Abstract
Objective: To investigate the relationship between Tiam1 gene expression and the invasion and metastasis of esophageal cancer. Methods: By RT-PCR technique, Tiam1 mRNA expression levels in 49 cases of esophageal cancer tissues and normal esophageal tissues were detected. Results: Average Tiam1 mRNA expression level in 49 cases of esophageal cancer tissues (1.83 ± 0.73) was significantly higher than that in normal esophageal tissues (0.87 ± 0.45) (P < 0.01); High Tiam1 mRNA expression rate in esophageal cancer tissues was positively correlated with clinical stage and T stage; Tiam1 mRNA expression rate was 59.38% (19/32) in patients with lymph node metastasis, and in patients without lymph node metastasis it was 23.53% (4/17), with statistically significant differences (P < 0.05). Conclusion: Tiam1 gene expression in esophageal cancer tissues was significantly higher than that in normal esophageal tissues, and its overexpression was positively correlated with invasion and metastasis of esophageal cancer.
Keywords: Esophageal cancer, Tiam1 gene, tumor invasion, tumor metastasis
Introduction
Esophageal cancer is one of the common gastrointestinal cancer in China [1,2]; lymph node metastasis is one of the most important biological characteristics, and it is also an independent prognostic factor of esophageal cancer [3]. Tiam1 gene (T lymphoma invasion and metastasis gene) is a gene isolated from mouse T lymphocytes, expressed in a variety of tumor cell lines [4]; high expression of Tiam1 can induce the proliferation and metastasis of tumor cells. But its role in the invasion and metastasis of human esophageal cancer is still unclear. This study used RT-PCR to detect Tiam1 mRNA expression in esophageal cancer, in order to explore its relationship with the invasion and metastasis of esophageal cancer.
Material and methods
Material
Samples were collected from resected fresh specimens from thoracic surgery department attached to our hospital between August 2004 and August 2005 hospitalized esophageal patients with a total of 49 cases and without preoperative radiation therapy and chemotherapy. Cancer tissue and normal esophageal tissue from 5 cm of lesions were taken and then they were stored in liquid nitrogen immediately. In 49 cases of esophageal cancer, 35 cases were male patients, and 14 cases were females, aging between 52 and 76 with mean age 65 ± 3.2; 32 cases were mid-esophageal cancer and 17 cases were lower-esophageal cancer; they were all underwent esophagectomy, regional lymph node dissection, and stomach substituting esophageal anastomosis. Intraoperative esophageal lesions were cut and then immediately took normal and esophageal tissue for cryopreservation. All the esophageal specimens were pathologically confirmed squamous cell carcinoma. Lymph node was used to identify pathological metastasis.
Immunohistochemistry (IHC) staining of Tiam1 gene
Expression of the Tiam1 gene was investigated by streptavidin-biotin-peroxidase complex method according to a previouspublication [5].
Extraction and identification of total RNA
1 ml of Trizol reagent (Invitrogen products) was added to 100 mg frozen tissue in a homogenized ice bath. The total RNA was extracted according to the operating instructions. A260 value was measured by using double beam spectrophotometer. A260/280 ratio was measured to detect its purity. 1.98% denaturing gel electrophoresis was used for analysis the integrity of total RNA.
cDNA preparation
Reverse transcription kit (RevertAid TM Fermentas) was used. Proceed as follows: 2.0 μg of total RNA was collected, and 1 μl random primer was added; then add water to 12 μl. 5 × buffer 4 μl, RNA inhibitors 1 μl and 10 mm old NTP 2 μl were added, 25°C water bathing for 5 min; reverse transcriptase 1 μl was added, water bathing at 25°C for 101 1 min, at 42°C for 60 min, and then at 70°C for 10 min to stop the reaction. Ice bath for spare.
PCR primer sequences
Tiam1 primer: F1: 5’-AAGACGTACTCAGGCCATGTCC-3’, F2: 5’-GACCCAAATGTCGCAGTCAG-3’. Internal reference β2-microglobulin (β2 M) primer: F1: 5’-ACCCCCACTGAAAAAGATGA-3’, F2: 5’-ATCTTCAAACCTCCATGATG-3’. Both were synthesized by Shanghai Sangon Company (Shanghai, China).
