activity and related recombination rate in the process of strand exchange; and the sequence variation might even cause distinctions in DNA topology, structure of chromatin, or chromosome domain organization [52,53]. The rs10505477 polymorphism investigated in the present study resides within a recombination hotspot and LD exists between the locus and other three locus (rs6983267, rs7014346, rs7837328) in the 8q24.21 region identified by GWAS, thus the four genetic variants might constitute part of hidden dangers for carcinogenesis. Additionally, the usage of recombination hotspots differs among populations [51], partly explaining the ethnic distribution of genetic variations. However, the possible effects of the four locus on the function of this hotspot and susceptibility to CRC demands more investigations.
The ultimately mentioned rs1982766, the tag SNP on chromosome 8q23.3, resides in an extended region with an anomalous and high LD [54]. The rs1982766 polymorphism has been demonstrated to display notable associations with CRC risk by means of repressing the expression of the eukaryotic translation initiation factor 3 subunit H (EIF3H) gene when interacting with the EIF3H promoter [55]. However, a succeeding assessment of ENCODE data and eQTLs implies that the expression level of the neighboring UTP23 [small subunit (SSU) processome component, homologue (yeast)], instead of EIF3H, was markedly influenced by the rs1982766 variant and UTP23 has become the most potential candidate gene associated with CRC in the 8q23.3 region [13,54]. As reported by Lu et al., UTP23 is a gene encoding UTP23, which is a conserved protein factor involved in the early assembly of ribosomal small subunit, affecting the precise identification of mRNA [56]. Since both UTP23 and EIF3H share related functions in mRNA translation, it also appears to be conceivable that the double genes are collaborately regulated by the rs1982766 polymorphism in terms of susceptibility to CRC [54]. To elucidate how EIF3H or UTP23, or both, are associated with CRC etiology, additional work is necessitated [13].
Although some puzzles have been made more unambiguous by this meta-analysis, along with previous studies, several limitations should also be noted. First, the selection of case groups followed heterogeneous inclusion criteria for parameters, such as tumor stage and site. Second, the coverage of the clinical data (sex, age group, etc.) of the case and control groups was not all-sided. Third, MAF or OR (95% CI), rather than genotype and allele frequencies, were reported in some studies. Fourth, most of the studies did not identify whether people recruited in the control group are healthy or affected to other disease. Fifth, no enough case-control studies on Asian and African studies could be included in the meta-analysis, restraining further analysis of the association between 8q23-24 and CRC among more ethnicities. Despite these deficiencies, to the best of our knowledge, this is the first study to perform a systematic meta-analysis of 5 locus on the 8q23-24 genetic stripe which have been identified by GWAS, suggesting that the rs6983267 polymorphism was CRC associated locus in the Caucasians, Asians and Africans; rs7014346 and rs10505477 genetic variations showed positive results in both the Caucasians and the Asians; and rs16892766 served as risk locus in double (Caucasian and African) ethnicities while the rs7837328 variant was a risky factor in a multinational group. However, for in-depth understanding about the biological and clinical role of five polymorphisms seated in the 8q23.3 and 8q24.21 region in CRC risk, further investigation would be in urgent need.