Abstract
Acute ischemic stroke (AIS) is one of the leading causes of death and disability worldwide. Circulating microRNAs (miRNAs) have been identified as potential biomarkers in the diagnosis and prognosis of multiple human diseases including AIS. In this study, serum expression levels of miR-15a, miR-16, and miR-17-5p were detected in AIS patients (n = 106) and healthy controls (n = 120) using quantitative real-time polymerase chain reaction (qRT-PCR). Statistical analyses were performed to investigate the associations between miRNA levels and AIS risk. The serum expression levels of miR-15a, miR-16, and miR-17-5p increased by 8.3 fold (P = 0.0104), 42 fold (P < 0.0001), and 9.9 fold (P = 0.0002) in AIS patients compared to controls, respectively. Multivariate logistic regression demonstrated that serum miR-17-5p level was a significant and independent predictor for determining the presence of AIS. Receiver operating characteristic (ROC) analysis revealed areas under the curves (AUCs) of 0.698 (95% confidence interval [CI]: 0.559-0.837, P = 0.01), 0.82 (95% CI: 0.71-0.931, P < 0.001), and 0.784 (95% CI: 0.666-0.903, P < 0.001) for miR-15a, miR-16, and miR-17-5p, respectively, while the AUC increased to 0.845 (95% CI: 0.74-0.949, P < 0.001) for the combination of these three micoRNAs. Our findings indicate that elevated serum expression of miR-15a, miR-16, and miR-17-5p is strongly associated with AIS and that the combination of these three microRNAs may be a promising serum biomarker for AIS.
Keywords: Acute ischemic stroke (AIS), MiR-15a, MiR-16, MiR-17-5p, Biomarker
Introduction
Stroke is the second leading cause of death worldwide and the leading cause of years of life lost due to premature mortality in East Asia; the predominant type is acute ischemic storke (AIS) [1]. It is estimated that someone has a stroke every 40 s, and a person dies of a stroke every 4 min in the U.S. [2]. The diagnosis of AIS still relies on time-consuming processes including medical history, clinical examination, and neuroimaging [3]. There is a need for rapid, accurate, specific, and sensitive biomarker-based testing to identify AIS patients; such a test might improve risk predication, early diagnosis, and prognostic assessments and could ultimately impact clinical outcomes and quality of life [4]. Although several blood biomarker candidates have been reported to be associated with AIS onset, none has been widely used in clinical practice [5].
MicroRNAs (miRNAs) are endogenous non-coding RNA molecules that play important gene-regulatory roles in plants and animals by pairing with the 3’ untranslated regions (3’UTRs) of specific protein-coding genes to repress their translation or induce their degradation [6]. It is estimated that more than one-third of human genes are conserved miRNA targets [7]. MiRNAs play key regulatory roles in various physiological processes and the pathogeneses of several diseases including cancer [8]. Recent studies have demonstrated altered miRNA expression profiles in AIS patients [9] and rodent models [10]. Specific serum miRNAs might serve as potential biomarkers of AIS [11]; for example, miR-15a and miR-16 are increased in the serum of patients with critical limb ischemia [12], and miR-15a was identified as a key factor of both myocardial ischemia [13] and ischemic stroke [14]. In addition, miR-17-5p may be a critical factor in post-stroke adult neurogenesis [15]. Taking these findings together, we hypothesized that the combination of miR-15a, miR-16, and miR-17-5p may be a useful AIS biomarker.
The aim of this study was to investigate the expressions of miR-15, miR-16, and miR-17-5p in the peripheral blood of AIS patients and healthy controls to evaluate the utility of these three miRNAs as AIS serum biomarkers.
Subjects and methods
Subject recruitment and sample collection
A total of 106 AIS patients admitted to Emergency Department of the Second Affiliated Hospital of Nanjing Medical University between August 2012 and February 2015 were enrolled in this study. The cohort included 55 men and 51 women with a mean age of 64.8 years (range, 39-88 years). AIS was confirmed with brain computed tomography (CT) or magnetic resonance imaging (MRI). Patients with the following disorders were excluded from the study: (1) symptoms indicative of subarachnoid hemorrhage, even if no imaging findings of hemorrhage were found on CT or MRI; (2) intracranial hemorrhage; (3) acute myocardial infarction; and (4) critical limb ischemia. Age- and sex-matched healthy controls without symptoms of AIS and history of cerebrovascular diseases were recruited from the physical examination center.
