Abstract
This paper describes a framework for learning a statistical model of non-rigid deformations induced by interventional procedures. We make use of this learned model to perform constrained non-rigid registration of pre-procedural and post-procedural imaging. We demonstrate results applying this framework to non-rigidly register post-surgical computed tomography (CT) brain images to pre-surgical magnetic resonance images (MRIs) of epilepsy patients who had intra-cranial electroencephalography electrodes surgically implanted. Deformations caused by this surgical procedure, imaging artifacts caused by the electrodes, and the use of multi-modal imaging data make non-rigid registration challenging. Our results show that the use of our proposed framework to constrain the non-rigid registration process results in significantly improved and more robust registration performance compared to using standard rigid and non-rigid registration methods.
Keywords: Non-rigid registration, Statistical deformation model, Surgical navigation, Magnetic resonance imaging (MRI), Computed tomography (CT), Epilepsy
Graphical abstract
Highlights
-
•
Accurate localization of implanted intra-cranial electroencephalography electrodes in epilepsy patients
-
•
A framework for training a statistical model of intra-subject non-rigid deformations induced by surgical procedures
-
•
The statistical deformation model captures the main modes of deformation from clinical pre-op and post-op training images
-
•
Apply our learned statistical deformation model to directly co-register post-operative CT to pre-operative MRI
-
•
Significant improvement in registration performance, with increased robustness and reduced worst-case performance
1. Introduction
Non-rigid image registration of longitudinal images acquired before and after (or during) an interventional procedure is a challenging task. While a large number of image registration algorithms have been proposed to estimate the transformation that best aligns two or more images (Sotiras et al., 2013), registering pre-procedural imaging with intra- or post-procedural imaging is particularly difficult due to a number of factors that may include: (i) procedure-induced deformation; (ii) missing structural correspondences; or (iii) highly non-linear intensity relationships, especially when using multi-modal imaging. These factors, along with the estimated deformation's potentially large number of degrees of freedom, makes finding the globally optimal transformation a difficult, ill-posed problem. In this paper, we present a framework to train a statistical deformation model (SDM) to capture intra-subject anatomical deformations that result from surgical interventions in epilepsy patients.
Epilepsy is a neurological disorder in which brain function is temporarily disturbed by electrophysiologic events, or seizures. For certain patients diagnosed with epilepsy who do not favorably respond to medication, surgical resection of ictal tissue, i.e. tissue involved in the generation of the seizure, is an effective method to reduce or to eliminate seizure activity (Cascino, 2004). The current “gold-standard” for localizing the focus of seizure activity involves intra-cranial electroencephalography (iEEG), in which surgeons perform a craniotomy and implant electrodes throughout the brain (Spencer et al., 1998). Following implantation, and after constant monitoring of the electrodes for several days, clinicians must determine if the localized ictal tissue corresponds to functionally eloquent areas of the brain, e.g. motor, sensory, and language regions. Pre-operative imaging, which may include magnetic resonance imaging (MRI) and functional imaging modalities such as functional MRI (fMRI), positron emission tomography (PET), and single-photon emission computed tomography (SPECT), can help identify these functionally eloquent regions of brain tissue and guide electrode placement (Nowell et al., 2015). Therefore, accurate spatial registration of electrodes with respect to these pre-implantation images is critical for planning the surgical resection, if feasible.
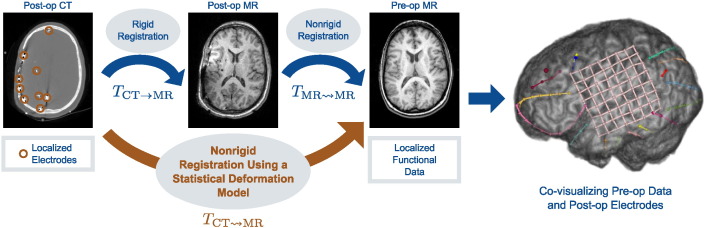
A variety of methods have been proposed to localize the implanted iEEG electrodes within the pre-implantation imaging data. Using intra-operative digital photography to identify implanted electrodes, the electrodes may then be manually localized with the pre-op MRI brain surface (Wellmer et al., 2002, Dalal et al., 2008). Alternatively, a post-implantation computed tomography (CT) image can be acquired in which the electrodes can easily be identified (see Fig. 1). Surface electrodes identified in the post-op CT can then be projected to the nearest point on the pre-op MRI brain surface (Hermes et al., 2010), however, this projection is done with respect to rigid registration and does not accurately account for non-rigid tissue deformations nor does it allow for localization of sub-dural electrodes. These sub-dural electrode displacements can be estimated, for example, using a kernel-based averaging of the surface electrode displacements (Taimouri et al., 2014). A post-implant anatomical MRI may also be acquired, but the electrodes cause significant imaging artifacts and distortions (see Fig. 1). While it is straightforward to visualize iEEG electrodes within post-implant MRIs by co-registration with the post-op CT using rigid intensity-based registration methods (Azarion et al., 2014), we ultimately want to visualize the electrodes with respect to each patient's pre-implant imaging in order to integrate this multi-modal imaging information into the surgical plan. At our institution, a trained technologist performs a series of multi-modal registrations using both the post-implant anatomical MRI and the post-implant CT images. Initially, a simple intensity thresholding algorithm automatically segments the electrodes in the post-op CT image. A technologist then inter-actively labels each electrode. The electrodes are subsequently projected to the pre-op MRI space by first rigidly registering the CT image to the post-op MRI (Wells et al., 1996) and then non-rigidly registering the post-op MRI to the pre-op MRI to account for post-surgical deformations (Studholme et al., 2001). Once the electrodes have been transformed to the pre-op imaging space, they may be co-visualized with any other functional image studies.
Fig. 1.
To co-visualize post-implant iEEG electrodes with pre-implant imaging data, as shown on the right, the current clinical registration framework (in blue) rigidly registers the post-op CT image and the segmented electrodes to a post-op MRI, and then non-rigidly registers the post-op MRI to the pre-op MRI. Our proposed method (in orange) directly registers the post-op CT image to the pre-op MRI using a statistical deformation model that captures the non-rigid deformations induced by the surgical procedure.
The post-op MRI, in this case, serves as a registration link between the post-op CT and pre-op MRIs. The non-rigid registration of pre- and post-op MRIs accounts for brain deformations caused by the craniotomy and implantation of electrodes. By combining this transformation with the rigid post-op MR-CT registration, the CT image is non-rigidly warped into MRI space. Without this intermediate step, the non-rigid registration of the CT to pre-op MRI would not be accurate, especially with respect to sub-cortical structures. CT's poor soft tissue contrast lacks salient structural information to account for non-rigid brain deformation. Furthermore, missing anatomical correspondences, such as the removal of the skull during surgery, as well as both imaging artifacts and intensity inhomogeneities caused by the presence of the electrodes, exacerbate the problem of registering post-op images to pre-op ones. Nevertheless, while the post-op MRI does provide a crucial link between the post-operative CT and pre-op MRI spaces, it offers little novel, clinical information relevant to the surgery.
In this work, we propose to obviate the use of the post-op MR image by learning the nonrigid deformation from post-op CT to pre-op MR. Fig. 1 illustrates the proposed method. Phasing out the intermediate post-op MR image could potentially reduce patient discomfort by eliminating one trip in and out of the MR scanner with the attached iEEG electrodes, reduce the opportunity for infection incurred by moving the patient to and from different locations in the hospital, and remove even potential minor risks to the patient such as the MR scanner causing electrode induction heating (Bhavaraju et al., 2002). In addition, eliminating one scan would provide a cost savings to the patient.
