Abstract
Background and Aims Ecological differentiation is recognized as an important factor for polyploid speciation, but little is known regarding whether the ecological niches of cytotypes differ between areas of sympatry and areas where single cytotypes occur (i.e. niche displacement).
Methods Ecological niches of four groups of Senecio carniolicus sensu lato (s.l.) (western and eastern diploid lineages, tetraploids and hexaploids) were characterized via Landolt indicator values of the accompanying vascular plant species and tested using multivariate and univariate statistics.
Key Results The four groups of S. carniolicus s.l. were ecologically differentiated mainly with respect to temperature, light and soil (humus content, nutrients, moisture variability). Niche breadths did not differ significantly. In areas of sympatry hexaploids shifted towards sites with higher temperature, less light and higher soil humus content as compared with homoploid sites, whereas diploids and tetraploids shifted in the opposite direction. In heteroploid sites of tetraploids and the western diploid lineage the latter shifted towards sites with lower humus content but higher aeration.
Conclusions Niche displacement can facilitate the formation of stable contact zones upon secondary contact of polyploids and their lower-ploid ancestors and/or lead to convergence of the cytotypes’ niches after they have attained non-overlapping ranges. Niche displacement is essential for understanding ecological consequences of polyploidy.
Keywords: autopolyploidy, biodiversity, coexistence, contact zones, ecological differentiation, range-wide niche displacement, Senecio carniolicus s.l., speciation.
INTRODUCTION
Polyploidy is a major force in plant diversification and speciation (Wood et al., 2009; Weiss-Schneeweiss et al., 2013). Polyploidization can confer instantaneous reproductive isolation and is therefore regarded as the most frequent mode of sympatric speciation (Otto and Whitton, 2000). However, reproductive isolation is rarely complete, hampering independent evolution of the polyploid. It is well recognized, especially from modelling results, that dispersal limitation (Baack, 2005), high levels of selfing (Levin, 1983) and particularly ecological differentiation (Fowler and Levin, 1984; Levin, 2002; Johnson et al., 2003; Oswald and Nuismer, 2011; Ramsey and Ramsey, 2014) are important factors allowing polyploids to persistently survive in sympatry with their lower-ploid ancestors, thus contributing to polyploid speciation.
Although habitat segregation is recognized as a major factor allowing different cytotypes to coexist (e.g. Lumaret et al., 1987; Thompson and Lumaret, 1992; Felber-Girard et al., 1996; Ståhlberg and Hedrén, 2009; Duchoslav et al., 2010), little is known regarding whether the ecological niches of cytotypes differ between areas of sympatry and areas where single cytotypes occur. Habitat requirements of ecologically similar taxa might differ more pronouncedly when occurring in sympatry than in allopatry (Grace and Wetzel, 1981; Peers et al., 2013), as is well known for phenotypic characters (i.e. character displacement: Dayan and Simberloff, 2005; Pfennig and Pfennig, 2009). In fact, the resulting niche displacement (Peers et al., 2013) is expected to reflect a complex pattern of displacement in morphological, ecophysiological and/or behavioural characters. There is limited evidence suggesting niche displacement in polyploid complexes (Felber-Girard et al., 1996; Ståhlberg and Hedrén, 2009) but these studies were based on geographically restricted sampling with a low number of populations harbouring diploids and polyploids. Range-wide tests of niche displacement in polyploid complexes are lacking so far.