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Haerian MS, Haerian BS, Rooki H, Molanaei S, Kosari F, Obohhat M, Hosseinpour P, Azimzadeh P, Mohebbi SR, Akbari Z, Zali MR. Association of 8q24.21 rs10505477-rs6983267 haplotype and age at diagnosis of colorectal cancer. Asian Pac J Cancer Prev. 2014;15:369–74. doi: 10.7314/apjcp.2014.15.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 4.Lascorz J, Forsti A, Chen B, Buch S, Steinke V, Rahner N, Holinski-Feder E, Morak M, Schackert HK, Gorgens H, Schulmann K, Goecke T, Kloor M, Engel C, Buttner R, Kunkel N, Weires M, Hoffmeister M, Pardini B, Naccarati A, Vodickova L, Novotny J, Schreiber S, Krawczak M, Broring CD, Volzke H, Schafmayer C, Vodicka P, Chang-Claude J, Brenner H, Burwinkel B, Propping P, Hampe J, Hemminki K. Genome-wide association study for colorectal cancer identifies risk polymorphisms in German familial cases and implicates MAPK signalling pathways in disease susceptibility. Carcinogenesis. 2010;31:1612–1649. doi: 10.1093/carcin/bgq146. [DOI] [PubMed] [Google Scholar]
- 5.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, Edlund CK, Haile RW, Gallinger S, Zanke BW, Lemire M, Rangrej J, Vijayaraghavan R, Chan AT, Hazra A, Hunter DJ, Ma J, Fuchs CS, Giovannucci EL, Kraft P, Liu Y, Chen L, Jiao S, Makar KW, Taverna D, Gruber SB, Rennert G, Moreno V, Ulrich CM, Woods MO, Green RC, Parfrey PS, Prentice RL, Kooperberg C, Jackson RD, Lacroix AZ, Caan BJ, Hayes RB, Berndt SI, Chanock SJ, Schoen RE, Chang-Claude J, Hoffmeister M, Brenner H, Frank B, Bezieau S, Kury S, Slattery ML, Hopper JL, Jenkins MA, Le Marchand L, Lindor NM, Newcomb PA, Seminara D, Hudson TJ, Duggan DJ, Potter JD, Casey G. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Hum Genet. 2012;131:217–234. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang B, Jia WH, Matsuda K, Kweon SS, Matsuo K, Xiang YB, Shin A, Jee SH, Kim DH, Cai Q, Long J, Shi J, Wen W, Yang G, Zhang Y, Li C, Li B, Guo Y, Ren Z, Ji BT, Pan ZZ, Takahashi A, Shin MH, Matsuda F, Gao YT, Oh JH, Kim S, Ahn YO, Chan AT, Chang-Claude J, Slattery ML, Gruber SB, Schumacher FR, Stenzel SL, Casey G, Kim HR, Jeong JY, Park JW, Li HL, Hosono S, Cho SH, Kubo M, Shu XO, Zeng YX, Zheng W. Large-scale genetic study in East Asians identifies six new loci associated with colorectal cancer risk. Nat Genet. 2014;46:533–542. doi: 10.1038/ng.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiffin N, Hosking FJ, Farrington SM, Palles C, Dobbins SE, Zgaga L, Lloyd A, Kinnersley B, Gorman M, Tenesa A, Broderick P, Wang Y, Barclay E, Hayward C, Martin L, Buchanan DD, Win AK, Hopper J, Jenkins M, Lindor NM, Newcomb PA, Gallinger S, Conti D, Schumacher F, Casey G, Liu T, Campbell H, Lindblom A, Houlston RS, Tomlinson IP, Dunlop MG. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet. 2014;23:4729–4737. doi: 10.1093/hmg/ddu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, Spain S, Lubbe S, Walther A, Sullivan K, Jaeger E, Fielding S, Rowan A, Vijayakrishnan J, Domingo E, Chandler I, Kemp Z, Qureshi M, Farrington SM, Tenesa A, Prendergast JG, Barnetson RA, Penegar S, Barclay E, Wood W, Martin L, Gorman M, Thomas H, Peto J, Bishop DT, Gray R, Maher ER, Lucassen A, Kerr D, Evans DG, Schafmayer C, Buch S, Volzke H, Hampe J, Schreiber S, John U, Koessler T, Pharoah P, van Wezel T, Morreau H, Wijnen JT, Hopper JL, Southey MC, Giles GG, Severi G, Castellvi-Bel S, Ruiz-Ponte C, Carracedo A, Castells A, Forsti A, Hemminki K, Vodicka P, Naccarati A, Lipton L, Ho JW, Cheng KK, Sham PC, Luk J, Agundez JA, Ladero JM, de la Hoya M, Caldes T, Niittymaki I, Tuupanen S, Karhu A, Aaltonen L, Cazier JB, Campbell H, Dunlop MG, Houlston RS. