Abstract
Background: The frequencies of Cytochrome P450 2C9 (CYP2C9) genotypes were various between populations. The aim of this study was to investigate the frequencies of the major variants of the CYP2C9 in Chinese Li minority populations. Methods: The promoter, exons and surrounding introns, and 3’-untranslated region of the CYP2C9 gene was detected by DNA sequencing to investigate in 100 unrelated healthy Chinese Li subjects. The protein function prediction was used the online tools: Sorting Intolerant From Tolerant (SIFT) and Phenotyping Version 2 (PolyPhen-2). The comparison of CYP2C9 allele frequencies in different populations were analyzed by Chi-square (χ2) test. Linkage disequilibrium (LD) analysis was performed using Haploview software. Results: We identified 17 different CYP2C9 single nucleotide polymorphisms (SNPs) in the Li population, including two missense mutations (3549 G > A and 42614 A > C) and two silent mutations (3514 T > Cand 50298A > T). The protein function prediction revealed the two missense mutations result in protein damaging. In addition, we detected the allele frequencies of CYP2C9*1, CYP2C9*3 and CYP2C9*42 were 98%, 1%, and 1%, respectively. Finally, we compared three major allelic frequency (CYP2C9*1, CYP2C9*2, and CYP2C9*3) between Li and other populations. We found that our results were similar to East Asians and Africans.
Keywords: CYP2C9, allele frequencies, Chinese Li minority populations, CYP2C9*1, CYP2C9*3, CYP2C9*42
Introduction
Cytochrome P450 genes is a super family of cysteine-heme enzymes, which catalyze the oxidation of various drugs and endogenous substrates, such as vitamin D, steroids, and fatty acids, including arachidonic acid (AA) [1]. CYP enzymes of the P450 2C9 (CYP2C9) subfamily are found in the liver, vascular smooth muscle, endothelial cells of human aorta and coronary artery [2-4]. Cytochrome P450 2C (CYP2C) subfamily of enzymes form 18-30% of human CYPs and metabolism nearly 20% of all therapeutic drugs commonly prescribed in clinical practice [5]. CYP2C gene is made up of four isoforms, CYP2C8, CYP2C9, CYP2C18 and CYP2C19 which are located together on chromosome 10q24 [6]. For example, CYP2C9 polymorphisms have been associated with an increased risk of bleeding in patients treated with standard doses of warfarin while phenytoin toxicity has also been reported in some patients [7].
The population of China consists of Han Chinese and 55 ethnic minorities currently recognized by the People’s Republic of China. The Li population, which is one among the 55 minority ethnic groups in China, exceeds 1.3 million and resides primarily in the Li and Miao Autonomous Prefecture in the center and southwest regions of the Hainan Province. So far, the data of CYP2C9 polymorphisms is lacked in the Li population, the main aim of this study was to investigate the CYP2C9 genetic polymorphisms in Li populations, and the secondary aim was to compare their allelic frequencies with previous observations of other studies. Our results will add some data to support the understanding of CYP2C9 variants and expect to promote personalized medicine in Li patients.
Materials and methods
Subjects
The study subjects consisted of 100 unrelated healthy Li subjects (including 50 males and 50 females) from Hainan province of China. All participants were recruited between October and December, 2014 from the People’s Hospital of Hainan Province. All participants were Li Chinese residing in the Hainan Provincial which is located southwest of China, and they had at least three generations of Li paternal ancestry. All subjects were deemed healthy based on medical history and a physical examination. The purpose and experimental procedures of the study were explained to all individuals, and written informed consent was obtained from all participants prior to sample donation. The study protocol was performed in accordance with the Declaration of Helsinki and was approved by the Ethics Committees of People’s Hospital of Hainan Province.
DNA sequencing of CYP2C9 variants
Genetic polymorphisms of CYP2C9 were screened by DNA sequencing. Briefly, 5 ml blood samples in tubes containing ethylene diaminetetraacetic acid (EDTA) were collected from the subjects and kept under -20°C until DNA was extracted. We used the GoldMag® nanoparticles method (GoldMag® Ltd. Xi’an, China) to extracted DNA from blood according to the manufacturer’s instructions genomic. The DNA samples were stored at -80°C until use. When used, the DNA was diluted to 50 ng/ul concentration. Primers for polymerase chain reaction (PCR) were designed to amplify the promoter, exons and the 3’-untranslated region of CYP2C9, and their sequences are provided in Table 1. Thermal cycling conditions were as follows: 95°C for 5 min; followed by 35 cycles of denaturation at 95°C for 30 s; annealing at 60°C for 1 min; and extension at 72°C for 1 min. A final extension step was performed at 72°C for 10 min. Preparation of DNA for sequencing included incubation of PCR products with 0.1 U of shrimp alkaline phosphatase (Roche, Basel, Switzerland) and 0.5 U of exonuclease I (New England Biolabs, Inc., Beverly, MA, USA) at 37°C for 45 min, followed by heat inactivation at 85°C for 20 min. The PCR products were sequenced using the ABI PrismBigDye Terminator Cycle Sequencing Kit version 3.1 (Applied Biosystems) on an ABI Prism3100 sequencer (Applied Biosystems).
Table 1.