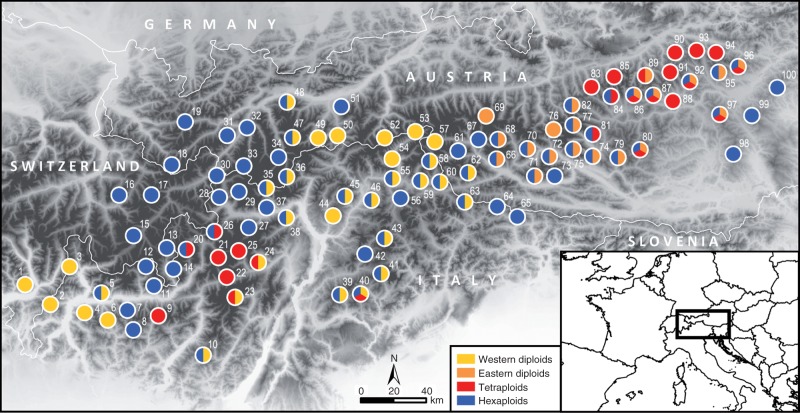
An excellent system to study ecological differentiation on a range-wide scale is Senecio carniolicus sensu lato (s.l.) (syn. Jacobaea c.; Asteraceae), a common plant in various alpine to nival habitats on siliceous bedrock in the eastern European Alps. This alpine polyploid complex consists of two geographically non-overlapping diploid lineages (Escobar García et al., 2012) as well as one tetraploid and one hexaploid cytotype (Sonnleitner et al., 2010; Fig. 1) that originated from the eastern diploid lineage (Sonnleitner et al., 2013). Although recently described as four separate species (Flatscher et al., 2015), we here refer to them as lineages or cytotypes to facilitate comparisons with previous studies (Schönswetter et al., 2007; Sonnleitner et al., 2010, 2013; Hülber et al., 2015). Based on coarse estimates of site conditions, cytotypes are ecologically segregated, but overlap broadly (Sonnleitner et al., 2010). The degree of separation and the discriminative ecological gradients, however, remain unknown. Throughout the distribution range of S. carniolicus s.l. in the eastern Alps, sites inhabited by a single cytotype (hereafter ‘homoploid sites’) as well as sites harbouring two or three cytotypes in sympatry (hereafter ‘heteroploid sites’) occur frequently (Suda et al., 2007; Sonnleitner et al., 2010; Fig. 1). The widespread co-occurrence of cytotypes might be facilitated by niche displacement, but this has not yet been tested. Here, we aim (1) to identify the ecological factors responsible for the previously identified cytotype differentiation (Sonnleitner et al., 2010) and (2) to test whether realized niches in heteroploid sites differ from those in homoploid sites as expected under a model of niche displacement. To this end, we used Landolt indicator values for vascular plant species surrounding target individuals of S. carniolicus to characterize the ecological niches of the four S. carniolicus groups (two diploid lineages, tetraploids, hexaploids) on the microhabitat level. Although originally developed for the Swiss flora (Landolt, 1977), Landolt indicator values have subsequently been extended to the entire Alps (Landolt et al., 2010). They represent qualitative assessments of ecological variables and hence are appropriate to characterize environmental conditions when their explicit measurement is not feasible because of, for instance, the remoteness and/or high number of sampling sites. Furthermore, they are less prone to between-year fluctuations in environmental conditions that may yield biased measurements, especially in cases of long-lived perennial plants of alpine regions (Diekmann, 2003).
Fig. 1.
Distribution of western diploids, eastern diploids, tetraploids and hexaploids of Senecio carniolicus in the eastern European Alps. Numbers correspond to site codes in Table S1 (Supplementary Information).
MATERIALS AND METHODS
Fieldwork
Ecological niches of western and eastern diploid lineages and of the polyploids (tetraploids and hexaploids) were analysed based on samples from 99 sites (i.e. mountains) across the entire distribution area in the European Alps (Fig. 1) as described previously (Sonnleitner et al., 2010). Individuals were selected to represent the entire local altitudinal range and all occupied habitat types. As at the time of the fieldwork recognition of cytotypes based on morphological characters was not possible, cytotypes are expected to be included without bias. Cytotype assignment of each sampled individual was taken from Sonnleitner et al. (2010). Individuals showing secondary ploidy levels (triploid, pentaploid, heptaploid, octoploid and enneaploid, together comprising 1·07 % of analysed individuals) were not considered in this study, resulting in a total data set of 2826 individuals (i.e. on average 29 individuals per site; Supplementary Information Table S1).