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 9.Cui R, Okada Y, Jang SG, Ku JL, Park JG, Kamatani Y, Hosono N, Tsunoda T, Kumar V, Tanikawa C, Kamatani N, Yamada R, Kubo M, Nakamura Y, Matsuda K. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60:799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haerian MS, Baum L, Haerian BS. Association of 8q24.21 loci with the risk of colorectal cancer: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2011;26:1475–1484. doi: 10.1111/j.1440-1746.2011.06831.x. [DOI] [PubMed] [Google Scholar]
- 11.Yang B, Thyagarajan B, Gross MD, Goodman M, Sun YV, Bostick RM. Genetic variants at chromosome 8q24, colorectal epithelial cell proliferation, and risk for incident, sporadic colorectal adenomas. Mol Carcinog. 2014;53(Suppl 1):E187–92. doi: 10.1002/mc.22047. [DOI] [PubMed] [Google Scholar]
- 12.Kupfer SS, Anderson JR, Hooker S, Skol A, Kittles RA, Keku TO, Sandler RS, Ellis NA. Genetic heterogeneity in colorectal cancer associations between African and European americans. Gastroenterology. 2010;139:1677–85. 85.e1–8. doi: 10.1053/j.gastro.2010.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, Nan H, Lemire M, Rangrej J, Figueiredo JC, Jiao S, Harrison TA, Liu Y, Chen LS, Stelling DL, Warnick GS, Hoffmeister M, Kury S, Fuchs CS, Giovannucci E, Hazra A, Kraft P, Hunter DJ, Gallinger S, Zanke BW, Brenner H, Frank B, Ma J, Ulrich CM, White E, Newcomb PA, Kooperberg C, LaCroix AZ, Prentice RL, Jackson RD, Schoen RE, Chanock SJ, Berndt SI, Hayes RB, Caan BJ, Potter JD, Hsu L, Bezieau S, Chan AT, Hudson TJ, Peters U. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72:2036–2044. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kupfer SS, Torres JB, Hooker S, Anderson JR, Skol AD, Ellis NA, Kittles RA. Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis. 2009;30:1353–1357. doi: 10.1093/carcin/bgp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lubbe SJ, Di Bernardo MC, Broderick P, Chandler I, Houlston RS. Comprehensive evaluation of the impact of 14 genetic variants on colorectal cancer phenotype and risk. Am J Epidemiol. 2012;175:1–10. doi: 10.1093/aje/kwr285. [DOI] [PubMed] [Google Scholar]
- 16.Xiong F, Wu C, Bi X, Yu D, Huang L, Xu J, Zhang T, Zhai K, Chang J, Tan W, Cai J, Lin D. Risk of genome-wide association study-identified genetic variants for colorectal cancer in a Chinese population. Cancer Epidemiol Biomarkers Prev. 2010;19:1855–1861. doi: 10.1158/1055-9965.EPI-10-0210. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Zhou Y, Chen P, Yang H, Yuan X, Tajima K, Cao J, Wang H. Genetic variants on chromosome 8q24 and colorectal neoplasia risk: a case-control study in China and a meta-analysis of the published literature. PLoS One. 2011;6:e18251. doi: 10.1371/journal.pone.0018251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho JW, Choi SC, Lee YF, Hui TC, Cherny SS, Garcia-Barcelo MM, Carvajal-Carmona L, Liu R, To SH, Yau TK, Chung CC, Yau CC, Hui SM, Lau PY, Yuen CH, Wong YW, Ho S, Fung SS, Tomlinson IP, Houlston RS, Cheng KK, Sham PC. Replication study of SNP associations for colorectal cancer in Hong Kong Chinese. Br J Cancer. 2011;104:369–375. doi: 10.1038/sj.bjc.6605977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JP, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339:b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtin K, Lin WY, George R, Katory M, Shorto J, Cannon-Albright LA, Bishop DT, Cox A, Camp NJ. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer Epidemiol Biomarkers Prev. 2009;18:616–621. doi: 10.1158/1055-9965.EPI-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daraei A, Salehi R, Salehi M, Emami MH, Janghorbani M, Mohamadhashem F, Tavakoli H. Effect of rs6983267 polymorphism in the 8q24 region and rs4444903 polymorphism in EGF gene on the risk of sporadic colorectal cancer in Iranian population. Med Oncol. 2012;29:1044–1049. doi: 10.1007/s12032-011-9980-2. [DOI] [PubMed] [Google Scholar]