Clinical characteristics and biochemical parameters
Peripheral blood samples were collected from all participants before treatment and transferred to separation gel vacuum procoagulant collective tubes. Serum samples were collected by centrifugation at 3500 r/min for 10 min and stored at -70°C for subsequent experiments. Total cholesterol, triglycerides, low-density lipoprotein (LDL), high-density lipoprotein (HDL), apolipoprotein A1 (ApoA1), apolipoprotein B (ApoB), and lipoprotein a (Lpa) were measured from samples collected after at least 12 h of fasting, using the Toshiba 200FR Neo chemistry autoanalyser (Toshiba Medical Systems, Tokyo, Japan). Body mass index (BMI) was calculated as weight divided by height squared (kg/m2).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from serum samples using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s instructions. Two chloroform purification steps were performed to remove proteins. RNA concentration and purity were determined with a NanoDrop™ ND-2000c Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and RNA quality was assessed using denaturing gels (15% polyacrylamide and 1% agarose).
The miRNAs assessed in the present study included hsa-miR-15a (miRBase aAccession number: MIMAT0000068), hsa-miR-16 (miRBase accession number: MIMAT0000069), and has-miR-17-5p (miRBase accession number: MIMAT0000070). The expression level of each miRNA relative to U6 snRNA was confirmed. Total RNA (1000 ng) was reverse-transcribed to cDNA using TaqMan microRNA RT Kit and stem-loop RT primers (Applied Biosystems, Foster City, CA, USA) with a final volume of 20 μL (added to 5 μL double-distilled water) according to the manufacturer’s protocol. QRT-PCR was performed with 1 μL reverse-transcribed product added into each well of a 384-well optical plate using the TaqMan PCR kit on the ABI 7900 Real-Time PCR System (Applied Biosystems). The reaction process was initiated at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min as described in the protocol. An equal number of samples from AIS patients and controls were arranged on each plate. All reactions were performed in triplicate. After the reactions, the Ct values were determined using the fixed threshold settings, and mean Ct was estimated from triplicate PCR results. The expression levels of the three target miRNAs relative to U6 snRNA were determined using the 2-ΔCt method.
Ethical consideration
This study was approved by the Institutional Ethics Committee of the Second Affiliated Hospital of Nanjing Medical University and the Institutional Review Board of Nanjing Medical University. All procedures involved in human subjects were managed in accordance with national law and regulation and the Helsinki Declaration. Written informed consent was obtained from all participants after they received a detailed description of the potential benefits of the study.
Statistical analysis
Statistical analysis was performed using SPSS statistical software (version 19.0, IBM Corp., Armonk, NY, USA). Normally distributed variables are expressed as mean ± standard deviation (SD), non-normally distributed variables are expressed as medians (25th, 75th percentiles), and categorical variables are expressed as number (%). All miRNA expression levels were non-normally distributed. Mann-Whitney U tests were used to compare cases and controls. Comparisons for all proportions were performed using Pearson’s χ2 test. Spearman correlation coefficients were calculated to determine associations between miRNA levels and various laboratory markers. We used unconditional logistic regression analyses to assess the association between AIS and the miRNAs while simultaneously controlling for multiple covariates including age, sex, BMI, smoking, alcohol consumption, hypertension, diabetes mellitus, and hyperlipidemia. Receiver operating characteristic (ROC) curves were plotted for each miRNA and the combination of the three miRNAs. All tests were two-sided, and significance was set at P < 0.05.
Results
Baseline participant characteristics
The baseline characteristics of the 226 participants are listed in Table 1. There was no difference in age, BMI, sex ratio, smoking, or drinking between the two groups. The proportions of hypertension and diabetes mellitus were higher in the AIS group. Among the laboratory markers, AIS patients had higher levels of ApoB and lower levels of HDL and ApoA1. Total cholesterol, triglycerides, LDL, and Lpa did not differ between the two groups.