Reaction system PCR amplification kit (SK2492, Shanghai Sangon Company) was used. The reaction system was 50 μl, including 2 mmol DNTP 5 μl, 10 × PCR buffer 5 μl, 10 μmol/L Tiam1 gene primers (upstream and downstream) each 18 μl, internal reference β2 M primer each 1 μl, 25 mmol/L MgCl2 5 μl, 5 U/μl Taq enzyme 016 μl, template DNA 5 μl, double distilled water 25.8 μl. Circulation was performed on MJ PTC2220 PCR instrument. Tiam1 gene and internal reference β2 M gene were amplified simultaneously: 94°C denaturation for 45 s, 55°C annealing for 1 min, 72°C extension for 1 min 30 s, 72°C extension for 5 min in the last cycle.
Optimal conditions
Optimal PCR cycle number for Tiam1 gene and optimal amount of total RNA template for reverse transcription were determined by one case of laryngeal carcinoma specimen; 1.5, 2.0 and 2.5 μl of total RNA template is subjected to reverse transcription, and 5 μl of each product was collected for PCR respectively with 29, 32, 35, 38 and 41 cycles; absorbance values of products were analyzed to determine the optimal PCR conditions for simultaneous amplification of Tiam1 and the internal reference β2 M gene.
Semi-quantitative PCR
5 μl PCR product was added in 23 g/L agarose gel containing ethidium bromide for electrophoresis, and the results were observed under ultraviolet light. The amplified fragments of Tiam1 and β2 M were respectively 253 and 118 bp. A value in each band was measured with automatic UVP gel image analysis system. The ratios of A values of Tiam1 and β2 M were the relative Tima1 mRNA levels of samples. Detection of each specimen was repeated three times, and the mean was calculated.
Statistical analysis
SPSS 17.0 software was used for analysis. Measurement data using t test, and count data using chi-square test. Logistic regression analysis was performed to identify the prognostic factor of esophageal cancer. A P < 0.05 was considered significant.
Results
The expression of Tiam1 protein in esophageal cancer tissues
Tiam1 protein expression was investigated in these esophageal cancer samples by immunohistochemistry method. In esophageal cancer tissues exhibited positive Tiam1 staining. In contrast the normal esophageal tissues displayed negative Tiam1 staining (Figure 1).
Figure 1.
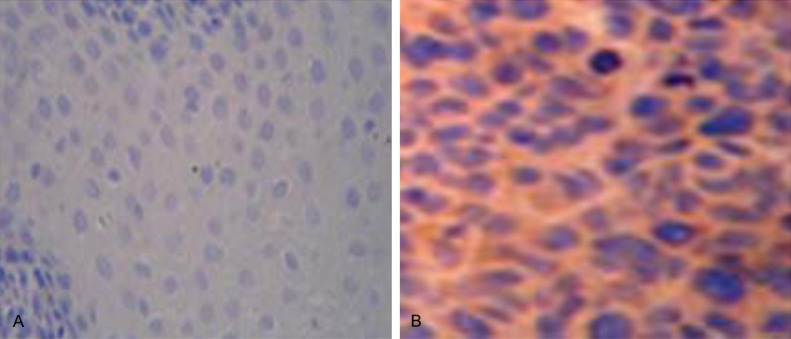
Immunohistochemical staining of Tiam1 protein in esophageal cancer and normal esophageal tissues. A. No Tiam1 protein staining was presentin the normal esophageal tissues. B. Stronger positive Tiam1 immunostaining in esophageal cancer tissues.
Optimization of semi-quantitative RT-PCR
Between the cycle numbers of 32 and 38, Tiam1 gene PCR product amount was linearly related with the cycle numbers. Increase of the cycle number could reduce the linear correlation; reducing the number of cycles would reduce the sensitivity of the PCR reaction. This linear relationship was observed when the amount of total RNA template was 2.0, 1.5 and 1.0 μg, and the PCR linear relationship of the cDNA prepared by 1.5 μg template was the best. In this experiment, 1.5 μg of total RNA template and 35 cycles were used to determine Tiam1 gene expression. Simultaneously, amplification of the internal reference β2-microglobulin was performed.
Tiam1 mRNA expression
Tiam1 mRNA was expressed in varying degrees in esophageal cancer and esophageal normal tissues. Electrophoresis maps of RT-PCR products were shown in Figure 2. Sequencing results confirmed that RT-PCR product was Tiam1 gene, and the base pairing accuracy was 97.5%.
Figure 2.