Statistical analyses
Environmental conditions were characterized by mean Landolt indicator values of accompanying vascular plant species (Landolt et al., 2010) calculated for circular plots of 0·2 m radius around each target individual. Landolt indicator values describe ecological requirements of species in terms of climate parameters (temperature, T; continentality, K; light, L) and of soil parameters (moisture, F; reaction, R; nutrients, N; humus content, H; aeration, D; moisture variability, W), all ranging from 1 (low) to 5 (high). For species indifferent to particular indicator values, we used the mean of the concerned indicator values at the respective sites; S. carniolicus was omitted from the calculation.
Multivariate analyses
Canonical correspondence analysis (CCA; ter Braak, 1986) was used to test for shifts of niche optima and differences in niche breadth among groups of S. carniolicus. CCA allows for non-linear and even non-monotonic (unimodal) relationships between species occurrences and environmental gradients, and performs well with skewed species distributions, with situations where not all environmental factors determining species composition are known, and even with highly correlated variables (Palmer, 1993). We constrained a matrix of Landolt indicator values, which were scaled to zero mean and unit variance prior to the ordination, by one of two design matrices. These design matrices, where columns represent the membership of a given individual to one of the groups without and with differentiation into homoploid and heteroploid sites, were used to test for overall niche differences among western diploids, eastern diploids, tetraploids and hexaploids, and for niche differences between homoploid and heteroploid sites of the same group (western diploids, eastern diploids, tetraploids and hexaploids), respectively. Because of only two sites inhabited exclusively by eastern diploids – one of which was only represented by six individuals – no comparisons between homoploid and heteroploid sites of this lineage were made.
Additionally, we identified effects of co-occurring cytotypes (the two diploid lineages do not co-occur: Escobar García et al., 2012) by using design-matrices representing the presences/absences of cytotypes within sites. The target cytotype and each potentially co-occurring cytotype were represented by one column each in the design matrix. The target cytotype was defined as present only in homoploid sites (i.e. the target cytotype co-occurs only with itself). This analysis was performed separately for individuals of each target cytotype except for eastern diploids.
Following Treier et al. (2009) we defined niche shifts as differences in niche optima among western diploids, eastern diploids, tetraploids and hexaploids, and niche breadth as the area of an ellipse representing a graphical summary of the cloud of individuals of each group (western diploids, eastern diploids, tetraploids and hexaploids). Niche shifts were calculated as the Euclidean distance between the centroids of the cloud of individuals of each group weighted by the Eigenvalues of ordination axes. Differences in niche breadth were calculated as differences in the area of ellipses defined by the niche optimum and the standard deviation of the projections of points on the ellipse axis, which were drawn orthogonally with maximal dispersion (Treier et al., 2009). CCAs were computed with the maximum number of axes, but only the first and second axes were considered for the calculation of niche shift and breadth. Ten thousand Monte Carlo randomizations were applied by randomly assigning individuals to environmental conditions (i.e. by randomizing the rows in the design matrix). The significance of niche shifts and differences in niche breadth were evaluated by comparing the observed values with the distribution of simulated values in one-sided (observed >simulated) and two-sided tests (observed <> simulated), respectively.
To avoid the inflation of Type I errors due to multiple comparisons we adjusted the threshold for P values to be interpreted as significant according to the Dunn–Šidák correction: P = 1 – 0·95(1/n), where n is the number of comparisons. Thus, for six pairwise comparisons among eastern diploids, western diploids, tetraploids and hexaploids, and three comparisons between homoploid and heteroploid sites for western diploids, tetraploids and hexaploids the threshold was set to 0·008 and 0·017, respectively.
Univariate analyses
To test for significant differences among western diploids, eastern diploids, tetraploids and hexaploids, linear mixed-effects models optimizing the restricted maximum-likelihood (following Laird and Ware, 1982) were applied separately to each Landolt indicator value. We used each indicator value as response, the groups as categorical predictor and the site as grouping variable to calculate random intercepts. With this approach we accounted for potential spatial clustering among individuals from the same site (Bolker et al., 2009; Dullinger and Hülber, 2011). For all models we assumed a Gaussian error distribution. Degrees of freedom of the models were defined as the number of observations minus the number of estimated parameters (i.e. 4) minus 1, assuming that the grouping variable consumed only one degree of freedom.