- 22.Ghazi S, von Holst S, Picelli S, Lindforss U, Tenesa A, Farrington SM, Campbell H, Dunlop MG, Papadogiannakis N, Lindblom A. Colorectal cancer susceptibility loci in a population-based study: Associations with morphological parameters. Am J Pathol. 2010;177:2688–2693. doi: 10.2353/ajpath.2010.100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruber SB, Moreno V, Rozek LS, Rennerts HS, Lejbkowicz F, Bonner JD, Greenson JK, Giordano TJ, Fearson ER, Rennert G. Genetic variation in 8q24 associated with risk of colorectal cancer. Cancer Biol Ther. 2007;6:1143–1147. doi: 10.4161/cbt.6.7.4704. [DOI] [PubMed] [Google Scholar]
- 24.He J, Wilkens LR, Stram DO, Kolonel LN, Henderson BE, Wu AH, Le Marchand L, Haiman CA. Generalizability and epidemiologic characterization of eleven colorectal cancer GWAS hits in multiple populations. Cancer Epidemiol Biomarkers Prev. 2011;20:70–81. doi: 10.1158/1055-9965.EPI-10-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong SN, Park CH, Kim JI, Kim DH, Kim HC, Chang DK, Rhee PL, Kim JJ, Rhee JC, Son HJ, Kim YH. Colorectal Cancer-Susceptibility Single Nucleotide Polymorphisms in Korean Population. J Gastroenterol Hepatol. 2015;30:849–857. doi: 10.1111/jgh.12331. [DOI] [PubMed] [Google Scholar]
- 26.Hutter CM, Slattery ML, Duggan DJ, Muehling J, Curtin K, Hsu L, Beresford SA, Rajkovic A, Sarto GE, Marshall JR, Hammad N, Wallace R, Makar KW, Prentice RL, Caan BJ, Potter JD, Peters U. Characterization of the association between 8q24 and colon cancer: gene-environment exploration and meta-analysis. BMC Cancer. 2010;10:670. doi: 10.1186/1471-2407-10-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Plummer SJ, Thompson CL, Merkulova A, Acheson LS, Tucker TC, Casey G. A common 8q24 variant and the risk of colon cancer: a population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17:339–342. doi: 10.1158/1055-9965.EPI-07-0713. [DOI] [PubMed] [Google Scholar]
- 28.Matsuo K, Suzuki T, Ito H, Hosono S, Kawase T, Watanabe M, Shitara K, Komori K, Kanemitsu Y, Hirai T, Yatabe Y, Tanaka H, Tajima K. Association between an 8q24 locus and the risk of colorectal cancer in Japanese. BMC Cancer. 2009;9:379. doi: 10.1186/1471-2407-9-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Middeldorp A, Jagmohan-Changur S, van Eijk R, Tops C, Devilee P, Vasen HF, Hes FJ, Houlston R, Tomlinson I, Houwing-Duistermaat JJ, Wijnen JT, Morreau H, van Wezel T. Enrichment of low penetrance susceptibility loci in a Dutch familial colorectal cancer cohort. Cancer Epidemiol Biomarkers Prev. 2009;18:3062–3067. doi: 10.1158/1055-9965.EPI-09-0601. [DOI] [PubMed] [Google Scholar]
- 30.Nan H, Morikawa T, Suuriniemi M, Imamura Y, Werner L, Kuchiba A, Yamauchi M, Hunter DJ, Kraft P, Giovannucci EL, Fuchs CS, Ogino S, Freedman ML, Chan AT. Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J Natl Cancer Inst. 2013;105:1852–1861. doi: 10.1093/jnci/djt331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pittman AM, Broderick P, Sullivan K, Fielding S, Webb E, Penegar S, Tomlinson I, Houlston RS. CASP8 variants D302H and -652 6N ins/del do not influence the risk of colorectal cancer in the United Kingdom population. Br J Cancer. 