Primers used to amplify regions in CYP2C9
| Primer name | Primer sequence (5’-3’) | DNA size for PCR (bp) |
|---|---|---|
| PromoterF | GCAGTGATGGAGAAGGGAGA | 924 |
| PromoterR | AATTCGGTGTGTGCCTCTTT | |
| Exon1_F | GAACCATCTGGGTTAACATTTG | 925 |
| Exon1_R | AGTGCATTCTTGGCACCAT | |
| Exon2_3_F | TGCCTTGAACATCACAGGCCATC | 826 |
| Exon2_3_R | TGGCTCTCAGCTTCAAACCCCC | |
| Exon4_F | TCTTGCCCTTTCCATCTCAG | 911 |
| Exon4_R | TGCCACATAATACGACAAAGTG | |
| Exon5_F | TGCTGTCATCTACAAAACGTGA | 908 |
| Exon5_R | CAGGGATTTGACTTCTTCCTTG | |
| Exon6_F | AATCCCAGGATGGGGTCTAC | 880 |
| Exon6_R | CAATTGATTCCAGTGCCTCA | |
| Exon7_F | TTTGATTGGAGATTTTATTCCATTT | 925 |
| Exon7_R | GATTCAGTTCTTTCCAAACTAGCC | |
| Exon8_F | TACTGCCCTTCTTTGGAACGGGAT | 809 |
| Exon8_R | ACCTCCCAACCCCCAACAGC | |
| Exon9_F | AGAATGTGAGGGTCCAGATCA | 913 |
| Exon9_R | TTCAGGGAAGGGAAAATGTG | |
| 3’-UTR_F | TGTGGGAGAAGCCCTGGCCG | 737 |
| 3’-UTR_R | AGAGCTGCCCCTGGACACAG |
PCR: polymerase chain reaction.
Data analysis
The Human Cytochrome P450 (CYP) Allele Nomenclature Database describes CYP2C9 variants according to the National Center of Biotechnology Information (NCBI) reference sequence NG_008385.1. Allelic frequency comparisons between Li and other populations were performed using the Chi-squared test with a significance level set at P = 0.05 [8]. Hardy-Weinberg equilibrium calculations were performed using the Arlequin program (http://anthropologie.unige.ch/arlequin). Linkage disequilibrium (LD) between loci pairs were assessed using Haploview software (version 4.2) (Mark Daly’s Laboratory, Massachusetts Institute of Technology/Harvard Broad Institute, Cambridge, MA, USA) [9]. Furthermore, LD was investigated across all single nucleotide polymorphisms (SNPs) and selected haplotypes. Haplotype blocks were defined based on the Gabriel definition (D’ > 0.9; minimum allele frequency > 5%) [10].
Transcriptional protein function prediction
We used the online tools UniProt Knowledgebase (UniProtKB) [11] (http://www.uniprot.org/uniprot/), Polymorphism Phenotyping v2 (PolyPhen-2) (http://genetics.bwh.harvard.edu/pph2/) and Sorting Intolerant From Tolerant (SIFT) (http://sift.bii.a-star.edu.sg/) to predict protein function of non-synonymous SNPs in CYP2C9 exon regions. Each variant was given a score based on the impact of its mutation on protein function. PolyPhen-2 results were divided into three categories: benign, potentially damaging, and probably damaging [12]. We referenced the previous reported that the SIFT output was then divided into four categories based on these scores: tolerant (0.201-1.00), borderline (0.101-0.20), potentially intolerant (0.051-0.10) and intolerant (0.00-0.05) [13].
Result
Genetic variants
In our study, 17 different SNPs were identified among Li subjects. We detected two missense mutations (3549 G > A and 42614 A > C) and two silent mutations (3514 T > C, and 50298 A > T). The two missense variants were identified in exon 3 and 7, respectively. And the two silent mutations located on exon 3 and 9, respectively (Table 2). However, these mutations have been reported in the NCBI database or in the Human CYPAllele Nomenclature Committee tables previously [14].
Table 2.
Frequency distribution of CYP2C9 polymorphisms in 100 Li subjects
| SNP | Region | Position | Nucleotide change | Amino-acid effect | Frequency % (n) |
|---|---|---|---|---|---|
| rs148342296 | Promoter | -477 | A > G | No translateda | 11/99 |
| rs9332105 | Intron 1 | 486 | G > C | No translated | 52/98 |
| rs201856860 | Exon 3 | 3514 | T > C | Ile112 = silentb | 1/100 |
| rs12414460 | Exon 3 | 3549 | G > A | Arg124Glnmissensec | 2/100 |
| rs9332127 | Intron 3 | 9032 | G > C | No translated | 3/99 |
| rs9332172 | Intron 5 | 33349 | A > G | No translated | 5/100 |
| rs148303924 | Intron 5 | 33622 | T > C | No translated | 1/100 |
| rs1057910 | Exon 7 | 42614 | A > C | Ile359Leumissensec | 2/99 |
| rs17847029 | Intron 7 | 42726 | C > T | No translated | 7/99 |
| rs93321230 | Intron 8 | 47545 | A > T | No translated | 2/100 |
| rs2298037 | Intron 8 | 47639 | C > T | No translated | 54/100 |
| rs147375734 | Intron 8 | 47947 | T > C | No translated | 3/98 |
| rs28371688 | Intron 8 | 50053 | G > A | No translated | 2/100 |
| rs1934669 | Intron 8 | 50056 | A > T | No translated | 56/100 |
| rs1057911 | Exon 9 | 50298 | A > T | Gly475 = silentb | 2/100 |
| rs9332244 | 3’-UTR | 50566 | A > G | No translated | 9/100 |
| rs9332245 | 3’-UTR | 50742 | T > A | No translated | 2/100 |
SNP, single nucleotide polymorphism;
Non translated: These synonymous SNP mutations have no effect on protein sequence;
silent mutation;
missense mutation.