To identify ecological gradients along which niches are displaced between homoploid and heteroploid sites a randomization procedure was applied. In pairwise comparisons of cytotypes (western diploids, tetraploids, hexaploids), for each cytotype 500 individuals from homoploid sites and 500 individuals from heteroploid sites, where the two compared cytotypes co-occur, were randomly chosen (with replacement) and differences in Landolt indicator values between individuals from homoploid and heteroploid sites were calculated. Statistical significance was assessed using two-sided t-tests adjusting for multiple comparisons by using Dunn–Šidák correction.
All analyses were performed with R 2.13.0 (http://www.R-project.org/). Mixed-effects models were calculated using the function lmer included in the package lme4 (http://cran.r-project.org/web/packages/lme4/lme4.pdf). Niche shifts and niche breadth were evaluated using the ade4 package (Dray and Dufour, 2007).
RESULTS
Ecological differentiation among groups
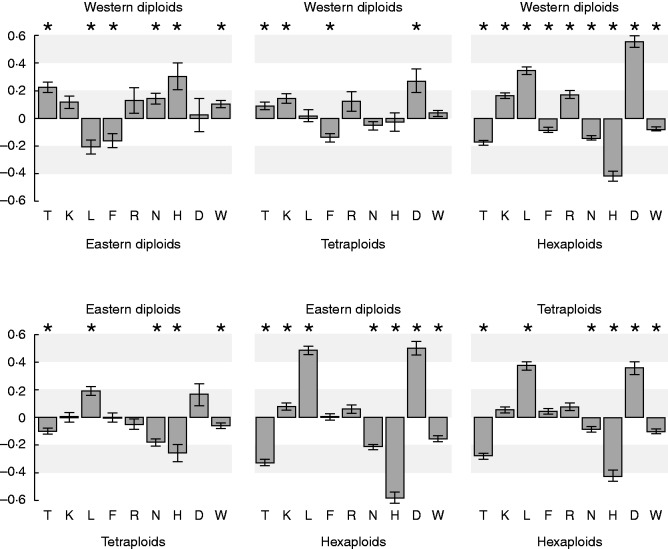
The four groups of S. carniolicus (western diploids, eastern diploids, tetraploids and hexaploids) were ecologically differentiated. Linear mixed-effects models revealed significantly different positions of western diploids, eastern diploids, tetraploids and hexaploids along the temperature (T) gradient, with eastern diploids occupying the coldest sites, followed by tetraploids and western diploids at intermediate sites, and hexaploids at the warmest sites (Fig. 2). The same order of these four groups – eastern diploids and hexaploids occupying opposite ends of a gradient with western diploids and tetraploids in intermediate positions – was found for humus content (H), soil nutrients (N) and soil moisture variability (W), with differences among groups being stronger for humus content (H) than for soil nutrients (N) and soil moisture variability (W). A similar differentiation but in reverse order was found along the light (L) gradient, with tetraploids and western diploids occupying the same range along the gradient. For soil aeration (D) hexaploids showed the lowest values, followed by tetraploids and both diploids at equally high values. The four groups showed similar requirements for soil moisture (F) except western diploids, which inhabited drier micro-habitats. This is in line with high continentality (K) values for western diploids. No clear differentiation of groups was found for soil reaction (R) except between western diploids and hexaploids.
Fig. 2.
Niche differentiation among western diploids (n = 612), eastern diploids (n = 305), tetraploids (n = 499) and hexaploids (n = 1410) of Senecio carniolicus derived from regression analyses of mean Landolt indicator values (Landolt et al., 2010) for temperature (T), continentality (K), light (L), soil moisture (F), soil reaction (R), soil nutrients (N), humus content (H), soil aeration (D) and soil moisture variability (W). Bars show fixed-effects coefficients (±SE) of linear mixed-effects models representing differences in mean indicator values between two groups and point towards the group with the higher value; asterisks indicate significant differences.