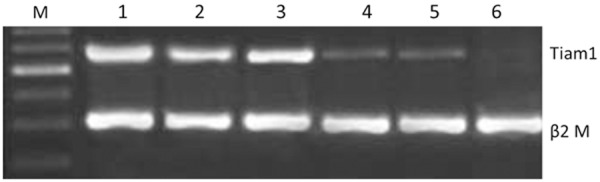
Relative expression analysis of Tiam1 mRNA in ESCC tissues and theadjacent normal esophageal tissues. M: DNA marker; 1-3: ESCC; 4-6: normal esophageal tissues.
Between the two groups of Esophageal carcinoma tissues and normal esophageal tissues, no statistically significant differences had been found in age and gender (P > 0.05); average Tiam1 mRNA expression levels in esophageal carcinomna was 1.83 ± 0.73, significantly higher than that in normal esophageal tissues (0.87 ± 0.45; P < 0.01).
Relationship between Tiam1 mRNA expression in esophageal carcinoma and clinical stages and T stages and N grades
The critical value of Tiam1 mRNA expression rate was based on the relative average A value (1.83) of esophageal Mta1RT PCR products, A > 1.83 for high expression and A < 1.83 for low expression. No statistically significant differences had been found in age and gender between two groups of Tiam1 mRNA highly-expressed and lowly-expressed in esophageal tissues (all P > 0.05); In various clinical stages, high Tiam1 mRNA expression respectively accounted for 18.2%, 43.2%, and 71.3% in I~II stages, III stage and IV stage, and the differences between the clinical stages were statistically significant (all P < 0. 05); and high expression rates increased with the increase of clinical stage (P < 0.05); in T staging, high Tiam1 mRNA expression rate respectively accounted for 0%, 46.7%, 58.3%, and 83.8% in T1, T2, T3 and T4; the difference was statistically significant between T stages (P < 0.05), and the high expression rate increased with T stage increasing (P < 0.05); high Tiam1 mRNA expression rate in patients with lymph node metastasis accounted for 59.3%, significantly higher than that in patients without lymph node metastasis, accounting for 21.4% (P < 0.05) (Table 1).
Table 1.
Relationship between Tiam1 mRNA expression and clinical outcomes
| Clinical indicators | Cases | Relative A values of RT-PCR products | P value | |
|---|---|---|---|---|
|
|
||||
| Highly-expressed | Lowly-expressed | |||
| Age (Years) | 49 | 62 ± 5.8 | 63 ± 5.6 | 0.183 |
| Gender | ||||
| Male | 28 | 12 | 16 | 0.572 |
| Female | 21 | 11 | 10 | |
| TNM | ||||
| 1~2 | 14 | 2 | 12 | 0.010 |
| 3 | 19 | 10 | 9 | |
| 4 | 16 | 11 | 5 | |
| T stages | ||||
| T1 | 8 | 1 | 7 | 0.025 |
| T2 | 20 | 9 | 11 | |
| T3 | 18 | 11 | 7 | |
| T4 | 3 | 2 | 1 | |
| Lymph node metastasis | ||||
| Yes | 17 | 4 | 13 | 0.017 |
| No | 32 | 19 | 13 | |
Logistic regression analysis
All subjects in the esophageal cancer group and the control group were analyzed as a single group. The stepwise logistic regression analysis was performed with multiple variables. The results showed that the Tiam1 gene expressionwas an independent risk factor of poor prognosis of esophageal cancer. The detailed data are shown in Table 2.
Table 2.
Logistic regression of the relation between Tiam1 mRNA expression and clinical outcomes
| Parameters | OR | 95% CI | P value |
|---|---|---|---|
| Tiam1 expression | 3.11 | 1.920~5.114 | 0.016 |
| Lymphnode metastasis | 2.133 | 1.104~3.254 | 0.011 |
| TNM | 4.231 | 2.012~8.443 | 0.022 |
Discussion
Tiam1 gene is a gene isolated from mouse T lymphoma cells by Habets et al [6], which is closely associated with tumor invasion and metastasis. The whole name of Tiam1 gene is cell lymphoma invasion and metastasis-inducing protein 1, which is a ubiquitousconversion factor for diphosphate guanine nucleotide (GDP) and triphosphate guanine nucleotide (GTP) [7]. It is mainly expressed in brain and testis and not or lowly expressed in other normal tissues. It belongs to Rho triphosphate guanine nucleotide hydrolase (GTPases) family [8]. Tiam1 is located in 21q22.1, encoding a protein containing 1591 amino acids and one DH homologous functional area involved in GDP-GTP exchange; Tiam1 gene is highly expressed in tumor cells of different tissue origins. Tiam1 genecauses tumor invasion and metastasis by Tiam1-Rac signaling pathway, regulating the transcription factor c-myc and LPA (lysophosphatidic acid), and affecting adhesion molecules. It has been currently reported that Tiam1 gene was closely related with the invasion and metastasis of renal cell carcinoma, breast cancer and colorectal cancer [9-11].