Allele and genotype frequency and non-synonymous mutation protein function predicted
As Table 3 showed, we identified three CYP2C9 alleles: the wild-type CYP2C9 allele, CYP2C9*1, which found in 98% of the population. One common allele, CYP2C9*3, comprised only 1% of the variants, while no subjects with CYP2C9*2 were identified. Furthermore, the rare allele CYP2C9*42, which was first reported in Li Chinese population, was present in 1% of our study subjects. We also calculated three CYP2C9 genotypes in the Li population. We found 96 individuals with the wild-type (*1*1) genotype have normal enzyme activity. Other identified genotypes included 2 individuals with the heterozygous genotype (*1*3), which leads to decreased enzyme activity. Additionally, we also found 2 individuals with the rare genotype (*1*42). Furthermore, we used the online tools PolyPhen-2 and SIFT to predict protein function of non-synonymous SNPs in CYP2C9 exon regions. PolyPhen-2 utilized two models (HumDiv and HumVar), in which the latter is more rigorous in its false discovery rate (Figure 1). PolyPhen-2 results revealed that rs12414460 (Arg124Gln) was probably damaging in both models. The SIFT protein function prediction results (score = 0 intolerant) were consistent with PolyPhen-2 which also revealed probably damaging (Figure 1A). However, the protein function prediction results of the rs1057910 (Ile359Leu) was inconsistentwithPolyPhen-2 and SIFT. The result of SIFT prediction was scored 0.04, which indicted the protein function was intolerant, while PolyPhen-2 results showed benign (Figure 1B).
Table 3.
CYP2C9 allele and genotype frequencies in 100 Li individuals
| Allele | Number | Phenotype | Frequency |
|
| |||
| *1 | 196 | Normal | 98.00% |
| *3 | 2 | Decreased | 1.00% |
| *42 | 2 | / | 1.00% |
| Total | 200 | 100.00% | |
|
| |||
| Genotype | Number | Phenotype | Frequency |
|
| |||
| *1/*1 | 96 | Normal | 96.00% |
| *1/*3 | 2 | Decreased | 2.00% |
| *1/*42 | 2 | / | 2.00% |
| Total | 100 | 100.00% | |
*indicated genotype typewhich according database of “Human Cytochrome P450 Allele Nomenclature”.
Figure 1.
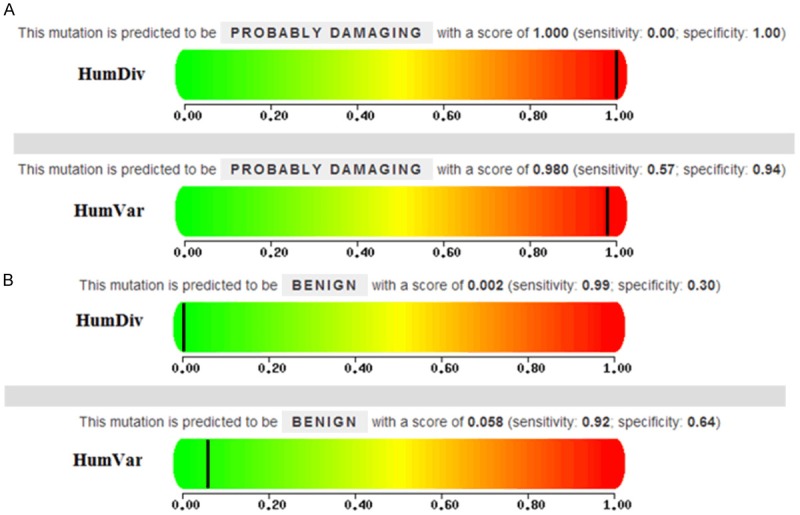
Protein function predicted by PolyPhen-2. A. Prediction of the mutation 3549 G > A. B. Prediction of the mutation 42614 A > C. HumDiv: is Mendelian disease variants vs divergence from close mammalian homologs of human proteins (5564 deleterious + 7539 neutral mutations from the same set of 978 human proteins). HumVar: is all human variants associated with some disease (except cancer mutations) or loss of activity/function vs common human polymorphism with no reported association with a disease of other effect (22196 deleterious + 21119 neutral mutations in 9679 human proteins).
Comparisons of inter-population
We further analyzed the distribution patterns of CYP2C9 allele between Li and other populations (involved East Asian, Middle East Arab, Africans, and Caucasian) around world (Table 4). Three major alleles, the wild-type allele CYP2C9*1, the prevalent allele CYP2C9*2 and CYP2C9*3 were compared. We found that the allele frequency of CYP2C9*1, CYP2C9*2 and CYP2C9*3 in our study was close to majority East Asian populations. In summary, the frequency of CYP2C9*1 was significant (P < 0.05) higher than Uyghur Chinese populations and Tamil Nadu. In contrast, CYP2C9*2 was significant (P < 0.05) lower than Uyghur Chinese, CYP2C9*3 was no significant with other East Asians. In Middle East Arab, the frequency of CYP2C9*1, CYP2C9*2 and CYP2C9*3 were significant different with all most Middle East Arabs. In Africans, we found the allele frequency of CYP2C9*1, CYP2C9*2 and CYP2C9*3 in our study was very similar to our study, the allele frequency of CYP2C9*1 was higher than African-American (P < 0.05). In Caucasians, we found that the allele frequency of CYP2C9*1 was significantly higher (P < 0.05) in Li compared with almost all Caucasian populations except Ecuadorian. In contrast, the allele frequency CYP2C9*2 in Li was significantly lower (P < 0.05) than all Caucasians which we involved in this study. The allele frequency of CYP2C9*3 was also obvious lower (P < 0.05) than Caucasian populations except for Brazilian, Mexican, Ecuadorian, Russian, Bolivian and American.