The proposed method leverages the mono-modality post-op MRI to pre-op MRI non-rigid registrations used in clinical practice to learn a statistical deformation model (SDM). While brain deformation may be caused by a variety of factors, such as gravity, tissue swelling, loss of cerebrospinal fluid (CSF), pharmacological response, surgical manipulation, and breathing (Nabavi et al., 2001, Dumpuri et al., 2007), our SDM is surgical-site dependent because larger brain deformations generally occur ipsilateral to the craniotomy site (Hartkens et al., 2003). We also assume that patients with craniotomies in similar locations will experience similar deformation characteristics. We perform a principal component analysis (PCA) of non-rigid MRI deformations to construct our SDM. Our SDM models intra-subject deformations that result from surgical intervention, and not inter-subject anatomical variability. To the best of our knowledge, training an SDM on a mono-modality registration task and using that SDM to perform a multi-modality registration is a novel application of SDMs. Using such high-quality training data is effective for training a PCA-based SDM (Onofrey et al., 2015). By doing so, the SDM can model sub-cortical deformations in the CT image that would otherwise not be possible with an intensity-only registration to the pre-op MRI. We present results showing the SDM significantly reduces registration error compared to rigid and non-rigid intensity-only MR-CT registrations.
This paper expands upon a previous conference paper (Onofrey et al., 2013). Sec. 2 begins with a review of prior work using statistical deformation models for deformable image registration. Sec. 3 then describes our proposed method to learn a model of procedure-induced non-rigid deformation and how we use that statistical deformation model to non-rigidly register pre-implantation MRI and post-implantation CT images. We present results in Sec. 4 and summarize the contributions of this paper in Sec. 5.
2. Background
2.1. Statistical deformation models
Parametric shape models attempt to model the variation of shape within a particular class of objects (Heimann and Meinzer, 2009). Point distribution models (PDMs) may in turn be used to constrain realizations of that shape to be from the distribution of plausible solutions defined by the probabilistic model. In particular, active shape models use a principal component analysis (PCA) of this point distribution to constrain the segmentation of shapes from images (Cootes et al., 1995). However, manual identification of corresponding landmark points for PDMs is a difficult, time-consuming task. While hierarchical models of shape have been proposed (Xue et al., 2006, Cerrolaza et al., 2012), PDMs do not easily allow modeling of more than one object. Statistical models of deformation, rather than shape, avoid the need for explicit segmentation of the object of interest, and instead model how the object or objects of interest deform according to a dense deformation field.
Statistical deformation models (SDMs) of dense deformation fields, which originated from work on computational anatomy (Grenander and Miller, 1998), statistically analyze the dense deformations between corresponding anatomical structures across a sample population. A PCA of the dense deformation fields can provide an orthonormal basis of dense non-rigid deformation (Joshi et al., 1997, Gee and Bajcsy, 1999, Cootes et al., 2004). Alternatives to PCA-based analysis and modeling of non-rigid deformation have also been proposed, which include a statistical manifold framework (Twining and Marsland, 2008) and partial least-squares regression (Singh et al., 2010). Parameterizing a high-dimensional, dense displacement field with a lower-dimensional free-form deformation (FFD) model (Rueckert et al., 1999) and then performing a PCA of the FFD's B-spline control point displacements (Rueckert et al., 2003) is attractive because we can control the dimensionality of the transformation's representation from the start, and the resulting SDM is potentially smoother and easier to optimize.
PCA-based SDMs can also be used to drive the image registration process itself (Rueckert et al., 2003). This method performs low-dimensional non-rigid registration by optimizing the linear combination of the SDM's eigenvectors to maximize an objective function measuring image similarity. This method has been used to model and register chest radiographs (Loeckx et al., 2003) and brain images (Wouters et al., 2006, Kim et al., 2008). The SDM may also serve as a regularization prior for high-dimensional non-rigid registrations to penalize deformations that differ from the statistical model (Xue et al., 2006, Berendsen et al., 2013). Given high-quality training data or a sufficient number of training samples, PCA is effective for learning an SDM of anatomical brain variation (Onofrey et al., 2015).
While PCA-based SDMs have difficulty modeling high-dimensional deformations (Onofrey et al., 2015), we hypothesize that the post-surgical brain deformation is of low enough dimension that an SDM can capture the gross deformations observed after surgery. Other authors have made use of PCA SDMs to register mono-modality medical images, primarily for the task of inter-subject registration (Rueckert et al., 2003, Xue et al., 2006, Kim et al., 2008) and for intra-subject motion compensation (Loeckx et al., 2003, He et al., 2010). Ehrhardt et al. (2011) learned models of intra-subject respiratory motion and co-registered these models to each other to create an inter-patient model of respiration. All of these works use their respective deformation models for mono-modality image registration. While Hu et al. (2012) perform multi-modal fusion of MR and ultrasound imaging for prostate biopsy, they utilize a statistical shape model to align a surface extracted from the MR space to image features in the ultrasound, which contrasts with our method to perform image-to-image non-rigid registration constrained by the SDM. We use our SDM, which was trained on mono-modal registration (pre-op MRI and post-op MRI) for multi-modal, intra-subject registration of each patient's post-op CT image to their pre-op MRI.
2.2. Manifold-learning for non-rigid registration
Manifold learning techniques differ from linear PCA in that they seek to find a non-linear mapping of high-dimensional data to a low-dimensional space. The Isomap algorithm (Tenenbaum et al., 2000), for example, uses the k-nearest neighbors (k-NN) of the training data to construct a manifold. This method has been applied within the medical imaging field to learn the manifolds of non-rigid image deformation (Gerber et al., 2010, Hamm et al., 2010, Jia et al., 2010, Ye et al., 2012). Here, the k-NN of the training data define the manifold, and manifold-based registration successively registers images that are most similar to each other along the manifold. However, in contrast to linear PCA, it is difficult for manifolds to function as generative models, e.g. construct novel transformations on the space defined by the manifold that could be used to register unseen images.
2.3. Biomechanical models for non-rigid brain registration
Incorporating biomechanical computational models into the image registration process can constrain the non-rigid deformation to be physically realistic. Such models have been used to track procedure-induced deformations over time in brain imaging (Hagemann et al., 1999, Paulsen et al., 1999, Ferrant et al., 2001, Sˇkrinjar et al., 2002, Wittek et al., 2007, DeLorenzo et al., 2012). These often, however, require tuned parameters to best approximate the elastic tissue properties of the brain. In contrast, our proposed method creates a statistical model of brain deformation that learns the procedure-induced tissue deformations directly from actual clinical data. In this paper, we learn a model of the deformation by computing the statistical distribution of the displacements of B-spline control points of a free-form deformation model. The inverse of the covariance matrix of this distribution is analogous to the stiffness tensor of a linear elastic model (Papademetris, 2000). We use the eigenvectors of this matrix to constrain a model-based deformation along preferential directions. In this sense, our method implicitly learns the biomechanical properties of the brain from observed clinical training data.
In order to account for variability in the sources of brain deformation within biomechanical models, Dumpuri et al., 2007, Dumpuri et al., 2010 create a series of deformations that includes gravity-induced brain shift, pharmacological-induced volume changes, and tissue swelling due to edema to create what they term an atlas of deformations. These model deformations act as training samples for an inverse model, which when combined with sparse intra-operative data, can compensate for intra-operative brain shift. Whereas Dumpuri et al.'s method generates intra-subject synthetic biomechanical model-based deformation samples for training, our method uses actual inter-subject deformation samples observed from clinical cases as training samples. A point of dissimilarity is that, in our problem setup, we have a volumetric image data which allow us to use image similarity as a driving force instead of the sparse measurements used in that work which in some sense aims to address the more challenging problem of predicting deformation during the procedure in the absence of volumetric intraoperative imaging.