Table 1.
Baseline participant characteristics
| Control | AIS | ||
|---|---|---|---|
|
|
|
||
| Characteristic | n = 120 | n = 106 | P |
| Age (years) | 61.5 ± 9.0 | 64.8 ± 11.1 | 0.224 |
| Male sex n (%) | 58 (48.3) | 55 (51.9) | 0.085 |
| BMI (kg/m2) | 23.5 ± 3.6 | 23.9 ± 3.0 | 0.723 |
| Smoking n (%) | 25 (20.8) | 36 (34.0) | 0.203 |
| Drinking n (%) | 54 (45.0) | 51 (48.1) | 0.862 |
| Hypertension n (%) | 43 (35.8) | 64 (60.4) | 0.000 |
| Diabetes mellitus n (%) | 38 (31.7) | 55 (51.9) | 0.002 |
| Total cholesterol (mmol/L) | 4.61 ± 1.15 | 4.62 ± 1.04 | 0.964 |
| Triglycerides (mmol/L) | 1.24 (0.83, 1.85) | 1.31 (0.94, 1.99) | 0.493 |
| LDL (mmol/L) | 2.83 ± 0.90 | 2.93 ± 0.95 | 0.706 |
| HDL (mmol/L) | 1.23 ± 0.39 | 0.99 ± 0.20 | 0.009 |
| ApoA1 (g/L) | 1.47 (1.17, 1.68) | 1.14 (1.0, 1.41) | 0.013 |
| ApoB (g/L) | 0.78 ± 0.29 | 0.96 ± 0.23 | 0.030 |
| Lpa (g/L) | 0.14 (0.07, 0.54) | 0.22 (0.12, 0.46) | 0.501 |
AIS, acute ischemic stroke; ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein; Lpa, lipoprotein(a). Data are presented as mean ± standard deviation or medians (25th percentile, 75th percentile).
Serum miR-15a, miR-16, and miR-17-5p levels
Serum levels of miR-15a, miR-16, and miR-17-5p were significantly higher in AIS patients compared to control subjects (Figure 1). The expression of miR-15a, miR-16, and miR-17-5p were increased by 8.3 fold (P = 0.0104), 42 fold (P < 0.0001), and 9.9 fold (P = 0.0002) in the serum of AIS patients relative to controls.
Figure 1.
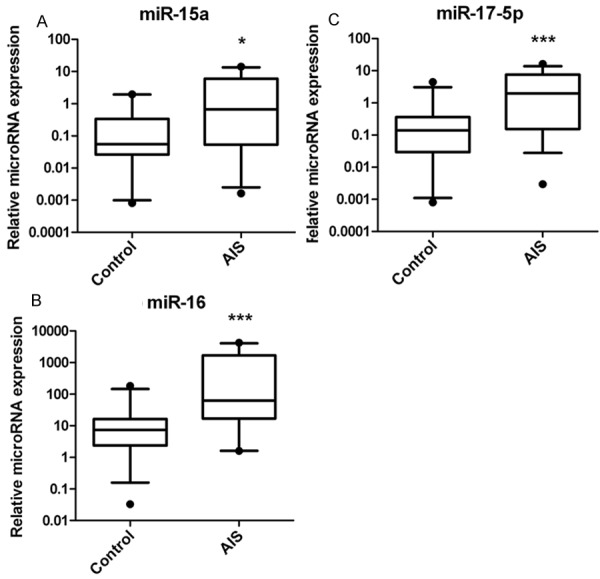
Quantitative real-time polymerase chain reaction (qRTPCR) was performed to measure miR-15a, miR-16, and miR-17-5p expression levels in AIS patients and healthy controls. The serum expression levels of miR-15a (A), miR-16 (B), and miR-17-5p (C) were increased by 8.3 fold (P = 0.0104), 42 fold (P < 0.0001), and 9.9 fold (P = 0.0002) in AIS patients (n = 106) as compared to healthy controls (n = 120). *P < 0.05, ***P < 0.001.