Table 4.
Three types of CYP2C9 allele frequencies in different populations
| Races | N | CYP2C9*1 | p | CYP2C9*2 | p | CYP2C9*3 | p | Ref |
|---|---|---|---|---|---|---|---|---|
| Li | 100 | 0.98 | / | 0 | / | 0.01 | / | Current study |
| East Asians | ||||||||
| Tibetan Chinese | 96 | 0.938 | 0.259 | 0 | / | 0.057 | 0.150 | [13] |
| Tibetan Chinese | 107 | 0.972 | 0.938 | 0 | / | 0.028 | 0.664 | [19] |
| Han Chineese | 2127 | 0.945 | 0.128 | 0.001 | / | 0.029 | 0.416 | [16] |
| Korean | 358 | 0.934 | 0.077 | 0 | / | 0.06 | 0.074 | [20] |
| Korean | 574 | 0.98 | 0.69 | 0 | / | 0.011 | 0.664 | [21] |
| Japanese | 218 | 0.979 | 0.714 | 0 | / | 0.021 | 0.815 | [22] |
| Uyghur Chinese | 214 | 0.902 | 0.013* | 0.096 | 0.001* | 0 | 0.318 | [23] |
| Bai Chinese | 132 | 0.955 | 0.501 | 0 | / | 0.045 | 0.246 | [19] |
| Hui Chinese | 164 | 0.954 | 0.448 | 0.046 | 0.073 | 0 | 0.379 | [23] |
| Mongolian Chinese | 560 | 0.97 | 0.820 | 0 | / | 0.03 | 0.422 | [24] |
| Vietnamese | 157 | 0.978 | 0.737 | 0 | / | 0.022 | 0.819 | [25] |
| Malay | 209 | 0.957 | 0.488 | 0.019 | 0.397 | 0.024 | 0.694 | [26] |
| Tamil Nadu | 135 | 0.907 | 0.022* | 0.026 | 0.280 | 0.067 | 0.070 | [27] |
| Middle East Arab | ||||||||
| Saudi (Al-Ahsa) | 131 | 0.844 | < 0.001* | 0.13 | < 0.001* | 0.023 | 0.809 | [28] |
| Saudi (Riyadh) | 192 | 0.792 | < 0.001* | 0.117 | < 0.001* | 0.091 | 0.007* | [29] |
| Egyptian | 247 | 0.820 | < 0.001* | 0.12 | < 0.001* | 0.06 | 0.082 | [30] |
| Jordanian | 263 | 0.797 | < 0.001* | 0.135 | < 0.001* | 0.068 | 0.026* | [31] |
| Lebanese | 161 | 0.792 | < 0.001* | 0.112 | < 0.001* | 0.096 | 0.005* | [32] |
| Omani | 189 | 0.897 | 0.01* | 0.074 | 0.012* | 0.029 | 0.535 | [33] |
| Africans | ||||||||
| Ethiopian | 150 | 0.934 | 0.171 | 0.043 | 0.090 | 0.023 | 0.785 | [34] |
| African-American | 600 | 0.867 | 0.001* | 0.028 | 0.180 | 0.02 | 0.775 | [35] |
| African-American | 490 | / | / | 0.011 | 1 | 0.018 | 0.888 | [36] |
| Beninese | 111 | 0.955 | 0.530 | 0 | / | 0 | 0.474 | [37] |
| Ghanaian | 204 | / | / | 0 | / | 0 | 0.329 | [6] |
| Iranian | 200 | / | / | 0.13 | < 0.001* | 0 | 0.333 | [38] |
| Caucasians | ||||||||
| Turkish | 499 | 0.794 | < 0.001* | 0.106 | < 0.001* | 0.1 | 0.003* | [39] |
| Brazilian | 103 | 0.83 | < 0.001* | 0.097 | 0.004* | 0.073 | 0.059 | [40] |
| Mexican | 98 | 0.86 | 0.002* | 0.08 | 0.012* | 0.06 | 0.125 | [41] |
| Ecuadorian | 194 | 0.93 | 0.069 | 0.054 | 0.042* | 0.015 | 0.855 | [42] |
| Swedish | 430 | 0.819 | < 0.001* | 0.107 | < 0.001* | 0.074 | 0.017* | [43] |
| Russian | 352 | 0.831 | < 0.001* | 0.119 | < 0.001* | 0.05 | 0.136 | [44] |
| Italian | 157 | 0.796 | < 0.001* | 0.112 | < 0.001* | 0.092 | 0.007* | [34] |
| British | 100 | 0.79 | < 0.001* | 0.125 | < 0.001* | 0.085 | 0.031* | [45] |
| Portuguese | 135 | 0.788 | < 0.001* | 0.132 | 0.0002* | 0.08 | 0.015* | [46] |
| Spanish | 1076 | 0.766 | < 0.001* | 0.156 | < 0.001* | 0.078 | 0.012* | [47] |
| French | 151 | 0.77 | < 0.001* | 0.15 | < 0.001* | 0.08 | 0.015* | [48] |
| Bolivian | 778 | 0.922 | 0.034* | 0.048 | 0.048* | 0.03 | 0.410 | [49] |
| Cuban | 132 | 0.72 | < 0.001* | 0.17 | < 0.001* | 0.11 | 0.002* | [50] |
| American | 100 | / | / | 0.08 | 0.011* | 0.06 | 0.124 | [51] |
| Croatian | 200 | 0.74 | < 0.001* | 0.165 | < 0.001* | 0.095 | 0.005* | [52] |
| German | 118 | 0.81 | < 0.001* | 0.14 | < 0.001* | 0.05 | 0.196 | [53] |
| Greek | 283 | / | / | 0.129 | < 0.001* | 0.081 | 0.012* | [54] |
| Belgian | 121 | 0.822 | < 0.001* | 0.1 | 0.001* | 0.074 | 0.050* | [37] |
Indicated P < 0.05.