3. Methods
3.1. Training the statistical deformation model
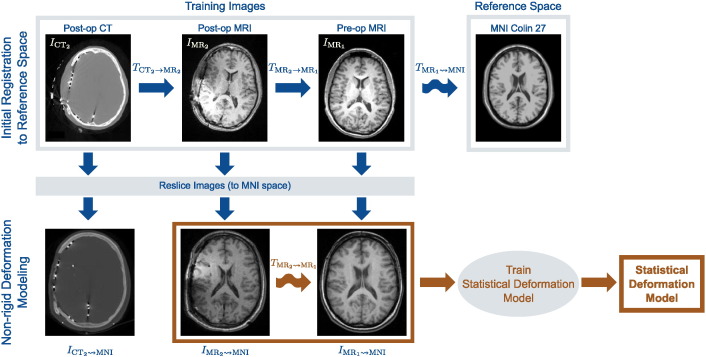
Given a database of N surgical epilepsy patients with craniotomies at similar locations and size, we train a statistical deformation model (SDM) to capture the non-rigid deformation of the brain due to implantation of iEEG electrodes. Fig. 2 illustrates our procedure for training this SDM. Each patient's dataset consists of a pre-implantation MR image , a post-implantation MR image , and a post-implantation CT image , where Imt denotes pre-op images acquired at time t = 1 and post-op images at time t = 2 for imaging modality m = {MR, CT}. To create the SDM, we transform all images from each of the N patients into a common reference space. For each patient, we perform the following: (i) rigidly register the post-op MR and CT images by maximizing their normalized mutual information (NMI) (Studholme et al., 1999), and obtain the transformation ; (ii) rigidly register the pre- and post-op MR images using NMI to produce the transformation (we use i → j to denote linear transformations from space i to space j); and (iii) non-rigidly register the pre-op MR images to the MNI Colin 27 brain, IMNI, using a free-form deformation (FFD) (Rueckert et al., 1999) and maximizing NMI, and write this transformation (we use i ⇝ j to denote non-rigid transformations from space i to space j). Once we compute all transformations, we reslice all images from each of the N patients into MNI space (181 × 217 × 181 volume with 1 mm3 resolution) by concatenating the transformations:
| (1) |
where ∘ is the transformation operator.
Fig. 2.
Our proposed framework to model intra-subject non-rigid deformations caused by surgical intervention. All training images are first resliced to a common reference space, e.g. MNI Colin 27 image space. We rigidly register the post-op and pre-op training images to each other, and then non-rigidly register the pre-op MR training image to the reference image. After reslicing all training images to this common reference, we non-rigidly register the post-op MR image to the pre-op MR image to account for interventional deformations. We then use these transformations, , to learn a statistical deformation model of non-rigid deformation. Here, i → j and indicates rigid and i ⇝ j indicates non-rigid transformation from image space i to space j.
The non-rigid registration of the pre-op MR image to the MNI brain in (1) spatially normalizes each patient's set of images to MNI space. However, non-rigid deformation between the post-op and pre-op imaging remains (in MNI space) as the post-op MR has yet to be non-rigidly registered to the pre-op MR. In clinical practice, this registration task uses skull-stripped brains to mitigate the effects of missing anatomical correspondences after surgery. However, since we plan to register the CT images directly to the pre-op MR, the skull is actually one of the most informative structures to register, even if it is lacking some correspondence. Therefore, to accurately register the MR images containing the skull, we first create a brain surface mask (Smith, 2002) in both the pre-op and post-op images. If the brain segmentation is inaccurate, a trained technician can manually correct the segmentation using a voxel-wise painting tool (Joshi et al., 2011). We then utilize an integrated intensity and point-feature registration algorithm (Papademetris et al., 2004) to register the two MR images. This algorithm uses an FFD transformation model and maximizes the NMI similarity metric and proceeds in a multi-resolution manner. We parameterize this FFD with a relatively large B-spline control point spacing, with a final isotropic grid resolution of 15 mm, because we seek a gross alignment of the head to this reference image rather than accurate, high-dimensional, non-rigid inter-subject registration with much lower grid spacing. With the point-weighting parameter set to 0.1, the brain surface points constrain the algorithm to align the cortical surface that otherwise has difficulty being accurately registered using intensity registration by itself. We denote the resulting transformations . It is these transformations that we use to train the SDM.
For each patient i = 1,…,N, we rewrite the transformation as a column vector of P concatenated FFD control point displacements in 3D, di ∈ R3P. We use a principal component analysis (PCA) to linearly approximate the deformation distribution (Rueckert et al., 2003).
| (2) |
where is the mean deformation of the N training registrations, Φ = (φ1,…,φK)i = 1 ∈∈ R3P × K is the matrix of K = min{N,3P} orthonormal deformation displacement vectors, and w ∈∈ RK is the vector of model variation coefficients. We sort the eigenvectors φk according to their corresponding eigen-values in decreasing order λ1≥. . . ≥ λk≥. . . ≥ λK The eigenvectors with the k largest eigenvalues define an SDM using k principal modes of variation, with 1 ≤ k ≤ K.
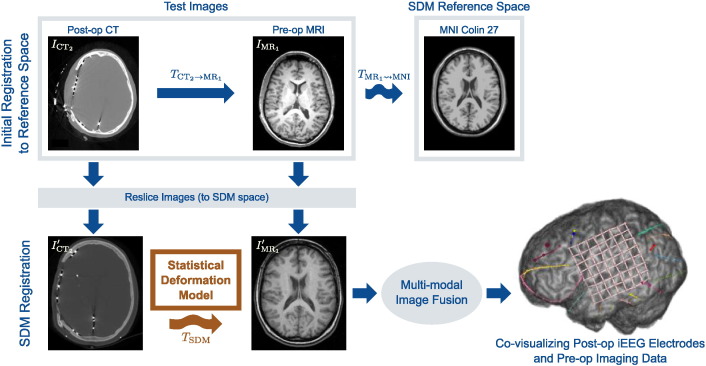
3.2. Non-rigid SDM MR-CT registration
Given a previously unseen pre-op MR and post-op CT image pair for a new patient, we use the SDM from Sec. 3.1 to non-rigidly register the post-op CT image to the pre-op MR without the use of a post-op MR image, as shown in Fig. 3. Before we non-rigidly register the two images, we must first transform the images into the SDM's reference space (MNI space) in which we define our model of deformation. We do so by rigidly registering the post-op CT, , to the pre-op MR, , and then non-rigidly registering to the MNI brain, IMNI. In both cases, we register the images by maximizing the NMI similarity metric. The non-rigid registration uses a FFD transformation with the same B-spline control point spacing (15 mm isotropic grid) used to register the pre-op MR images in Sec. 3.1. We use the resulting transformations to reslice both images into MNI space such that
| (3) |
where ∘ is the transformation operator and It indicates the resliced image.
Fig. 3.
Multi-modal, non-rigid registration using a statistical deformation model (SDM) of intervention-induced deformation. In order to apply the SDM for registration, we first transform the original images to the SDM reference space by first rigidly registering the post-op CT image to the pre-op MR and then non-rigidly registering the pre-op MR image to the reference template image (MNI image). Once brought into the SDM space, the resliced post-op CT image is then non-rigidly registered to the resliced pre-op MR image using the SDM to constrain the deformation. The iEEG electrodes identified in the post-op CT image may than be co-visualized with the pre-op imaging data.
With the images now transformed to the SDM reference space, we non- rigidly register to The SDM model coefficients w in (2) are a low-dimensional parameterization (k degrees of freedom) of a high-dimensional FFD d, and we denote this transformation TSDM(x; w) for points in the reference image domain x ∈ ΩMNI ⊂ R3. We register and by optimizing the objective function
| (4) |
where J is the NMI similarity metric evaluated at all points x ∈ ΩMNI. We use a multi-resolution image pyramid and conjugate gradient optimization to solve (4), using w = 0 as an initial solution, which corresponds to the mean training deformation in (2). We also constrain the SDM deformation to be within 3 standard deviations of the mean, .
3.3. Co-visualizing electrodes in pre-op MR image space
In the previous section, we non-rigidly registered the post-implant CT to the pre-op MR using our proposed SDM of interventional deformation within the space of the SDM (MNI image space). While each patient's full set of pre-op and post-op data may be co-visualized within the SDM image space by applying the transformations from (3), it is more natural to visualize the implanted iEEG electrodes within the pre-implantation imaging data's native space. To visualize the CT imaging data in pre-op MR space, we apply the following transformations
| (5) |
where we combine the transformations from (3) and (4), and is the inverse transformation moving the image from the SDM image space back to native pre-op MR image space. While FFD transformations are not necessarily diffeomorphic, our use of relatively smooth transformation with large B-spline control point spacing (15 mm) makes approximation of this inverse transformation possible.