Engerset al [12] found that Tiam1 protein was not or lowly expressed in renal tubular epithelial-derived renal clear cell carcinoma, which was consistent with the biological behaviors and clinical characteristics of the tumor, such as low growth and rare metastasis. Malignant tumor invasion and metastasis are caused by a series of complex and multi-step interactions between tumor cells and host cells and interstitial. This process involves the roles of a variety of genes and their products. Tiam1 Tiam1 gene is a gene isolated from mouse T lymphoma cells by Habets et al [13]; it is a ubiquitous conversion factor for diphosphate guanine nucleotide (GDP) and triphosphate guanine nucleotide (GTP), belonging to Rho-GTPases family. Tiam1 gene is located in the q22 band of chromosome 21 and the centromeric end of Aml 21 gene, containing two exons separated by one intron; two exons were about 7.3 kb, and the intron was 14 kb. This gene encodes a protein with a molecular weight of 177,000, consisting of 1591 amino acid residues, with DH and PH homologous functional regions. DH homologous functional area is involved in GDP-GTP conversion to promote the release of GDP and the binding of GTP. PH homologous functional region not only is involved in subcellular localization, but also directly regulates the nucleotide exchange in DH region. Meanwhile, Tiam1 is a kind of GDP isolated stimulator (GDS) and a activity-regulating factor of Rac and Rho, affecting cytoskeleton, cell adhesion and movement by participating in Tiam1 Rac information transfer pathways, and playing an important role in tumor development, invasion and metastasis [3,4]. Therefore, Tiam1 disorder in this pathway may lead to oncogenic transformation, tumor progression and metastasis.
Human Tiam1 is mainly expressed in normal brain and testis tissues, but not or lowly expressed in other normal tissues; it is highly expressed in tumor cells of different tissue origins [13]. Engers et al [12] found that Tiam1 protein was not or lowly expressed in renal tubular epithelial-derived renal clear cell carcinoma, which was consistent with the biological behaviors and clinical characteristics of the tumor, such as low growth and rare metastasis. A locus mutation of Tiam1 plays an important role in the progression of human renal cell carcinoma. Adam L et al detected the Tiam1 expression levels in breast tumor cells and found that Tiam1 interacting with the cytoskeletal protein and ankyrin resulted inoncogenic signal transduction and metastasis of breast tumor. Sun Qing et al detected the Tiam1 gene expression in metastatic colorectal cancer cell lines and tissue samples; semi-quantitative RT-PCR showed that, Tiam1 in metastatic colorectal carcinoma cell line showed a positive rate of 100%; Northern dot hybridization and situ hybridization indicated that Tiam1 gene was highly expressed in metastatic cell lines and invasive and metastatic cancer.
The role of Tiam1 gene in human esophageal cancer invasion and metastasis is rarely reported at home and abroad. This study focused on Tiam1, the key gene in tumor invasion and metastasis, used RT-PCR technique to detect Tiam1 gene expression at the transcriptional level and investigated its relationship with the invasion and metastasis of esophageal cancer; the results showed that the average level of Tiam1 mRNA in esophageal cancer tissues was significantly higher than that in normal esophageal tissues (P < 0.05), prompting that Tiam1 gene may provide a reference for the discrimination of esophageal cancer and normal tissues; this study also found that the expression rates of Tiam1 mRNA in esophageal carcinoma significantly increased with the increase of clinical stage and grade (P < 0.05); Tiam1 mRNA expression rates in esophageal carcinoma with lymph node metastasis was significantly higher than those in esophageal carcinoma without lymph node metastasis (P < 0.05). High expression rates to some extent reflected the local invasion and distant metastasis of esophageal cancer, providing potential information for comprehensive treatment of esophageal cancer. Studies have shown that, Tiam1 gene may play an important role in the invasion and metastasis of esophageal cancer. Presumably inhibiting the expression of Tiam1 gene by antisense technology and cutting off TiamlRac information transfer pathway may down-regulate the invasiveness of esophageal cancer cells and inhibit the invasion and metastasis of esophageal cancer.