Association of serum miRNA expression levels with clinical characteristics
Serum miR-15a levels showed a significant positive correlation with age (r = 0.276, P < 0.05; Table 2). There was a strong negative correlation between serum miR-16 levels and HDL (r = -0.376, P < 0.01) and ApoA1 (r = -0.301, P < 0.05) (Table 3). No significant correlation was observed between miR-17-5p and any clinical characteristic (Table 4).
Table 2.
Correlations between miR-15a levels and clinical parameters
| Characteristic | Correlation coefficient (r) | P |
|---|---|---|
| Age (years) | 0.276 | 0.040 |
| BMI (kg/m2) | 0.307 | 0.057 |
| Total cholesterol (mg/dL) | 0.032 | 0.821 |
| Triglycerides (mg/dL) | -0.091 | 0.515 |
| LDL (mg/dL) | 0.119 | 0.399 |
| HDL (mg/dL) | -0.250 | 0.074 |
| ApoA1 (g/L) | -0.171 | 0.263 |
| ApoB (g/L) | 0.204 | 0.179 |
| Lpa (g/L) | 0.001 | 0.996 |
ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; BMI, body mass index; HDL, high-density lipoprotein; LDL, lowdensity lipoprotein; Lpa, lipoprotein(a).
Table 3.
Correlation between miR-16 levels and clinical parameters
| Characteristic | Correlation coefficient (r) | P |
|---|---|---|
| Age (years) | 0.047 | 0.732 |
| BMI (kg/m2) | 0.065 | 0.693 |
| Total cholesterol (mg/dL) | -0.076 | 0.589 |
| Triglycerides (mg/dL) | -0.031 | 0.826 |
| LDL (mg/dL) | -0.067 | 0.636 |
| HDL (mg/dL) | -0.376 | 0.006 |
| ApoA1 (g/L) | -0.301 | 0.044 |
| ApoB (g/L) | 0.233 | 0.123 |
| Lpa (g/L) | -0.109 | 0.489 |
ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; BMI, body mass index; HDL, high-density lipoprotein; LDL, low-density lipoprotein; Lpa, lipoprotein(a).
Table 4.
Correlation between miR-17-5p levels and clinical parameters
| Characteristic | Correlation coefficient (r) | P |
|---|---|---|
| Age (years) | 0.101 | 0.461 |
| BMI (kg/m2) | -0.179 | 0.277 |
| Total cholesterol (mg/dL) | 0.097 | 0.491 |
| Triglycerides (mg/dL) | 0.234 | 0.091 |
| LDL (mg/dL) | -0.022 | 0.874 |
| HDL (mg/dL) | -0.207 | 0.140 |
| ApoA1 (g/L) | -0.077 | 0.614 |
| ApoB (g/L) | 0.154 | 0.314 |
| Lpa (g/L) | 0.079 | 0.617 |
ApoA1, apolipoprotein A1; ApoB, apolipoprotein B; BMI, body mass index; HDL, high-density lipoprotein; LDL, lowdensity lipoprotein; Lpa, lipoprotein(a).
Logistic regression analysis
Simple logistic regression analysis revealed that hypertension; diabetes mellitus; and HDL, ApoA1, and miR-15a, miR-16, and miR-17-5p levels were associated with the presence of AIS as diagnosed by the CT or MRI. These variables were entered into a backward, stepwise, multivariate logistic regression model. The results demonstrated that serum miR-17-5p level was a significant and independent predictor for AIS (Table 5).
Table 5.