4. Results
From a clinical dataset of surgical epilepsy patients who underwent intra-cranial electroencephalography (iEEG) at Yale, we manually identified 18 patients with lateral craniotomies (10 on the right side, 8 on the left) with large electrode grids (80 × 80 mm) implanted. In order to increase the dataset size, we flipped the left-side craniotomy images to be right-side craniotomies because we know that the gross brain deformation correlates to craniotomy location (Hartkens et al., 2003) and we assume here that the pattern of deformation is symmetric. For each patient in the database, we have the following images:
(i) a pre-implantation MR image (256 × 256 × 106 at 0.977 × 0.977 × 1.5 mm resolution); (ii) a post-implantation MR image (256 × 256 × 110 at 0.977 × 0.977 × 1.5 mm resolution); and (iii) a post-implantation CT image (512 × 512 × 137 at 0.488 × 0.488 × 1.25 mm resolution). We implemented and ran our algorithm as part of the BioImage Suite software package (Joshi et al., 2011).
4.1. Experimental setup
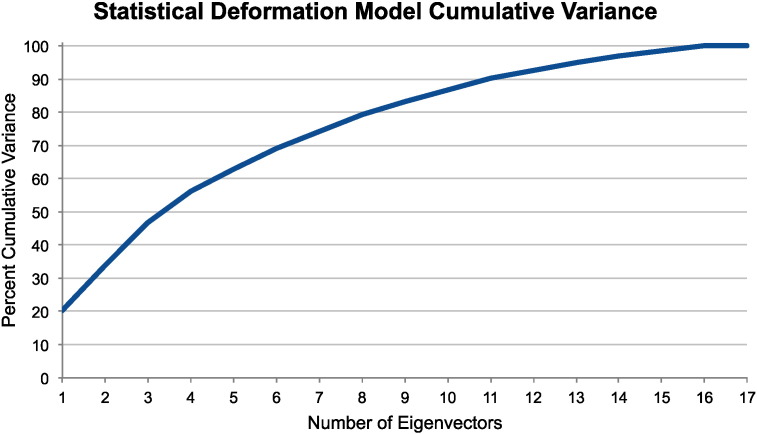
To demonstrate our proposed non-rigid statistical deformation model (SDM) registration approach, we performed leave-one-out testing. For each patient i = 1,. . .,18, we trained the SDM as described in Sec. 3.1 by omitting the i-th patient from the training set, which consisted of the N = 17 remaining samples. We then registered the i-th patient's post-op CT to their pre-op MR using the SDM (in MNI space) as described in Sec. 3.2. We repeated our registration method in (4) using different numbers of eigenvectors, i.e. using the first k = 1, 2, 3, 4, 5, 10, 15, and 17 eigenvectors. Fig. 4 shows that these first k eigenvectors account for approximately 20, 34, 47, 56, 63, 87, 98, and 100% of the SDM's cumulative variance, respectively. We compared our proposed method to directly register to with existing, standard registration methods: (i) intensity-based rigid registration, which in this case is the identity transformation from (3), and (ii) intensity-based, unconstrained non-rigid FFD . For both comparison methods, we optimized the registration by maximizing the normalized mutual information (NMI) similarity metric. For the FFD transformation model, we used 15 mm isotropic 3D B-spline control point spacing to match our SDM transformations from Sec. 3.1. An FFD with 15 mm control point spacing required P = 2, 535 control points to parameterize a transformation with 7,605 degrees of freedom in 3D.
Fig. 4.
Statistical deformation model (SDM) cumulative variance from an example leave-one-out test using N = 17 training deformations and parameterized by an FFD transformation with 15 mm control point spacing. The first eigenvector accounts for approximately 20% of the deformation variance.
To evaluate registration performance, we treated the non-rigid transformations generated during training with the post-op MR image in Sec. 3.1, and as used in current practice, as a ground-truth. Given an estimated transformation , we then calculated the magnitude of transformation error
| (6) |
which is the squared difference of displacement between the transformation estimate and our ground-truth transformation at all points in the image. While this metric evaluates performance against the current non-rigid gold-standard, through anecdotal evidence, surgeons generally only trust rigid registration for volumetric MR-CT registration. We therefore assessed MR-CT registration results with the rigid registration in mind. Furthermore, we evaluated maximum transformation error as surgeons are often interested in quantifying worst-case performance. In Sec. 4.2, we quantify this transformation error throughout the image volume, and in Sec. 4.3, we quantify it more specifically at the implanted electrodes locations.
4.2. Quantifying Transformation Error
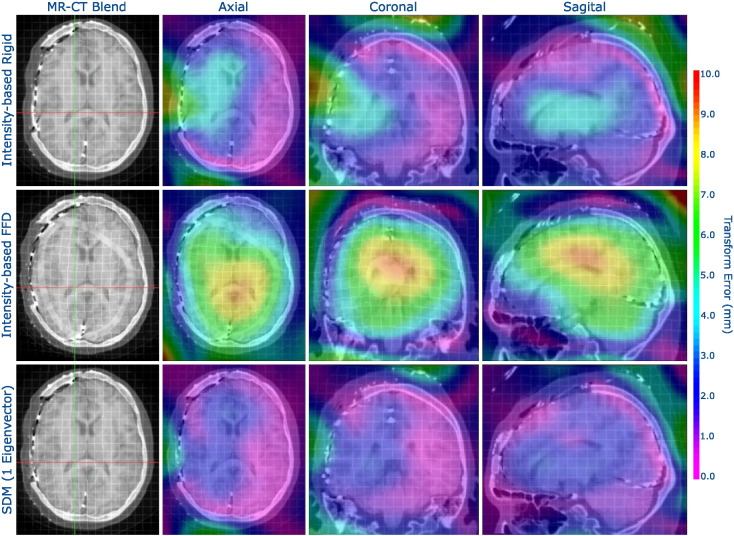
We spatially visualized ε(x), ∀x ∈∈ ΩMNI using a colormap overlay, as shown in Fig. 5, to compare the estimates of , and . In comparison to rigid registration, the proposed SDM method reduced error throughout the brain, and particularly so around the craniotomy and ventricles. These results also highlight the poor performance of the unconstrained FFD MR-CT registration, which performed worse than the rigid registration. Due to the poor soft-tissue contrast in the CT, the intensity-based FFD failed to accurately register the interior of the brain. Even though both the FFD and the SDM used the same NMI similarity metric, the SDM constrained the transformation to accurately mimic the interior deformations that resulted from the electrode implantation procedure.
Fig. 5.
Visualizing transformation error (ε from Eq. (6)) spatially using a colormap overlay for an example patient with a craniotomy (left side of the axial images). We show registration results using (i) intensity-based rigid registration, (ii) intensity-based, unconstrained FFD, and (iii) our proposed non-rigid SDM method using a single eigenvector, with the non-rigid registrations parameterized by an FFD transformation model with 15 mm isotropic control point spacing. The registration constrained by the SDM outperformed the other methods, particularly in the areas around the craniotomy and around the ventricles. Furthermore, unconstrained, intensity-only FFD registration performed noticeably worse than intensity-only rigid registration.
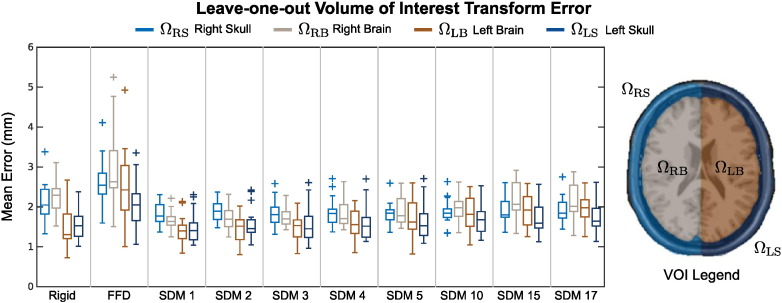
In Fig. 6, we quantified the mean transform error throughout 4 different volumes of interest (VOIs) in the brain: (i) the right skull ΩRS; (ii) the right brain hemisphere ΩRB; (iii) the left brain hemisphere ΩLB; and (iv) the left skull ΩLS, such that Ωi ⊂ ΩMNI and The ΩRB and ΩRS VOIs were of particular interest since they were ipsilateral to the craniotomy.