Ref: Reference.
Linkage disequilibrium analysis
We further used Haploview software to assess LD between pairs of loci. The overall LD across the CYP2C9 gene is depicted in Figure 2. We found 19 red LD points (17 bright red and 2 light red) between two SNPs. But there was no LD block within CYP2C9 among this Li population study.
Figure 2.
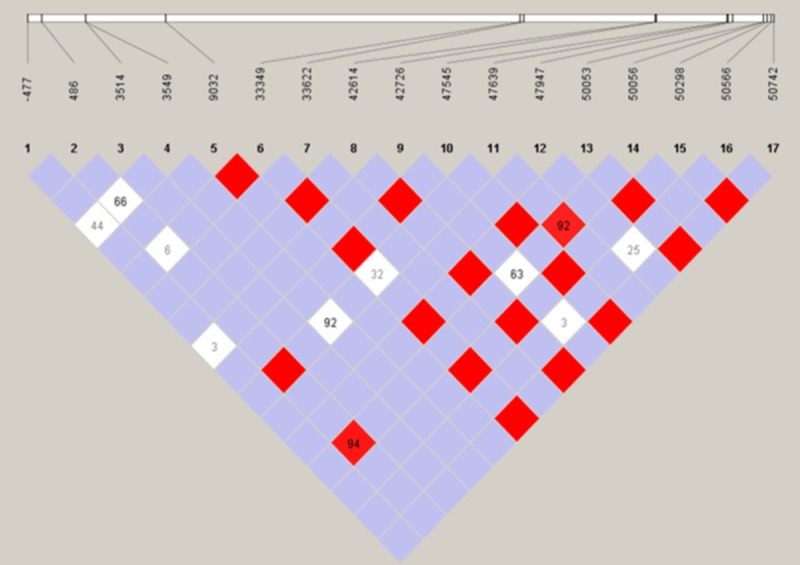
17 SNPs linkage disequilibrium (LD) analysis of CYP2C9 in Li population. Abbreviations: LD: Linkage disequilibrium; LOD: likelihood of odds; LD is indicated by bright red (very strong: LOD ≥ 2, D’ = 1), light red (LOD ≥ 2, D’ < 1) and blue (LOD < 2, D’ = 1) for intermediate LD, and white (none: LOD < 2, D’ < 1).
Discussion
This is the first study to screen the DNA sequence of CYP2C9 in Li populations, we found 17 different SNPs among 100 Li subjects. CYP2C9 genetic polymorphisms are highly relevant in the metabolism of clinically prescribed drugs and may influence patient responsiveness and adverse drug reactions. While previous studies have analyzed CYP2C9 genetic polymorphisms focus on Chinese Han, Tibetan, Bai, and Hui populations, butto date no study focused on Chinese Li population. Thus, our results provided a better understanding of CYP2C9 variants and added some database for promoting personalized medicine in Li patients.
Three major allelic polymorphisms (CYP2C9*1, CYP2C9*2, and CYP2C9*3) were compared between Li and other ethnic populations. In general, we found that allele frequencies identified in Li were most similar to East Asian and African populations but were different from those of Caucasian and Middle East Arab populations. For example, the frequencies of CYP2C9*2, and CYP2C9*3 among our Li were similar to East Asian and African populations which significantly lower than Middle East Arab and Caucasian, this was consistent with previous research [15]. The phenomenon of lacking CYP2C9*2 allele in the Li group is in accordance with its reported absence in the majority of East Asian populations. However, the variant of CYP2C9*2 was also identified in certain Asian populations, only found in Uyghur Chinese (P < 0.05). The Uyghur are an ethnic group of Central Asia. They are one of China’s 56 officially recognized ethnicities. Throughout the history of Central Asia, they left a lasting imprint on both the culture and tradition. Today in China, Uyghur live primarily in the Xinjiang Uygur Autonomous Region in the northwest of China. There are also existing Uygur communities in Kazakhstan, Kyrgyzstan, Mongolia, Uzbekistan, and Turkey. It is known that the genetic polymorphisms were influenced by environmental and genetic factors. We inferred that these differences may be attributed to the origin and geographical isolation experienced by different ethnic populations, as well as their dietary habits and lifestyles, or other factors, all of which may affect CYP2C9 polymorphisms. In addition, we also detected a rare mutations CYP2C9*42, which had be designated by the Human CYP Allele Nomenclature Committee [16].
We detected two missense mutations and two silent mutations in protein coding region. The non-synonymous mutation protein function predicted showed CYP2C9*42 (3549G > A result Arg124Gln amino acid alteration) result in protein damaging. The protein mutation prediction result of CYP2C9*3 (42614 A > Cinvolved Ile359Leu changed) was inconsistent between PolyPhen-2 and SIFT. SIFT scored 0.04 showed intolerant probably damaging but the PolyPhen-2 prediction result was benign. Although our result of CYP2C9*3 (42614 A > C result Ile359Leu amino acid alteration) protein function predicted was not uniform, but previous studies indicated that homozygous mutationshow 95% decreased the enzyme activity compared to the wild type [17]. However, the protein function prediction results of the variant CYP2C9*3 were highly inconsistent, since the accuracy of SIFT and PolyPhen-2 prediction typically reaches 63% and 75%, with false positive rates as high as 19% and 9%, respectively [18]. Therefore, the prediction results may be require and biased further experimental data to more reliably predict the effects of variants identified in our study.