Logistic regression analysis for presence of AIS in participants
| Univariate logistic regression | Multivariate logistic regression | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| OR | 95% CI | P | OR | 95% CI | P | |
| Hypertension | 6.900 | 2.10-22.57 | 0.001 | 34.200 | 1.845-634.1 | 0.018 |
| Diabetes mellitus | 7.527 | 2.21-25.60 | 0.001 | 19.820 | 1.27-351.61 | 0.033 |
| HDL (mg/dL) | 0.035 | 0.003-0.425 | 0.009 | 0.0250 | 0.00-4.82 | 0.17 |
| ApoA1 (g/L) | 0.730 | 0.172-5.155 | 0.936 | - | - | - |
| ApoB (g/L) | 26.863 | 1.428-505.12 | 0.028 | 5.328 | 0.008-3424 | 0.612 |
| miR-15a | 1.710 | 1.021-2.866 | 0.042 | 1.314 | 0.344-5.018 | 0.952 |
| miR-16 | 1.013 | 0.999-1.027 | 0.077 | - | - | - |
| miR-17-5p | 1.943 | 1.139-3.313 | 0.015 | 3.968 | 1.001-14.29 | 0.035 |
ApoA1, apolipoprotein A1; ApoB, apolipoprotein B CI, confidence interval; HDL, high-density lipoprotein; OR, odds ratio.
ROC analysis
ROC analysis was performed to evaluate whether the tested serum miRNAs were useful AIS biomarkers (Figure 2). The area under curve (AUC) was 0.698 (95% confidence interval [CI]: 0.559-0.837, P = 0.01), 0.82 (95% CI: 0.71-0.931, P < 0.001) and 0.784 (95% CI: 0.666-0.903, P < 0.001) for miR-15a, miR-16, and miR-17-5p, respectively, while the AUC increased to 0.845 (95% CI: 0.74-0.949, P < 0.001) for the combination of all three miRNAs.
Figure 2.
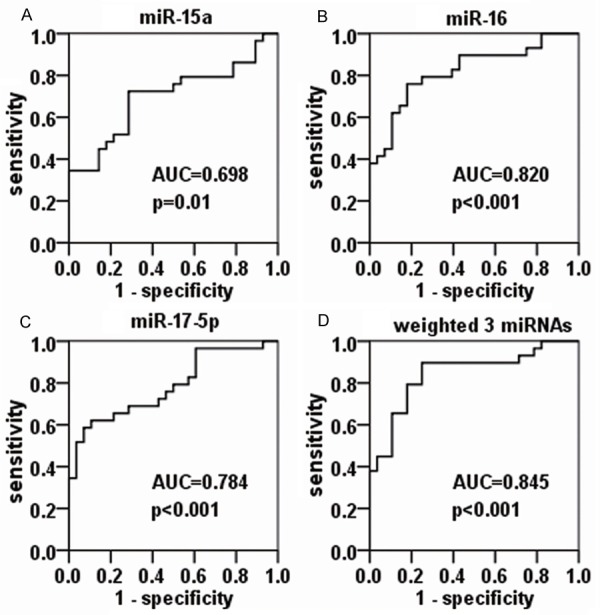
Receiver operating characteristic (ROC) analysis of miR-15a, miR-16, and miR-17-5p for AIS. The areas under curve (AUCs) are 0.698 (95% CI: 0.559-0.837, P = 0.01), 0.82 (95% CI: 0.71-0.931, P < 0.001), and 0.784 (95% CI: 0.666-0.903, P < 0.001) for miR-15a (A), miR-16 (B), and miR-17-5p (C), respectively, and the AUC increases to 0.845 (95% CI: 0.740-0.949, P < 0.001) for the combination of all three miRNAs (D).
Discussion
Circulating miRNAs have been identified as potential biomarkers for multiple conditions including cancer [16,17] and cardiovascular and cerebrovascular diseases [18,19]. The results of the present study demonstrate elevated serum levels of miR-15a, miR-16, and miR-17-5p in AIS patients compared to the controls. Moreover, miR-17-5p was an independent predictor for the presence of AIS. An ROC analysis revealed that the combination of miR-15a, miR-16, and miR-17-5p may be a potential AIS biomarker.