Fig. 6.
Mean transformation error for 18 patients in 4 different VOIs using (i) rigid, (ii) intensity-only FFD, and (iii) the proposed SDM registration methods, where SDM k denotes registration using the first k eigenvectors. The boxplots on the left show leave-one-out testing results for the SDM using different numbers of deformation eigenvectors. The boxplots show median, inner quartile extremes, and outlier values (outside 2.7 standard deviations).
We created these VOIs by dilating the MNI mask with a morphological filter enough times to cover the skull. Fig. 6 shows calculated within each of these VOIs. In general, both the right skull and right brain VOIs exhibited higher mean than in the left skull and left brain across all 18 leave-out-tests for all three registration methods, which was to be expected since this was the side of the craniotomy and largest deformation. We also noted that generally increased as we used more modes of variation in our SDM, which is most likely explained by the PCA over-fitting to the training set. The results shown in Fig. 6 also show that the unconstrained, non-rigid FFD registration performed significantly worse (two-tailed paired t-test p < 0.05) than either the rigid or our proposed non-rigid SDM registration methods in all VOIs, which supports the observations visualizing transformation error (Fig. 5). Our proposed SDM registration method significantly reduced (two-tailed paired t-test p < 0.05) in ΩRB using 1–5 eigenvectors and in ΩRS using 1–15 eigenvectors in comparison to rigid registration. Using the SDM registration, however, significantly increased in ΩLB when using 15 and 17 eigenvectors and in ΩLB when using 17 eigenvectors. Otherwise, there were no significant increases in in ΩLB or ΩLS with respect to rigid registration.
4.3. Quantifying electrode transformation error
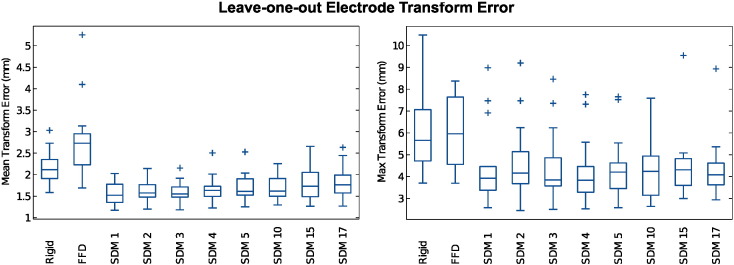
We quantified ε(xe) at each patient's electrode locations xe ∈ ΩE ⊂ ΩMNI, where ΩE is the set of all electrodes for each patient. The mean number of electrodes for a patient was 197. Fig. 7 summarizes the distributions of patient mean error and maximum error εmax over all xe ∈ ΩE. For all results, we evaluated statistical significance using two-tailed paired t-tests with p < 0.05.
Fig. 7.
Our proposed non-rigid SDM registration method significantly reduced transformation error (two-tailed paired t-test) compared to standard rigid and unconstrained FFD intensity registration methods. SDM k denotes registration using the first k = 1, 2, 3, … eigenvectors. We plot the distributions of both the mean (left plot) and the maximum (right plot) transformation error at electrode locations for the 18 leave-one-out MR-CT registrations for transformations. The boxplots show median, inner quartile, extremes, and outlier values.
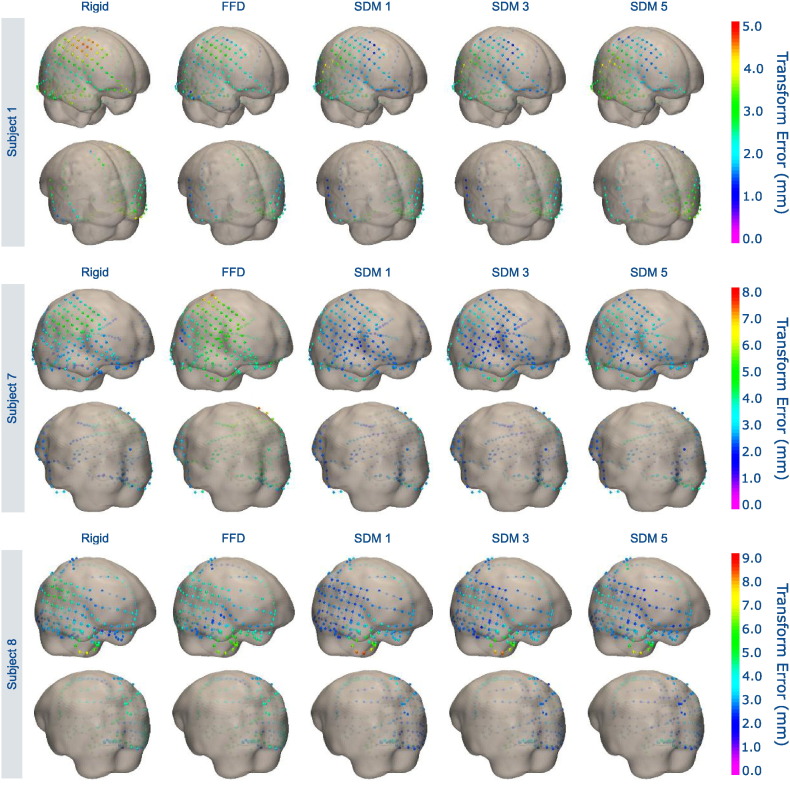
Our proposed SDM registration significantly reduced both and εmax with respect to both rigid registration and non-rigid, unconstrained FFD registration for all numbers of eigenvectors k tested (Table 1). The unconstrained FFD significantly increased with respect to rigid registration. Across all 18 leave-one-out tests, the mean electrode error using the SDM with 1 mode was 1.58 ± 0.24 mm, which compared to 2.12 ± 0.37 mm for rigid registration and 2.75 ± 0.85 mm and for unconstrained FFD (all reported values are mean ± standard deviation). Similarly, the maximum electrode error εmax values using the SDM with 1 mode was 4.39 ± 1.70 mm, which compared to 6.08 ± 1.84 mm for rigid registration and 5.95 ± 1.64 mm for FFD across all 18 tests. There was no significant difference in εmax between rigid and unconstrained FFD. Fig. 8 spatially visualizes these electrode transformation error for three different subjects, and shows that our proposed registration method reduces transformation error in most areas throughout the brain. Fig. 9 shows example results transforming the electrodes to pre-op MR imaging space using the respective transformation estimates and (5).
Table 1.
The distributions of mean electrode transformation error and maximum electrode transformation error εmax using rigid, non-rigid FFD, our proposed statistical deformation model (SDM) across all 18 leave-one-out registration tests. SDM k refers to the model using k eigenvectors. Reported values are Mean ± SD and maximum values.
| Method | (mm) | Max. (mm) | εmax (mm) | Max. εmax (mm) |
|---|---|---|---|---|
| Rigid | 2.12 ± 0.37 | 3.03 | 6.08 ± 1.84 | 10.48 |
| FFD | 2.75 ± 0.85 | 5.25 | 5.95 ± 1.64 | 8.37 |
| SDM 1 | 1.58 ± 0.24 | 2.03 | 4.39 ± 1.70 | 8.98 |
| SDM 2 | 1.63 ± 0.23 | 2.14 | 4.62 ± 1.62 | 9.20 |
| SDM 3 | 1.60 ± 0.23 | 2.16 | 4.39 ± 1.55 | 8.46 |
| SDM 4 | 1.65 ± 0.28 | 2.51 | 4.22 ± 1.43 | 7.75 |
| SDM 5 | 1.70 ± 0.29 | 2.53 | 4.38 ± 1.40 | 7.65 |
| SDM 10 | 1.68 ± 0.27 | 2.26 | 4.24 ± 1.23 | 7.59 |
| SDM 15 | 1.81 ± 0.40 | 2.66 | 4.43 ± 1.43 | 9.54 |
| SDM 17 | 1.82 ± 0.38 | 2.64 | 4.34 ± 1.31 | 8.93 |
Fig. 8.