Conclusions
This study provides the first pharmacogenomics information of CYP2C9 in Chinese Li population and the comparision of CYP2C9 allele frequencies with other populations worldwide. Our study provides a limited theoretical basis for safer drug administration and better therapeutic treatments in this unique population.
Acknowledgements
This study was funded by the Social Development Funding of Hainan Province (No. SF201402). We are grateful to the healthy individuals for their participation in this study. We also thank the clinicians and hospital staff who contributed to the sample and data collection.
Disclosure of conflict of interest
None.
References
- 1.Lee CR, Goldstein JA, Pieper JA. Cytochrome P450 2C9 polymorphisms: a comprehensive review of the in-vitro and human data. Pharmacogenetics. 2002;12:251–263. doi: 10.1097/00008571-200204000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Fleming I. Cytochrome p450 and vascular homeostasis. Circ Res. 2001;89:753–762. doi: 10.1161/hh2101.099268. [DOI] [PubMed] [Google Scholar]
- 3.Delozier TC, Kissling GE, Coulter SJ, Dai D, Foley JF, Bradbury JA, Murphy E, Steenbergen C, Zeldin DC, Goldstein JA. Detection of human CYP2C8, CYP2C9, and CYP2J2 in cardiovascular tissues. Drug Metab Dispos. 2007;35:682–688. doi: 10.1124/dmd.106.012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Z, Zhu Q, Ma Y, Huang D, Pan S, Xie X, Liu F, Cha E. Diplotypes of CYP2C9 gene is associated with coronary artery disease in the Xinjiang Han population for women in China. Lipids Health Dis. 2014;13:143. doi: 10.1186/1476-511X-13-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol. 2001;52:349–355. doi: 10.1046/j.0306-5251.2001.01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kudzi W, Dodoo AN, Mills JJ. Characterisation of CYP2C8, CYP2C9 and CYP2C19 polymorphisms in a Ghanaian population. BMC Med Genet. 2009;10:124. doi: 10.1186/1471-2350-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Weide J, Steijns LS, van Weelden MJ, de Haan K. The effect of genetic polymorphism of cytochrome P450 CYP2C9 on phenytoin dose requirement. Pharmacogenetics. 2001;11:287–291. doi: 10.1097/00008571-200106000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Adamec C. [Example of the Use of the Nonparametric Test. Test X2 for Comparison of 2 Independent Examples] . Cesk Zdrav. 1964;12:613–619. [PubMed] [Google Scholar]
- 9.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 10.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 11.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot. Methods Mol Biol. 2007;406:89–112. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 12.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin T, Geng T, He N, Shi X, Wang L, Yuan D, Kang L. Genetic polymorphism analysis of the drug-metabolizing enzyme CYP2C9 in a Chinese Tibetan population. Gene. 2015;567:196–200. doi: 10.1016/j.gene.2015.04.084. [DOI] [PubMed] [Google Scholar]
- 14.Suarez-Kurtz G. Pharmacogenomics in admixed populations. Trends Pharmacol Sci. 2005;26:196–201. doi: 10.1016/j.tips.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 15.Tabrizi AR, Zehnbauer BA, Borecki IB, McGrath SD, Buchman TG, Freeman BD. The frequency and effects of cytochrome P450 (CYP) 2C9 polymorphisms in patients receiving warfarin. J Am Coll Surg. 2002;194:267–273. doi: 10.1016/s1072-7515(01)01163-2. [DOI] [PubMed] [Google Scholar]
- 16.Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, Yang LP, Qian JC, Wang ZS, Zhu GH, Zhang XH, Ge RS, Hu GX, Cai JP. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. Pharmacogenomics J. 2014;14:85–92. doi: 10.1038/tpj.2013.2. [DOI] [PubMed] [Google Scholar]
- 17.King BP, Khan TI, Aithal GP, Kamali F, Daly AK. Upstream and coding region CYP2C9 polymorphisms: correlation with warfarin dose and metabolism. Pharmacogenetics. 2004;14:813–822. doi: 10.1097/00008571-200412000-00004. [DOI] [PubMed] [Google Scholar]
- 18.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 19.Zeng WT, Zheng QS, Huang M, Cen HJ, Lai Y, Chen WY, Zhao LZ, Leng XY. Genetic polymorphisms of VKORC1, CYP2C9, CYP4F2 in Bai, Tibetan Chinese. Pharmazie. 2012;67:69–73. [PubMed] [Google Scholar]
- 20.Bae JW, Kim HK, Kim JH, Yang SI, Kim MJ, Jang CG, Park YS, Lee SY. Allele and genotype frequencies of CYP2C9 in a Korean population. Br J Clin Pharmacol. 2005;60:418–422. doi: 10.1111/j.1365-2125.2005.02448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon YR, Shon JH, Kim MK, Lim YC, Lee HR, Park JY, Cha IJ, Shin JG. Frequency of cytochrome P450 2C9 mutant alleles in a Korean population. Br J Clin Pharmacol. 2001;51:277–280. doi: 10.1046/j.1365-2125.2001.00340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasu K, Kubota T, Ishizaki T. Genetic analysis of CYP2C9 polymorphism in a Japanese population. Pharmacogenetics. 1997;7:405–409. doi: 10.1097/00008571-199710000-00011. [DOI] [PubMed] [Google Scholar]