Both miR-15a and miR-16 are localized in the minimally deleted region at chromosome 13q14 and are highly expressed in CD5+ B cells [20]. Previously, miR-15a and miR-16 were predominantly studied in chronic lymphocytic leukemia (CLL) [21,22] and tumors [23,24]. MiR-15a and miR-16 act as tumor suppressor genes in pituitary tumors by directly targeting Sox5, imply that these miRNAs have potential as therapeutic targets for invasive pituitary tumors [25]. miR-15a and miR-16 induce apoptosis by targeting Bcl2 and control cell proliferation by targeting Akt3, nuclear factor kappa B (NF-κB), and various cell cycle regulators such as cyclins D1 (CCND1) and D2 (CCND2) [26]. Recent studies have shown miR-15a and miR-16 suppress angiogenesis by targeting vascular endothelial growth factor (VEGF) and fibroblast growth factor (FGF)-2 [26,27]. Gao et al. reported that miR-15a plays a significant role in the regulation of neuronal maturation, and its overexpression inhibits dendritic morphogenesis in immature neurons [28]. In addition, these two miRNAs have been demonstrated to play important roles in limb ischemia [12]. miR-15a was recently identified as a key factor involved in myocardial ischemia [13] and ischemic stroke [14]. Yin et al. reported that oxygen-glucose deprivation may activate miR-15a expression in cerebral vascular endothelial cells; decreases in miR-15a expression may protect against blood-brain barrier disruption and cerebral infarction in mice after transient focal cerebral ischemia [14]. These findings suggest that miR-15a may be a viable therapeutic target in addition to a diagnostic tool.
Results obtained in the last decade suggest that miR-17-5p could be a diagnostic biomarkers for breast cancer [29]. It was recently shown that miR-17-5p has more complex functions in cell cycling than miR-15a and miR-16 [30-32], and the miR17-92 cluster may be a critical factor in post-stroke adult neurogenesis [15]. Hong et al. showed that a synthetic miR-17-5p mimic is able to rescue the proliferation defect of Dicer1-null astrocytes, while an antisense inhibitor of miR-17-5p blocks lipopolysaccharide-induced astrocytic proliferation [33]. In this case-control study, we observed a significant positive association between serum miR-17-5p levels and the risk of AIS that persisted after adjusting for various AIS risk factors in multivariate models. This result suggests that miR-17-5p might be an independent predictor for AIS. Chen et al. reported that miR-17-5p may be a circulating biomarker for coronary atherosclerosis severity in patients with coronary artery disease [34]. We did not observe a significant correlation between miR-17-5p and serum lipid levels in the present study.
Individual miRNAs have been identified as biomarkers of AIS; however, they have not shown satisfactory sensitivity or specificity. MiR-15a, miR16-1, and the miR-17-92 cluster play important regulatory roles in cell proliferation, differentiation, and apoptosis. High levels of miR-15a, miR-16-1, and miR-17 are associated with shorter progression-free survival (PFS) in multiple myeloma, suggesting poor prognosis [35]. Integration of multiple miRNAs as blood biomarkers would be useful for the diagnosis, treatment, and management of AIS [36]. MiR-15a and miR-16 may influence angiogenesis, whereas miR-17-5p could affect neurogenesis, two critical processes for repairing cerebral ischemia injury. We observed higher levels of miR-15a, miR-16, and miR-17-5p in AIS patients and found that the combination of these three miRNAs showed great diagnostic value.
It is important to consider our findings in the context of several limitations. Firstly, the biological functions of miR-15a, miR-16 and miR-17-5p were not investigated as we did not examine their target genes. The biological function of an individual miRNA varies greatly because it can target multiple mRNA sites. Although a large number of miR-15a/16 and miR-17-5p target genes were identified previously, their exact roles in the brain after AIS onset remain to be clarified. Secondly, whether the expressions of miR-15a, miR-16, and miR-17-5p undergo dynamic changes requires further investigation and we did not perform any analyses to determine whether miRNA levels could be used to predict outcome. Thirdly, we assessed a relatively small number of subjects.
Conclusions
In conclusion, our findings show up-regulation of serum miR-15a, miR-16, and miR-17-5p levels in AIS patients compared to healthy controls. MiR-17-5p was an independent predictor of AIS. The findings suggest that the combination of miR-15a, miR-16, and miR-17-5p may be a promising AIS biomarker. Repeated miRNAs measurements in a larger-scale study and mechanistic experiments into an association between AIS and these three miRNAs are warranted to confirm their utility as biomarkers.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant no. 81270428, 81470501), Scientific Research Project of Jiangsu Provincial Department of Health (grant no. H201206), and Key Project of Nanjing Medical University (grant no. 2011NJMU252).