Visualizing electrode transformation error for three sample patients using (i) rigid registration, (ii) unconstrained, non-rigid FFD registration, and (iii) our proposed non-rigid SDM registration using k = 1, 3, 5 eigenvectors. We display the electrodes in their “gold-standard” locations registered using the post-implant MR images. We show the 3D pre-op MR brain surface as transparent in order to visualize the subdural electrodes. Our proposed method reduces transformation error both at the site of the craniotomy with the large electrode grids and at locations further away.
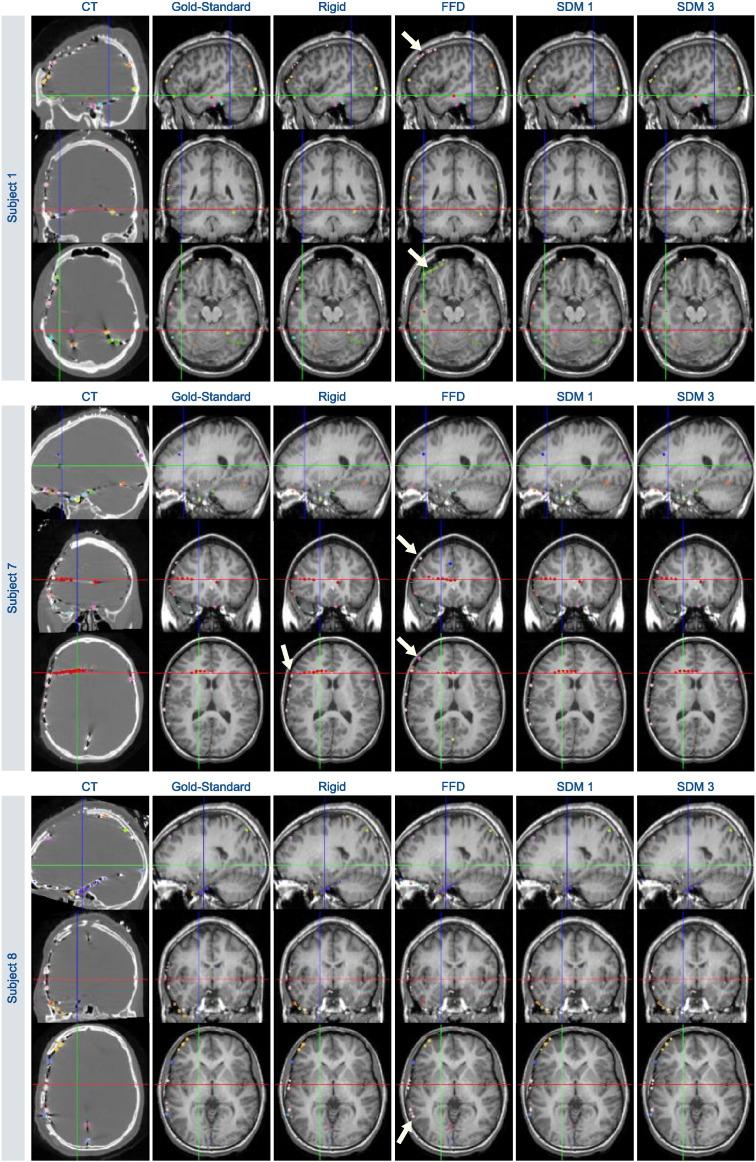
Fig. 9.
Visualizing electrodes for three patients in the post-op CT image space and transformed to pre-op MR imaging space using: (i) “gold-standard” non-rigid registration making use of the post-op MR image, (ii) rigid registration, (iii) unconstrained, non-rigid FFD registration, and (iv) our proposed non-rigid SDM registration using k = 1, 3 eigenvectors. Arrows highlight electrodes that are poorly localized with respect to the pre-op anatomy due to mis-registration. Crosshairs indicate the axial, coronal, and sagittal image slices displayed for each subject.
5. Discussion and conclusion
Our proposed method to model post-surgical non-rigid deformations with a statistical deformation model (SDM) significantly reduces both mean and maximum transformation errors in multi-modality non-rigid MR-CT registration compared to standard rigid and unconstrained non-rigid multi-modal registrations. As shown in Sec. 4, standard, unconstrained non-rigid registration methods perform worse than rigid methods, at least in this dataset where the CT images had both deformations and significant artifacts. Our proposed low-dimensional non-rigid registration method effectively compensates for brain deformations induced by the surgical craniotomy and implantation of iEEG electrodes. The proposed SDM does not model anatomical brain variability, but rather the deformation of a subject's brain as a result of craniotomy and electrode implantation, which is relatively smooth. Our method utilizes a PCA deformation model that, while simple, captures these low-dimensional intra-subject deformations and adds robustness to the image registration process. Our proposed registration method, using only the first few eigenvectors, exhibits better worst-case performance because the SDM transformation constrains the multi-modal registration to be from the space of observed deformations learned from a training set of clinical surgeries.
While the proposed PCA-based SDM registration method is simple, it effectively captures the low-dimensional deformations, even using a single eigenvector, as evidenced by the results using real, clinical data (Sec. 4). Surgeons can only place large grids (80 × 80 mm) in a small set of locations resulting in a constrained brain shift problem. This work is not an attempt to solve the general pre- to post-surgery registration problem, but rather account for the deformation caused by this well-defined (but common) procedure in epilepsy. The proposed method, when using 15 mm control point spacing, significantly reduced both average and worst-case registration transformation error throughout the brain volume (Fig. 6) and at electrode locations (Fig. 7) compared to rigid and unconstrained FFD registration methods using only the first few eigenvectors, specifically 1 ≤ k ≤ 5. This improved and more robust registration performance comes with a greater than 99% reduction in transformation degrees of freedom (DoF) (from 7605 DoF to 1 ≤ k ≤ 5 DoF). Constraining the non-rigid transformation by the low-dimensional SDM prohibits the registration optimization routine in (4) from seeking local minima in the similarity function that correspond to invalid deformations.
While our choice to parameterize the non-rigid SDM FFD transformation with 15 mm control point spacing was based upon the current Yale Epilepsy program workflow, we also experimented training and testing our proposed framework using an FFD model with a 30 mm isotropic control point spacing. This lower-dimensional FFD required P = 392 control points to parameterize a non-rigid transformation with 1,176 degrees of freedom. We reran our leave-one-out tests by performing SDM construction (Sec. 3.1) and SDM non-rigid registration (Sec. 3.2) using this alternate FFD parameterization. Similarly, we reran the experiments from Sec. 4 to compare these SDM results to both intensity-based rigid and intensity-based, unconstrained FFD registration with 30 mm isotropic control point spacing. The results using this alternative FFD parameterization were nearly identical to our results presented using the 15 mm control point spacing. Again, our constrained SDM outperformed both the rigid and the unconstrained, non-rigid registration methods, yielding results practically identical to those shown in Fig. 6, Fig. 7.
Although we present results for training a SDM at only a single craniotomy location, in the future, we aim to create craniotomy site-specific SDMs to model the corresponding deformations at different locations. Furthermore, we could improve training set registrations by including additional labeled anatomical structures, e.g. the ventricles, to improve PCA model construction (Onofrey et al., 2015). SDM training could also benefit from better registration to a common reference space as the use of a single reference subject, the MNI brain, biases any registrations towards that subject's particular anatomy. In this paper, our framework requires that all images be transformed to this common space in which the SDM was defined. In practice, it might be better to warp all the transformations to the test image's space, and then compute the SDM in this native space. However, this method also presents difficulties because it requires numerous non-rigid registrations of each training image to the test image. Alternative non-rigid transformation models Sotiras et al. (2013), other than our chosen FFD parameterization, could be used within our framework for SDM training and registration. However, a limitation of PCA-based SDMs is that, even if the non-rigid training deformations are diffeomorphic, the linear combination of deformation eigenvectors may not necessarily be diffeomorphic. However, this is less of an issue for deformations with large control point spacing, that are smooth and model gross intra-subject deformations, as are used in our method.
It is difficult, however, for us to make an appropriate comparison of our error results (1.58 ± 0.24 mm using only the first eigenvector) to other recent methods. Taimouri et al. reported electrode registration errors of 1.31 ± 0.69 mm (Taimouri et al., 2014) and Dalal et al. reported errors of 1.50 ± 0.50 mm (Dalal et al., 2008).