- 23.Zuo J, Xia D, Jia L, Guo T. Genetic polymorphisms of drug-metabolizing phase I enzymes CYP3A4, CYP2C9, CYP2C19 and CYP2D6 in Han, Uighur, Hui and Mongolian Chinese populations. Pharmazie. 2012;67:639–644. [PubMed] [Google Scholar]
- 24.Yang ZF, Cui HW, Hasi T, Jia SQ, Gong ML, Su XL. Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Mongolian population in China. Genet Mol Res. 2010;9:1844–1851. doi: 10.4238/vol9-3gmr938. [DOI] [PubMed] [Google Scholar]
- 25.Lee SS, Kim KM, Thi-Le H, Yea SS, Cha IJ, Shin JG. Genetic polymorphism of CYP2C9 in a Vietnamese Kinh population. Ther Drug Monit. 2005;27:208–210. doi: 10.1097/01.ftd.0000153402.91854.42. [DOI] [PubMed] [Google Scholar]
- 26.Ngow HA, Wan Khairina WM, Teh LK, Lee WL, Harun R, Ismail R, Salleh MZ. CYP2C9 polymorphism: prevalence in healthy and warfarintreated Malay and Chinese in Malaysia. Singapore Med J. 2009;50:490–493. [PubMed] [Google Scholar]
- 27.Adithan C, Gerard N, Vasu S, Balakrishnan R, Shashindran CH, Krishnamoorthy R. Allele and genotype frequency of CYP2C9 in Tamilnadu population. Eur J Clin Pharmacol. 2003;59:707–709. doi: 10.1007/s00228-003-0666-3. [DOI] [PubMed] [Google Scholar]
- 28.Alzahrani AM, Ragia G, Hanieh H, Manolopoulos VG. Genotyping of CYP2C9 and VKORC1 in the Arabic population of Al-Ahsa, Saudi Arabia. Biomed Res Int. 2013;2013:315980. doi: 10.1155/2013/315980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mirghani RA, Chowdhary G, Elghazali G. Distribution of the major cytochrome P450 (CYP) 2C9 genetic variants in a Saudi population. Basic Clin Pharmacol Toxicol. 2011;109:111–114. doi: 10.1111/j.1742-7843.2011.00692.x. [DOI] [PubMed] [Google Scholar]
- 30.Hamdy SI, Hiratsuka M, Narahara K, El-Enany M, Moursi N, Ahmed MS, Mizugaki M. Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br J Clin Pharmacol. 2002;53:596–603. doi: 10.1046/j.1365-2125.2002.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yousef AM, Bulatova NR, Newman W, Hakooz N, Ismail S, Qusa H, Zahran F, Anwar Ababneh N, Hasan F, Zaloom I, Khayat G, Al-Zmili R, Naffa R, Al-Diab O. Allele and genotype frequencies of the polymorphic cytochrome P450 genes (CYP1A1, CYP3A4, CYP3A5, CYP2C9 and CYP2C19) in the Jordanian population. Mol Biol Rep. 2012;39:9423–9433. doi: 10.1007/s11033-012-1807-5. [DOI] [PubMed] [Google Scholar]
- 32.Djaffar-Jureidini I, Chamseddine N, Keleshian S, Naoufal R, Zahed L, Hakime N. Pharmacogenetics of coumarin dosing: prevalence of CYP2C9 and VKORC1 polymorphisms in the Lebanese population. Genet Test Mol Biomarkers. 2011;15:827–830. doi: 10.1089/gtmb.2010.0248. [DOI] [PubMed] [Google Scholar]
- 33.Tanira MO, Al-Mukhaini MK, Al-Hinai AT, Al Balushi KA, Ahmed IS. Frequency of CYP2C9 genotypes among Omani patients receiving warfarin and its correlation with warfarin dose. Community Genet. 2007;10:32–37. doi: 10.1159/000096279. [DOI] [PubMed] [Google Scholar]
- 34.Scordo MG, Aklillu E, Yasar U, Dahl ML, Spina E, Ingelman-Sundberg M. Genetic polymorphism of cytochrome P450 2C9 in a Caucasian and a black African population. Br J Clin Pharmacol. 2001;52:447–450. doi: 10.1046/j.0306-5251.2001.01460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11:781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Limdi N, Goldstein J, Blaisdell J, Beasley T, Rivers C, Acton R. Influence of CYP2C9 Genotype on warfarin dose among African American and European Americans. Per Med. 2007;4:157–169. doi: 10.2217/17410541.4.2.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allabi AC, Gala JL, Desager JP, Heusterspreute M, Horsmans Y. Genetic polymorphisms of CYP2C9 and CYP2C19 in the Beninese and Belgian populations. Br J Clin Pharmacol. 2003;56:653–657. doi: 10.1046/j.1365-2125.2003.01937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zand N, Tajik N, Moghaddam AS, Milanian I. Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Iranian population. Clin Exp Pharmacol Physiol. 2007;34:102–105. doi: 10.1111/j.1440-1681.2007.04538.x. [DOI] [PubMed] [Google Scholar]