Disclosure of conflict of interest
None.
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA, Memish ZA. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, Huffman MD, Judd SE, Kissela BM, Kittner SJ, Lackland DT, Lichtman JH, Lisabeth LD, Mackey RH, Magid DJ, Marcus GM, Marelli A, Matchar DB, McGuire DK, Mohler ER 3rd, Moy CS, Mussolino ME, Neumar RW, Nichol G, Pandey DK, Paynter NP, Reeves MJ, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Wong ND, Woo D, Turner MB. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams HP Jr, del Zoppo G, Alberts MJ, Bhatt DL, Brass L, Furlan A, Grubb RL, Higashida RT, Jauch EC, Kidwell C, Lyden PD, Morgenstern LB, Qureshi AI, Rosenwasser RH, Scott PA, Wijdicks EF. Guidelines for the early management of adults with ischemic stroke: a guideline from the American Heart Association/American Stroke Association Stroke Council, Clinical Cardiology Council, Cardiovascular Radiology and Intervention Council, and the Atherosclerotic Peripheral Vascular Disease and Quality of Care Outcomes in Research Interdisciplinary Working Groups: the American Academy of Neurology affirms the value of this guideline as an educational tool for neurologists. Stroke. 2007;38:1655–1711. doi: 10.1161/STROKEAHA.107.181486. [DOI] [PubMed] [Google Scholar]
- 4.Saenger AK, Christenson RH. Stroke biomarkers: progress and challenges for diagnosis, prognosis, differentiation, and treatment. Clin Chem. 2010;56:21–33. doi: 10.1373/clinchem.2009.133801. [DOI] [PubMed] [Google Scholar]
- 5.Rothstein L, Jickling GC. Ischemic stroke biomarkers in blood. Biomark Med. 2013;7:37–47. doi: 10.2217/bmm.12.104. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 8.Jansson MD, Lund AH. MicroRNA and cancer. Mol Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4:e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee ST, Chu K, Jung KH, Yoon HJ, Jeon D, Kang KM, Park KH, Bae EK, Kim M, Lee SK, Roh JK. MicroRNAs induced during ischemic preconditioning. Stroke. 2010;41:1646–1651. doi: 10.1161/STROKEAHA.110.579649. [DOI] [PubMed] [Google Scholar]
- 11.Xu J, Zhao J, Evan G, Xiao C, Cheng Y, Xiao J. Circulating microRNAs: novel biomarkers for cardiovascular diseases. J Mol Med (Berl) 2012;90:865–875. doi: 10.1007/s00109-011-0840-5. [DOI] [PubMed] [Google Scholar]
- 12.Spinetti G, Fortunato O, Caporali A, Shantikumar S, Marchetti M, Meloni M, Descamps B, Floris I, Sangalli E, Vono R, Faglia E, Specchia C, Pintus G, Madeddu P, Emanueli C. MicroRNA-15a and microRNA-16 impair human circulating proangiogenic cell functions and are increased in the proangiogenic cells and serum of patients with critical limb ischemia. Circ Res. 2013;112:335–346. doi: 10.1161/CIRCRESAHA.111.300418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu LF, Liang Z, Lv ZR, Liu XH, Bai J, Chen J, Chen C, Wang Y. MicroRNA-15a/b are upregulated in response to myocardial ischemia/reperfusion injury. J Geriatr Cardiol. 2012;9:28–32. doi: 10.3724/SP.J.1263.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yin KJ, Fan Y, Hamblin M, Zhang J, Zhu T, Li S, Hawse JR, Subramaniam M, Song CZ, Urrutia R, Lin JD, Chen YE. KLF11 mediates PPARgamma cerebrovascular protection in ischaemic stroke. Brain. 2013;136:1274–1287. doi: 10.1093/brain/awt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu XS, Chopp M, Wang XL, Zhang L, Hozeska-Solgot A, Tang T, Kassis H, Zhang RL, Chen C, Xu J, Zhang ZG. MicroRNA-17-92 cluster mediates the proliferation and survival of neural progenitor cells after stroke. J Biol Chem. 2013;288:12478–12488. doi: 10.1074/jbc.M112.449025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, Hu Z, Zhuang R, Ning G, Zhang C, Yuan Y, Li Z, Zen K, Ba Y, Zhang CY. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–618. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 18.Weng H, Shen C, Hirokawa G, Ji X, Takahashi R, Shimada K, Kishimoto C, Iwai N. Plasma miR-124 as a biomarker for cerebral infarction. Biomed Res. 2011;32:135–141. doi: 10.2220/biomedres.32.135. [DOI] [PubMed] [Google Scholar]