The Monte Carlo approach based on CCA revealed significant differences in niche optima of the four groups except for western diploids compared with tetraploids (Table 1). Groups were separated along an environmental gradient combining light (L), soil aeration (D), soil nutrients (N), humus content (H), temperature (T) and soil moisture variability (W; Supplementary Information Figs S1 and S2). Eastern diploids and hexaploids were located at the extreme ends of this gradient, while western diploids and tetraploids occupied intermediate positions. The latter two were separated, albeit not significantly, along a moisture (F) and continentality (K) gradient. In contrast to niche optima, no significant differences were found for niche breadth (Table 1).
Table 1.
Comparison of niche optima (niche shift) and ecological tolerance (niche breadth) among groups of Senecio carniolicus in the Alps derived from canonical correspondence analysis
| Shift* | Breadth | |
|---|---|---|
| Western diploids – eastern diploids | 0·001 | 0·480 |
| Western diploids – tetraploids | 0·394 | 0·534 |
| Western diploids – hexaploids | 0·003 | 0·866 |
| Eastern diploids – tetraploids | 0·006 | 0·514 |
| Eastern diploids – hexaploids | <0·001 | 0·147 |
| Tetraploids – hexaploids | <0·001 | 0·508 |
* P values of significant differences among groups after Dunn–Šidák correction for multiple comparisons are given in bold.
Ecological differentiation between homoploid and heteroploid sites
Ecological niches of cytotypes in heteroploid sites were displaced compared with homoploid sites. In a CCA (Fig. 3; Supplementary Information Fig. S3) niche optima of cytotypes in homoploid and heteroploid sites were significantly different for western diploids (P = 0·005) and for tetraploids (P = 0·019) but not for hexaploids (P = 0·168; significance assessed via randomizations). However, compared with homoploid sites, niches of hexaploids in heteroploid sites tended towards higher temperature (T) and lower soil moisture (F), while tetraploids were displaced roughly in the opposite direction along the gradient. Western diploid individuals occurred at sites with higher light (L) and soil aeration (D) in heteroploid compared with homoploid sites. These shifts resulted in stronger niche separation in the presence of other cytotypes, i.e. the pattern predicted by niche displacement. When looking at mutual effects of two cytotypes, the niches of hexaploids were displaced in response to the presence of tetraploids as well as of western diploids (Table 2), both of which responded to the presence of hexaploids. In contrast, tetraploids and western diploids did not interfere with each other, but tetraploids showed a stronger differentiation if co-occurring with eastern diploids. No changes in niche breadth were detected among homoploid and heteroploid sites (western diploids: P = 0·169; tetraploids: P = 0·036; hexaploids: P = 0·682).
Fig. 3.