2008;98:1434–1436. doi: 10.1038/sj.bjc.6604314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poynter JN, Figueiredo JC, Conti DV, Kennedy K, Gallinger S, Siegmund KD, Casey G, Thibodeau SN, Jenkins MA, Hopper JL, Byrnes GB, Baron JA, Goode EL, Tiirikainen M, Lindor N, Grove J, Newcomb P, Jass J, Young J, Potter JD, Haile RW, Duggan DJ, Le Marchand L. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 33.Schafmayer C, Buch S, Volzke H, von Schonfels W, Egberts JH, Schniewind B, Brosch M, Ruether A, Franke A, Mathiak M, Sipos B, Henopp T, Catalcali J, Hellmig S, ElSharawy A, Katalinic A, Lerch MM, John U, Folsch UR, Fandrich F, Kalthoff H, Schreiber S, Krawczak M, Tepel J, Hampe J. Investigation of the colorectal cancer susceptibility region on chromosome 8q24.21 in a large German case-control sample. Int J Cancer. 2009;124:75–80. doi: 10.1002/ijc.23872. [DOI] [PubMed] [Google Scholar]
- 34.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, Barnetson RA, Theodoratou E, Cetnarskyj R, Cartwright N, Semple C, Clark AJ, Reid FJ, Smith LA, Kavoussanakis K, Koessler T, Pharoah PD, Buch S, Schafmayer C, Tepel J, Schreiber S, Volzke H, Schmidt CO, Hampe J, Chang-Claude J, Hoffmeister M, Brenner H, Wilkening S, Canzian F, Capella G, Moreno V, Deary IJ, Starr JM, Tomlinson IP, Kemp Z, Howarth K, Carvajal-Carmona L, Webb E, Broderick P, Vijayakrishnan J, Houlston RS, Rennert G, Ballinger D, Rozek L, Gruber SB, Matsuda K, Kidokoro T, Nakamura Y, Zanke BW, Greenwood CM, Rangrej J, Kustra R, Montpetit A, Hudson TJ, Gallinger S, Campbell H, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thean LF, Li HH, Teo YY, Koh WP, Yuan JM, Teoh ML, Koh PK, Tang CL, Cheah PY. Association of Caucasian-identified variants with colorectal cancer risk in Singapore Chinese. PLoS One. 2012;7:e42407. doi: 10.1371/journal.pone.0042407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 37.Tuupanen S, Niittymaki I, Nousiainen K, Vanharanta S, Mecklin JP, Nuorva K, Jarvinen H, Hautaniemi S, Karhu A, Aaltonen LA. Allelic imbalance at rs6983267 suggests selection of the risk allele in somatic colorectal tumor evolution. Cancer Res. 2008;68:14–17. doi: 10.1158/0008-5472.CAN-07-5766. [DOI] [PubMed] [Google Scholar]
- 38.von Holst S, Picelli S, Edler D, Lenander C, Dalen J, Hjern F, Lundqvist N, Lindforss U, Pahlman L, Smedh K, Tornqvist A, Holm J, Janson M, Andersson M, Ekelund S, Olsson L, Ghazi S, Papadogiannakis N, Tenesa A, Farrington SM, Campbell H, Dunlop MG, Lindblom A. Association studies on 11 published colorectal cancer risk loci. Br J Cancer. 2010;103:575–580. doi: 10.1038/sj.bjc.6605774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H, Haiman CA, Burnett T, Fortini BK, Kolonel LN, Henderson BE, Signorello LB, Blot WJ, Keku TO, Berndt SI, Newcomb PA, Pande M, Amos CI, West DW, Casey G, Sandler RS, Haile R, Stram DO, Le Marchand L. Finemapping of genome-wide association studyidentified risk loci for colorectal cancer in African Americans. Hum Mol Genet. 2013;22:5048–5055. doi: 10.1093/hmg/ddt337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wokolorczyk D, Gliniewicz B, Sikorski A, Zlowocka E, Masojc B, Debniak T, Matyjasik J, Mierzejewski M, Medrek K, Oszutowska D, Suchy J, Gronwald J, Teodorczyk U, Huzarski T, Byrski T, Jakubowska A, Gorski B, van de Wetering T, Walczak S, Narod SA, Lubinski J, Cybulski C. A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 2008;68:9982–9986. doi: 10.1158/0008-5472.CAN-08-1838. [DOI] [PubMed] [Google Scholar]
- 41.Zhou B, Shu B, Yang J, Liu J, Xi T, Xing Y. C-reactive protein, interleukin-6 and the risk of colorectal cancer: a meta-analysis. Cancer Causes Control. 2014;25:1397–1405. doi: 10.1007/s10552-014-0445-8. [DOI] [PubMed] [Google Scholar]