Both methods calculated electrode registration error using the visible surface electrodes only, and not the depth electrodes. We, on the other hand, calculated error using both the surface and depth electrodes. Furthermore, we cannot appropriately compare our error values to these prior works due to differences in our method for calculating error. Both of these works computed their errors with respect to 2D intra-operative photographs, in which they projected their 3D electrode location estimates onto the photograph's 2D space, and then computed errors in 2D. We, on the other hand, compute error in 3D space with respect to a reference transformation, using the clinical state-of-the-art non-rigid registration of the post-op MR image to pre-op MR transformation available at our institution as a reference.
While the goal of this work was to directly non-rigidly register post-op CT images to pre-op MR images of the same subject, the ideas presented here constitute a general framework to non-rigidly register multi-modal volumetric images to effectively compensate for deformations induced by interventional procedures. In particular, we demonstrate how we can use a small subset of high quality training data (in this case, the rare availability of post-electrode implantation MR images) to learn the properties of a deformation model in a given case of non-rigid deformation, and to subsequently use this knowledge to solve the non-rigid registration problem in the more general case with lesser quality data (the direct non-rigid, multi-modal registration of pre-op MR to post-implantation CT). Similar principles could be applied to non-rigidly register other multi-modal interventional images, e.g. interventional ultrasound images to pre-operative MR images.
Acknowledgments
This work was supported by National Institute of Biomedical Imaging and Bioengineering (NIBIB) R03 EB012969.
Contributor Information
John A. Onofrey, Email: john.onofrey@yale.edu.
Lawrence H. Staib, Email: lawrence.staib@yale.edu.
Xenophon Papademetris, Email: xenophon.papademetris@yale.edu.
References
- Azarion A.A., Wu J., Pearce A., Krish V.T., Wagenaar J., Chen W., Zheng Y., Wang H., Lucas T.H., Litt B., Gee J.C., Davis K.A. An open-source automated platform for three-dimensional visualization of subdural electrodes using CT-MRI coregistration. Epilepsia. 2014;55(12):2028–2037. doi: 10.1111/epi.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen F.F., van der Heide U.A., Langerak T.R., Kotte A.N., Pluim J.P. Free-form image registration regularized by a statistical shape model: application to organ segmentation in cervical MR. Comput. Vis. Image Underst. 2013;117(9):1119–1127. [Google Scholar]
- Bhavaraju N., Nagaraddi V., Chetlapalli S., Osorio I. Electrical and thermal behavior of non-ferrous noble metal electrodes exposed to MRI fields. Magn. Reson. Imaging. 2002;20(4):351–357. doi: 10.1016/s0730-725x(02)00506-4. [DOI] [PubMed] [Google Scholar]
- Cascino G.D. Surgical treatment for epilepsy. Epilepsy Res. 2004;60(2):179–186. doi: 10.1016/j.eplepsyres.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Cerrolaza J., Villanueva A., Cabeza R. Hierarchical statistical shape models of multiobject anatomical structures: application to brain MRI. IEEE Trans. Med. Imaging. 2012;31(3):713–724. doi: 10.1109/TMI.2011.2175940. [DOI] [PubMed] [Google Scholar]
- Cootes T., Taylor C., Cooper D., Graham J. Active shape models—their training and application. Comput. Vis. Image Underst. 1995;61(1):38–59. [Google Scholar]
- Cootes T., Marsland S., Twining C., Smith K., Taylor C. Groupwise diffeomorphic non-rigid registration for automatic model building. In: Pajdla T., Matas J., editors. Computer Vision — ECCV 2004. Vol. 3024 of Lecture Notes in Computer Science. Springer; 2004. pp. 316–327. [Google Scholar]
- Dalal S.S., Edwards E., Kirsch H.E., Barbaro N.M., Knight R.T., Nagarajan S.S. Localization of neurosurgically implanted electrodes via photograph-MRI-radiograph coregistration. J. Neurosci. Methods. 2008;174(1):106–115. doi: 10.1016/j.jneumeth.2008.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLorenzo C., Papademetris X., Staib L., Vives K., Spencer D., Duncan J. Volumetric intraoperative brain deformation compensation: model development and phantom validation. IEEE Trans. Med. Imaging. 2012;31(8):1607–1619. doi: 10.1109/TMI.2012.2197407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumpuri P., Thompson R.C., Dawant B.M., Cao A., Miga M.I. An atlas-based method to compensate for brain shift: preliminary results. Med. Image Anal. 2007;11(2):128–145. doi: 10.1016/j.media.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumpuri P., Thompson R., Cao A., Ding S., Garg I., Dawant B., Miga M. A fast and efficient method to compensate for brain shift for tumor resection therapies measured between preoperative and postoperative tomograms. Biomed. Eng.IEEE Trans. 2010;57(6):1285–1296. doi: 10.1109/TBME.2009.2039643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt J., Werner R., Schmidt-Richberg A., Handels H. Statistical modeling of 4D respiratory lung motion using diffeomorphic image registration. IEEE Trans. Med. Imaging. 2011;30(2):251–265. doi: 10.1109/TMI.2010.2076299. [DOI] [PubMed] [Google Scholar]
- Ferrant M., Nabavi A., Macq B., Jolesz F., Kikinis R., Warfield S. Registration of 3-D intraoperative MR images of the brain using a finite-element biomechanical model. IEEE Trans. Med. Imaging. 2001;20(12):1384–1397. doi: 10.1109/42.974933. [DOI] [PubMed] [Google Scholar]
- Gee J.C., Bajcsy R.K. Elastic matching: Continuum mechanical and probabilistic analysis. In: Toga A.W., editor. Brain Warping. Academic Press; 1999. pp. 183–197. [Google Scholar]
- Gerber S., Tasdizen T., Fletcher P.T., Joshi S., Whitaker R. Manifold modeling for brain population analysis. Med. Image Anal. 2010;14(5):643–653. doi: 10.1016/j.media.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenander U., Miller M. Computational anatomy: an emerging discipline. Q. Appl. Math. 1998;56(4):617–694. [Google Scholar]
- Hagemann A., Rohr K., Stiehl H., Spetzger U., Gilsbach J. Biomechanical modeling of the human head for physically based, nonrigid image registration. IEEE Trans. Med. Imaging. 1999;18(10):875–884. doi: 10.1109/42.811267. [DOI] [PubMed] [Google Scholar]
- Hamm J., Ye D.H., Verma R., Davatzikos C. Gram: a framework for geodesic registration on anatomical manifolds. Med. Image Anal. 2010;14(5):633–642. doi: 10.1016/j.media.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartkens T., Hill D., Castellano-Smith A., Hawkes D., Maurer C.R., Martin J., Hall A., Liu W., Truwit C. Measurement and analysis of brain deformation during neurosurgery. Medical Imaging. IEEE Trans. Med. Imaging. 2003;22(1):82–92. doi: 10.1109/TMI.2002.806596. [DOI] [PubMed] [Google Scholar]
- He T., Xue Z., Xie W., Wong S. Online 4-D CT estimation for patient-specific respiratory motion based on real-time breathing signals. In: Jiang T., Navab N., Pluim J., Viergever M., editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2010. Vol. 6363 of Lecture Notes in Computer Science. Springer; 2010. pp. 392–399. [DOI] [PubMed] [Google Scholar]
- Heimann T., Meinzer H.-P. Statistical shape models for 3D medical image segmentation: a review. Med. Image Anal. 2009;13(4):543–563. doi: 10.1016/j.media.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Hermes D., Miller K.J., Noordmans H.J., Vansteensel M.J., Ramsey N.F. Automated electrocorticographic electrode localization on individually rendered brain surfaces. J. Neurosci. Methods. 2010;185(2):293–298. doi: 10.1016/j.jneumeth.2009.10.005. [DOI] [PubMed] [Google Scholar]
- Hu Y., Ahmed H.U., Taylor Z., Allen C., Emberton M., Hawkes D., Barratt D. MR to ultrasound registration for image-guided prostate interventions. Med. Image Anal. 2012;16(3):687–703. doi: 10.1016/j.media.2010.11.003. [DOI] [PubMed] [Google Scholar]