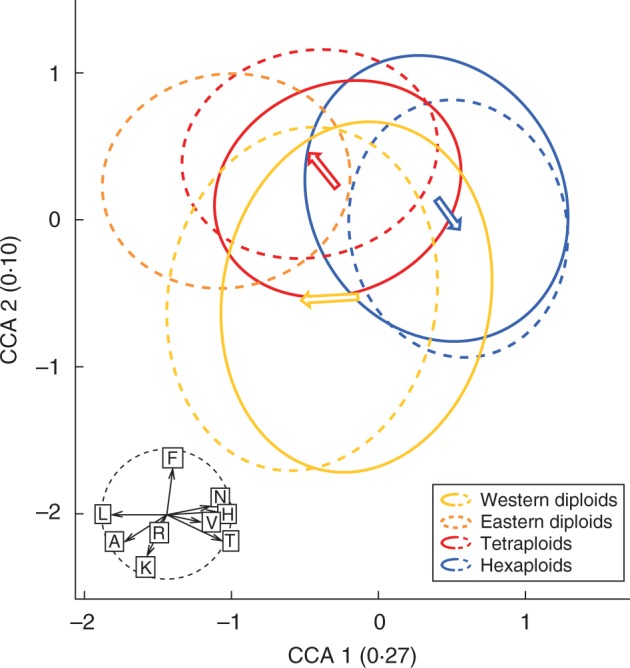
Niche displacement between homoploid (solid outline) and heteroploid sites (dashed outline) of western diploids (nhomoploid = 337, nheteroploid = 275), eastern diploids (nheteroploid = 270), tetraploids (nhomoploid = 294, nheteroploid =205) and hexaploids (nhomoploid = 856, nheteroploid = 554) of Senecio carniolicus derived from canonical correspondence analysis. Homoploid sites of eastern diploids were not included due to their low number. Differentiation of niche optima was defined as the Euclidean distance among centroids of ellipses weighted by axis inertia. The correlation circle (r = 1) shows the environmental variables used as constraints in the ordination, i.e. the Landolt indicator values for temperature (T), continentality (K), light (L), soil moisture (F), soil reaction (R), soil nutrient (N), humus content (H), soil aeration (D) and soil moisture variability (W) averaged for all vascular plant species surrounding S. carniolicus individuals. Axes inertias (i.e. eigenvalues of constrained axes) are given in parentheses. Arrows indicate the direction of shifts in niche optima from homoploid to heteroploid sites.
Table 2.
Comparison of niche optima (niche shift) and ecological tolerance (niche breadth) between homoploid and heteroploid sites for groups of Senecio carniolicus
| Shift* | Breadth | |
|---|---|---|
| Western diploids: | ||
| with tetraploids | 0·105 | 0·053 |
| with hexaploids | <0·001 | 0·581 |
| Tetraploids: | ||
| with western diploids | 0·493 | 0·594 |
| with eastern diploids | 0·001 | 0·765 |
| with hexaploids | <0·001 | 0·750 |
| Hexaploids: | ||
| with western diploids | 0·001 | 0·471 |
| with eastern diploids | 0·042 | 0·447 |
| with tetraploids | <0·001 | 0·601 |
* P values of significant differences after Dunn–Šidák correction for multiple comparisons are given in bold.
Significant differences in the direction of niche displacement were detected for single indicator values (Supplementary Information Fig. S4). In the presence of western diploid or tetraploid cytotypes, hexaploids shifted towards sites with higher temperature (T), less light (L) and higher humus content (H), whereas western diploids and tetraploids shifted towards lower temperature (T), more intensive light (L) and lower soil humus content (H). In heteroploid sites with tetraploids, western diploids shifted towards sites with lower humus content (H) but higher aeration (A).
DISCUSSION
Ecological differentiation among cytotypes
Niche differentiation is considered an important prezygotic isolation mechanism fostering polyploid establishment (Levin, 1983; Fowler and Levin, 1984; Thompson and Lumaret, 1992; Levin, 2003). Although climatic niches are often shared on a broad scale (Glennon et al., 2014; but see Thompson et al., 2014), ecological differentiation is amply documented on various scales (e.g. Johnson et al., 2003; Duchoslav et al., 2010; McIntyre, 2012). Using Landolt indicator values (Landolt et al., 2010) as a surrogate for fine-scale environmental parameters, we show that the four groups – corresponding to three cytotypes – constituting S. carniolicus s.l. are indeed differentiated along a complex ecological gradient (Fig. 2, Supplementary Information Figs S1, S2; Table 1). Eastern diploids are found in open, wind-exposed, cold habitats with well-drained substrates, whereas hexaploids occupy habitats with deeper, more humus- and nutrient-rich soils and higher temperatures; habitats of tetraploids show intermediate ecological characteristics. Western diploids occur in drier conditions than these three groups (Supplementary Information Fig. S1).