- 42.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O’Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous ME, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet. 2007;39:989–94. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 43.Jorde LB, Wooding SP. Genetic variation, classification and ‘race’. Nat Genet. 2004;36:S28–33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 44.Ioannidis JP. Non-replication and inconsistency in the genome-wide association setting. Hum Hered. 2007;64:203–213. doi: 10.1159/000103512. [DOI] [PubMed] [Google Scholar]
- 45.Ireland J, Carlton VE, Falkowski M, Moorhead M, Tran K, Useche F, Hardenbol P, Erbilgin A, Fitzgerald R, Willis TD, Faham M. Largescale characterization of public database SNPs causing non-synonymous changes in three ethnic groups. Hum Genet. 2006;119:75–83. doi: 10.1007/s00439-005-0105-x. [DOI] [PubMed] [Google Scholar]
- 46.Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, Kidd KK. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet. 2005;13:677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]
- 47.Weir BS, Cardon LR, Anderson AD, Nielsen DM, Hill WG. Measures of human population structure show heterogeneity among genomic regions. Genome Res. 2005;15:1468–1476. doi: 10.1101/gr.4398405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wasserman NF, Aneas I, Nobrega MA. An 8q24 gene desert variant associated with prostate cancer risk confers differential in vivo activity to a MYC enhancer. Genome Res. 2010;20:1191–1197. doi: 10.1101/gr.105361.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prokunina-Olsson L, Hall JL. No effect of cancer-associated SNP rs6983267 in the 8q24 region on co-expression of MYC and TCF7L2 in normal colon tissue. Mol Cancer. 2009;8:96. doi: 10.1186/1476-4598-8-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, Kivioja T, Bjorklund M, Wei G, Yan J, Niittymaki I, Mecklin JP, Jarvinen H, Ristimaki A, Di-Bernardo M, East P, Carvajal-Carmona L, Houlston RS, Tomlinson I, Palin K, Ukkonen E, Karhu A, Taipale J, Aaltonen LA. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat Genet. 2009;41:885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 51.Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in finescale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- 52.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 53.Spencer CC, Deloukas P, Hunt S, Mullikin J, Myers S, Silverman B, Donnelly P, Bentley D, McVean G. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carvajal-Carmona LG, Cazier JB, Jones AM, Howarth K, Broderick P, Pittman A, Dobbins S, Tenesa A, Farrington S, Prendergast J, Theodoratou E, Barnetson R, Conti D, Newcomb P, Hopper JL, Jenkins MA, Gallinger S, Duggan DJ, Campbell H, Kerr D, Casey G, Houlston R, Dunlop M, Tomlinson I. Finemapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1 and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum Mol Genet. 2011;20:2879–2888. doi: 10.1093/hmg/ddr190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pittman AM, Naranjo S, Jalava SE, Twiss P, Ma Y, Olver B, Lloyd A, Vijayakrishnan J, Qureshi M, Broderick P, van Wezel T, Morreau H, Tuupanen S, Aaltonen LA, Alonso ME, Manzanares M, Gavilan A, Visakorpi T, Gomez-Skarmeta JL, Houlston RS. Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cisacting regulator of EIF3H. PLoS Genet. 2010;6:e1001126. doi: 10.1371/journal.pgen.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu J, Sun M, Ye K. Structural and functional analysis of Utp23, a yeast ribosome synthesis factor with degenerate PIN domain. RNA. 2013;19:1815–1824. doi: 10.1261/rna.040808.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