- 39.Aynacioglu AS, Brockmoller J, Bauer S, Sachse C, Guzelbey P, Ongen Z, Nacak M, Roots I. Frequency of cytochrome P450 CYP2C9 variants in a Turkish population and functional relevance for phenytoin. Br J Clin Pharmacol. 1999;48:409–415. doi: 10.1046/j.1365-2125.1999.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lima MV, Ribeiro GS, Mesquita ET, Victer PR, Vianna-Jorge R. CYP2C9 genotypes and the quality of anticoagulation control with warfarin therapy among Brazilian patients. Eur J Clin Pharmacol. 2008;64:9–15. doi: 10.1007/s00228-007-0385-2. [DOI] [PubMed] [Google Scholar]
- 41.A LL, Dorado P, O’Kirwan F, Jepson R, Licinio J, Wong ML. Lower frequency of CYP2C9*2 in Mexican-Americans compared to Spaniards. Pharmacogenomics J. 2004;4:403–406. doi: 10.1038/sj.tpj.6500278. [DOI] [PubMed] [Google Scholar]
- 42.Dorado P, Beltrán LJ, Machín E, Peñas-Lledó EM, Terán E, Llerena A CEIBA. FP Consortium of the Ibero-American Network of Pharmacogenetics and Pharmacogenomics RIBEF. Losartan hydroxylation phenotype in an Ecuadorian population: influence of CYP2C9 genetic polymorphism, habits and gender. Pharmacogenomics. 2012;13:1711–1717. doi: 10.2217/pgs.12.160. [DOI] [PubMed] [Google Scholar]
- 43.Yasar U, Eliasson E, Dahl ML, Johansson I, Ingelman-Sundberg M, Sjoqvist F. Validation of methods for CYP2C9 genotyping: frequencies of mutant alleles in a Swedish population. Biochem Biophys Res Commun. 1999;254:628–631. doi: 10.1006/bbrc.1998.9992. [DOI] [PubMed] [Google Scholar]
- 44.Gra O, Mityaeva O, Berdichevets I, Kozhekbaeva Z, Fesenko D, Kurbatova O, Goldenkova-Pavlova I, Nasedkina T. Microarray-based detection of CYP1A1, CYP2C9, CYP2C19, CYP2D6, GSTT1, GSTM1, MTHFR, MTRR, NQO1, NAT2, HLA-DQA1, and AB0 allele frequencies in native Russians. Genet Test Mol Biomarkers. 2010;14:329–342. doi: 10.1089/gtmb.2009.0158. [DOI] [PubMed] [Google Scholar]
- 45.Stubbins MJ, Harries LW, Smith G, Tarbit MH, Wolf CR. Genetic analysis of the human cytochrome P450 CYP2C9 locus. Pharmacogenetics. 1996;6:429–439. doi: 10.1097/00008571-199610000-00007. [DOI] [PubMed] [Google Scholar]
- 46.Oliveira E, Marsh S, van Booven DJ, Amorim A, Prata MJ, McLeod HL. Pharmacogenetically relevant polymorphisms in Portugal. Pharmacogenomics. 2007;8:703–712. doi: 10.2217/14622416.8.7.703. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez-Diz P, Estany-Gestal A, Aguirre C, Blanco A, Carracedo A, Ibanez L, Passiu M, Provezza L, Ramos-Ruiz R, Ruiz B, Salado-Valdivieso I, Velasco EA, Figueiras A. Prevalence of CYP2C9 polymorphisms in the south of Europe. Pharmacogenomics J. 2009;9:306–310. doi: 10.1038/tpj.2009.16. [DOI] [PubMed] [Google Scholar]
- 48.Yang JQ, Morin S, Verstuyft C, Fan LA, Zhang Y, Xu CD, Barbu V, Funck-Brentano C, Jaillon P, Becquemont L. Frequency of cytochrome P450 2C9 allelic variants in the Chinese and French populations. Fundam Clin Pharmacol. 2003;17:373–376. doi: 10.1046/j.1472-8206.2003.00148.x. [DOI] [PubMed] [Google Scholar]
- 49.Bravo-Villalta HV, Yamamoto K, Nakamura K, Baya A, Okada Y, Horiuchi R. Genetic polymorphism of CYP2C9 and CYP2C19 in a Bolivian population: an investigative and comparative study. Eur J Clin Pharmacol. 2005;61:179–184. doi: 10.1007/s00228-004-0890-5. [DOI] [PubMed] [Google Scholar]
- 50.Llerena A, Alvarez M, Dorado P, Gonzalez I, Penas LE, Perez B, Cobaleda J, Calzadilla LR. Interethnic differences in the relevance of CYP2C9 genotype and environmental factors for diclofenac metabolism in Hispanics from Cuba and Spain. Pharmacogenomics J. 2014;14:229–234. doi: 10.1038/tpj.2013.28. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan-Klose TH, Ghanayem BI, Bell DA, Zhang ZY, Kaminsky LS, Shenfield GM, Miners JO, Birkett DJ, Goldstein JA. The role of the CYP2C9-Leu359 allelic variant in the tolbutamide polymorphism. Pharmacogenetics. 1996;6:341–349. doi: 10.1097/00008571-199608000-00007. [DOI] [PubMed] [Google Scholar]
- 52.Bozina N, Granic P, Lalic Z, Tramisak I, Lovric M, Stavljenic-Rukavina A. Genetic polymorphisms of cytochromes P450: CYP2C9, CYP2C19, and CYP2D6 in Croatian population. Croat Med J. 2003;44:425–428. [PubMed] [Google Scholar]
- 53.Burian M, Grosch S, Tegeder I, Geisslinger G. Validation of a new fluorogenic real-time PCR assay for detection of CYP2C9 allelic variants and CYP2C9 allelic distribution in a German population. Br J Clin Pharmacol. 2002;54:518–521. doi: 10.1046/j.1365-2125.2002.01693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arvanitidis K, Ragia G, Iordanidou M, Kyriaki S, Xanthi A, Tavridou A, Manolopoulos VG. Genetic polymorphisms of drug-metabolizing enzymes CYP2D6, CYP2C9, CYP2C19 and CYP3A5 in the Greek population. Fundam Clin Pharmacol. 2007;21:419–426. doi: 10.1111/j.1472-8206.2007.00510.x. [DOI] [PubMed] [Google Scholar]