- Jia H., Wu G., Wang Q., Shen D. Absorb: Atlas building by self-organized registration and bundling. NeuroImage. 2010;51(3):1057–1070. doi: 10.1016/j.neuroimage.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi S.C., Miller M.I., Grenander U. On the geometry and shape of brain sub-manifolds. Int. J. Pattern Recogn. Artificial Intell. 1997;11(8):1317–1343. [Google Scholar]
- Joshi A., Scheinost D., Okuda H., Belhachemi D., Murphy I., Staib L., Papademetris X. Unified framework for development, deployment and robust testing of neuroimaging algorithms. Neuroinformatics. 2011;9:69–84. doi: 10.1007/s12021-010-9092-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-J., Kim M.-H., Shen D. Computer Vision and Pattern Recognition Workshops, 2008. CVPRW′08. IEEE Computer Society Conference. 2008. Learning-based deformation estimation for fast non-rigid registration; pp. 1–6. [Google Scholar]
- Loeckx D., Maes F., Vandermeulen D., Suetens P. Temporal subtraction of thorax CR images using a statistical deformation model. IEEE Trans. Med. Imaging. 2003;22(11):1490–1504. doi: 10.1109/TMI.2003.819291. [DOI] [PubMed] [Google Scholar]
- Nabavi A., Black P.M., Gering D.T., Westin C.-F., Mehta V., Pergolizzi R.S., Ferrant M., Warfield S.K., Hata N., Schwartz R.B., Wells W.M., Kikinis R., Jolesz F.A. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery. 2001;48(4):787–798. doi: 10.1097/00006123-200104000-00019. [DOI] [PubMed] [Google Scholar]
- Nowell M., Rodionov R., Zombori G., Sparks R., Winston G., Kinghorn J., Diehl B., Wehner T., Miserocchi A., McEvoy A.W., Ourselin S., Duncan J. Utility of 3D multimodality imaging in the implantation of intracranial electrodes in epilepsy. Epilepsia. 2015;56(3):403–413. doi: 10.1111/epi.12924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofrey J.A., Staib L.H., Papademetris X. Learning nonrigid deformations for constrained multi-modal image registration. In: Mori K., Sakuma I., Sato Y., Barillot C., Navab N., editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2013. Vol. 8151 of Lecture Notes in Computer Science. Springer; 2013. pp. 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onofrey J., Papademetris X., Staib L. Low-dimensional non-rigid image registration using statistical deformation models from semi-supervised training data. IEEE Trans. Med. Imaging. 2015;34(7):1522–1532. doi: 10.1109/TMI.2015.2404572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papademetris X. Estimation of 3D Left Ventricular Deformation from Medical Images Using Biomechanical Models. Yale University; May 2000. Ph.D. thesis. [DOI] [PubMed] [Google Scholar]
- Papademetris X., Jackowski A.P., Schultz R.T., Staib L.H., Duncan J.S. Integrated intensity and point-feature nonrigid registration. In: Barillot C., Haynor D.R., Hellier P., editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2004. Vol. 3216 of Lecture Notes in Computer Science. Springer; 2004. pp. 763–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen K.D., Miga M.I., Kennedy F.E., Hoopens P.J., Hartov A., Roberts D.W. A computational model for tracking subsurface tissue deformation during stereotactic neurosurgery. Biomed. Eng. IEEE Trans. 1999;46(2):213–225. doi: 10.1109/10.740884. [DOI] [PubMed] [Google Scholar]
- Rueckert D., Sonoda L., Hayes C., Hill D., Leach M., Hawkes D. Nonrigid registration using free-form deformations: application to breast MR images. Med. Imaging IEEE Trans. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Rueckert D., Frangi A., Schnabel J. Automatic construction of 3-D statistical deformation models of the brain using nonrigid registration. Med. Imaging IEEE Trans. 2003;22(8):1014–1025. doi: 10.1109/TMI.2003.815865. [DOI] [PubMed] [Google Scholar]
- Sˇkrinjar O., Nabavi A., Duncan J. Model-driven brain shift compensation. Med. Image Anal. 2002;6(4):361–373. doi: 10.1016/s1361-8415(02)00062-2. [DOI] [PubMed] [Google Scholar]
- Singh N., Fletcher P., Preston J., Ha L., King R., Marron J., Wiener M., Joshi S. Multivariate statistical analysis of deformation momenta relating anatomical shape to neuropsychological measures. In: Jiang T., Navab N., Pluim J., Viergever M.A., editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2010. Vol. 6363 of Lecture Notes in Computer Science. Springer; 2010. pp. 529–537. [DOI] [PubMed] [Google Scholar]
- Smith S.M. Human Brain Mapping. 17 (3) 2002. Fast robust automated brain extraction; pp. 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiras A., Davatzikos C., Paragios N. Deformable medical image registration: a survey. IEEE Trans. Med. Imaging. 2013;32(7):1153–1190. doi: 10.1109/TMI.2013.2265603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer S.S., Sperling M., Shewmon A. Ch. Intracranial electrodes. Lippincott-Raven; Philadelphia: 1998. Epilepsy, a comprehensive textbook; pp. 1719–1748. [Google Scholar]
- Studholme C., Hill D., Hawkes D. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recogn. 1999;32(1):71–86. [Google Scholar]
- Studholme C., Novotny E., Zubal I., Duncan J. Estimating tissue deformation between functional images induced by intracranial electrode implantation using anatomical MRI. NeuroImage. 2001;13(4):561–576. doi: 10.1006/nimg.2000.0692. [DOI] [PubMed] [Google Scholar]
- Taimouri V., Akhondi-Asl A., Tomas-Fernandez X., Peters J., Prabhu S., Poduri A., Takeoka M., Loddenkemper T., Bergin A., Harini C., Madsen J., Warfield S. Electrode localization for planning surgical resection of the epileptogenic zone in pediatric epilepsy. Int. J. Comput. Assist. Radiol. Surg. 2014;9(1):91–105. doi: 10.1007/s11548-013-0915-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum J.B., Silva V.d., Langford J.C. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290(5500):2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- Twining C.J., Marsland S. Constructing an atlas for the diffeomorphism group of a compact manifold with boundary, with application to the analysis of image registrations. J. Comput. Appl. Math. 2008;222(2):411–428. [Google Scholar]
- Wellmer J., Von Oertzen J., Schaller C., Urbach H., König R., Widman G., Van Roost D., Elger C.E. Digital photography and 3D MRI-based multimodal imaging for individualized planning of resective neocortical epilepsy surgery. Epilepsia. 2002;43(12):1543–1550. doi: 10.1046/j.1528-1157.2002.30002.x. [DOI] [PubMed] [Google Scholar]
- Wells W.M., III, Viola P., Atsumi H., Nakajima S., Kikinis R. Multi-modal volume registration by maximization of mutual information. Med. Image Anal. 1996;1(1):35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- Wittek A., Miller K., Kikinis R., Warfield S.K. Patient-specific model of brain deformation: application to medical image registration. J. Biomech. 2007;40(4):919–929. doi: 10.1016/j.jbiomech.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Wouters J., D'Agostino E., Maes F., Vandermeulen D., Suetens P. Non-rigid brain image registration using a statistical deformation model. In: Reinhardt J., Pluim J., editors. Medical Imaging 2006: Image Processing. Vol. 6144. 2006. pp. 614411–614418. (Proc. of SPIE). [Google Scholar]
- Xue Z., Shen D., Davatzikos C. Statistical representation of high-dimensional deformation fields with application to statistically constrained 3D warping. Med. Image Anal. 2006;10(5):740–751. doi: 10.1016/j.media.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Ye D., Hamm J., Kwon D., Davatzikos C., Pohl K. Regional manifold learning for deformable registration of brain mr images. In: Ayache N., Delingette H., Golland P., Mori K., editors. Medical Image Computing and Computer-Assisted Intervention — MICCAI 2012. Vol. 7512 of Lecture Notes in Computer Science. Springer; 2012. pp. 131–138. [DOI] [PMC free article] [PubMed] [Google Scholar]