- 19.Gan CS, Wang CW, Tan KS. Circulatory microRNA-145 expression is increased in cerebral ischemia. Genet Mol Res. 2012;11:147–152. doi: 10.4238/2012.January.27.1. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and downregulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C, Bartucci M, Muto G, Peschle C, De Maria R. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 24.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, Lin CP, Ribatti D, Rollins BJ, Witzig TE, Anderson KC, Ghobrial IM. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–6680. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renjie W, Haiqian L. MiR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015;356:568–578. doi: 10.1016/j.canlet.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Sun CY, She XM, Qin Y, Chu ZB, Chen L, Ai LS, Zhang L, Hu Y. miR-15a and miR-16 affect the angiogenesis of multiple myeloma by targeting VEGF. Carcinogenesis. 2013;34:426–435. doi: 10.1093/carcin/bgs333. [DOI] [PubMed] [Google Scholar]
- 27.Yin KJ, Olsen K, Hamblin M, Zhang J, Schwendeman SP, Chen YE. Vascular endothelial cell-specific microRNA-15a inhibits angiogenesis in hindlimb ischemia. J Biol Chem. 2012;287:27055–27064. doi: 10.1074/jbc.M112.364414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao Y, Su J, Guo W, Polich ED, Magyar DP, Xing Y, Li H, Smrt RD, Chang Q, Zhao X. Inhibition of miR-15a Promotes BDNF Expression and Rescues Dendritic Maturation Deficits in MeCP2-Deficient Neurons. Stem Cells. 2015;33:1618–1629. doi: 10.1002/stem.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertoli G, Cava C, Castiglioni I. MicroRNAs: New Biomarkers for Diagnosis, Prognosis, Therapy Prediction and Therapeutic Tools for Breast Cancer. Theranostics. 2015;5:1122–1143. doi: 10.7150/thno.11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 31.Cloonan N, Brown MK, Steptoe AL, Wani S, Chan WL, Forrest AR, Kolle G, Gabrielli B, Grimmond SM. The miR-17-5p microRNA is a key regulator of the G1/S phase cell cycle transition. Genome Biol. 2008;9:R127. doi: 10.1186/gb-2008-9-8-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan SW, Lee DY, Deng Z, Shatseva T, Jeyapalan Z, Du WW, Zhang Y, Xuan JW, Yee SP, Siragam V, Yang BB. MicroRNA MiR-17 retards tissue growth and represses fibronectin expression. Nat Cell Biol. 2009;11:1031–1038. doi: 10.1038/ncb1917. [DOI] [PubMed] [Google Scholar]
- 33.Hong P, Jiang M, Li H. Functional requirement of dicer1 and miR-17-5p in reactive astrocyte proliferation after spinal cord injury in the mouse. Glia. 2014;62:2044–2060. doi: 10.1002/glia.22725. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Xu L, Hu Q, Yang S, Zhang B, Jiang H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int J Cardiol. 2015;197:123–124. doi: 10.1016/j.ijcard.2015.06.037. [DOI] [PubMed] [Google Scholar]
- 35.Gao X, Zhang R, Qu X, Zhao M, Zhang S, Wu H, Jianyong L, Chen L. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk Res. 2012;36:1505–1509. doi: 10.1016/j.leukres.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 36.Jickling GC, Sharp FR. Blood biomarkers of ischemic stroke. Neurotherapeutics. 2011;8:349–360. doi: 10.1007/s13311-011-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