Molecular mechanism of Tiam1 gene in tumor metastasis is not entirely clear, but the results of this study show that, Tiam1 gene was related with the invasion and metastasis of esophageal cancer; high expression rate of Tiam1 mRNA in esophageal cancer tissues can be used as an indicator of potential esophageal malignancy, and provide potential information to guide the treatment of esophageal cancer.
Disclosure of conflict of interest
None.
References
- 1.Tian X, Zhou JG, Zeng Z, Shuai T, Yi LJ, Ma L, Wang Y, Cao H, Song GM. Cetuximab in patients with esophageal cancer: a systematic review and meta-analysis of randomized controlled trials. Med Oncol. 2015;32:127. doi: 10.1007/s12032-015-0521-2. [DOI] [PubMed] [Google Scholar]
- 2.Zhang SK, Guo LW, Chen Q, Zhang M, Liu SZ, Quan PL, Lu JB, Sun XB. Prevalence of human papillomavirus 16 in esophageal cancer among the Chinese population: a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2014;15:10143–9. doi: 10.7314/apjcp.2014.15.23.10143. [DOI] [PubMed] [Google Scholar]
- 3.Xie S, Huang J, Kang G, Fan G, Wang W. Surgical treatment of synchronous gastric and esophageal carcinoma: case report and review of literature. Thorac Cardiovasc Surg Rep. 2013;2:35–7. doi: 10.1055/s-0033-1351357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu H, Jiang L, Liu X. Effect of downregulation of Tiam1 by siRNA on esophageal squamous cell carcinoma EC9706 cells. Zhonghua Zhong Liu Za Zhi. 2014;36:250–6. [PubMed] [Google Scholar]
- 5.Liu HT, Wang N, Wang X, Li SL. Overexpression of Pim-1 is associated with poor prognosis in patients with esophagealsquamous cell carcinoma. J Surg Oncol. 2010;102:683–688. doi: 10.1002/jso.21627. [DOI] [PubMed] [Google Scholar]
- 6.Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, Collard JG. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–49. doi: 10.1016/0092-8674(94)90216-x. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Lu L, Liu C, Shan L, Yuan D. MicroRNA-377 Suppresses Cell Proliferation and Invasion by Inhibiting TIAM1 Expression in Hepatocellular Carcinoma. PLoS One. 2015;10:e0117714. doi: 10.1371/journal.pone.0117714. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Wang B, Li W, Liu H, Yang L, Liao Q, Cui S, Wang H, Zhao L. miR-29b suppresses tumor growth and metastasis in colorectal cancer via downregulating Tiam1 expression and inhibiting epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1335. doi: 10.1038/cddis.2014.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao L, Liu Y, Sun X, He M, Ding Y. Overexpression of T lymphoma invasion and metastasis 1 predict renal cell carcinoma metastasis and overall patient survival. J Cancer Res Clin Oncol. 2011;137:393–8. doi: 10.1007/s00432-010-0895-7. [DOI] [PubMed] [Google Scholar]
- 10.Wu YQ, Xie YY, Peng G. Effects of Tiam1 on invasion and metastasis of breast carcinoma and its mechanisms. Zhonghua Zhong Liu Za Zhi. 2012;34:831–4. doi: 10.3760/cma.j.issn.0253-3766.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Yu LN, Zhang QL, Li X, Hua X, Cui YM, Zhang NJ, Liao WT, Ding YQ. Tiam1 transgenic mice display increased tumor invasive and metastatic potential of colorectal cancer after 1,2-dimethylhydrazine treatment. PLoS One. 2013;8:e73077. doi: 10.1371/journal.pone.0073077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engers R, Zwaka TP, Gohr L, Weber A, Gerharz CD, Gabbert HE. Tiam1 mutations in human renal-cell carcinomas. Int J Cancer. 2000;88:369–76. doi: 10.1002/1097-0215(20001101)88:3<369::aid-ijc8>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 13.Habets GG, van der Kammen RA, Stam JC, Michiels F, Collard JG. Sequence of the human invasion-inducing TIAM1 gene, its conservation in evolution and its expression in tumor cell lines of different tissue origin. Oncogene. 1995;10:1371–6. [PubMed] [Google Scholar]