The ecological differentiation of eastern diploids and their polyploid derivatives exhibits a clear directionality, reflecting the order of polyploidization events, i.e. tetraploids are autopolyploid derivatives of eastern diploids and hexaploids originated from tetraploids (western diploids were not involved in the ancestry of polyploids; Escobar García et al., 2012; Sonnleitner et al., 2013). Directional niche shift is probably the result of directional phenotypic change (e.g. taller growth of polyploids: Bennett, 1971), even though the relationship between phenotype and genome size (which correlates with ploidy level in S. carniolicus: Sonnleitner et al., 2010) is not always monotonic (Balao et al., 2011). The phenotypic traits underlying the directional ecological differentiation of polyploid S. carniolicus are unknown, but probably include stouter habit (Flatscher et al., 2015) conferring higher competitive ability (Levin, 2002; Parisod et al., 2010) in habitats with denser vegetation cover (Sonnleitner et al., 2010). In addition, polyploid S. carniolicus may have higher nutritional demands, which would be more easily met in habitats with deeper and more nutrient-rich soil (Supplementary Information Fig. S1). In polyploid S. carniolicus these same traits may be (co-)responsible for the lack of increased niche breadth, presumed to be common in polyploids (Parisod et al., 2010).
Displacement, but no narrowing of niches in sympatry
Although character displacement is recognized as an important mechanism for enabling closely related species to coexist (Dayan and Simberloff, 2005; Pfennig and Pfennig, 2009; Beans, 2014), it has rarely been invoked in the context of cytotype co-occurrence (Van Dijk et al., 1992; Van Dijk and Bijlsma, 1994). We show that the niches of cytotypes in a heteroploid species are affected by the presence/absence of other cytotypes, i.e. that niche displacement (Peers et al., 2013) occurs. Specifically, niche optima between homoploid and heteroploid sites were shifted roughly along the same environmental gradients that determine the cytotypes’ niches, leading to stronger ecological differentiation of co-occurring cytotypes (Fig. 3, Supplementary Information Fig. S3). Previously, patterns consistent with niche displacement have been found in the grass Anthoxanthum alpinum in a narrow contact zone of ecologically non-differentiated diploids and autotetraploids (Felber-Girard et al., 1996) as well as in contact zones of the diploid and autotetraploid orchid Dactylorrhiza maculata s.l. with evidence for wider ecological amplitudes in homoploid compared with heteroploid populations (Ståhlberg and Hedrén, 2009). However, both studies cover only one or two local contact zones.
Patterns consistent with niche displacement can also result from variation in resource availability between homo- and heteroploid populations, ecological sorting or chance (Grant, 1975; Arthur, 1982). In S. carniolicus, homoploid and heteroploid sites are found over large parts of the distribution range and are not geographically separated (Fig. 1), rendering variation in resource availability or processes of ecological sorting as the sole causes for the observed divergence unlikely. Using a randomization procedure (i.e. comparing the actual shift against a null distribution), we confirmed that niche shifts are stronger than expected by chance.
The occurrence of niche displacement in S. carniolicus depended on the cytotypes involved, but always was symmetrical, making competitive superiority of either partner (Cooley, 2007) as the underlying cause unlikely. Hexaploids showed significant displacement in sympatry with western diploids and tetraploids, but not with eastern diploids (Table 2; this may be responsible for the lack of statistical significance in the CCA) despite their frequent co-occurrence (Schönswetter et al., 2007; Hülber et al., 2009; Sonnleitner et al., 2010). As hexaploids and eastern diploids are reproductively isolated via strongly reduced seed set and seed viability (Sonnleitner et al., 2013) and showed well-differentiated niches (Fig. 2, Table 1), selective pressure for stronger niche differentiation may be low. Tetraploids showed significant displacement with hexaploids and eastern diploids, but not with western diploids (Table 2), despite having similar ecological requirements (Fig. 2), which might be due to their rare co-occurrence (Sonnleitner et al., 2010).
In contrast to previously studied plants (Felber-Girard et al., 1996; Ståhlberg and Hedrén, 2009), niche displacement in S. carniolicus includes shifts in niche optima but no significant change in niche breadth (Table 2). Niche breadths may remain constant if resource use expands into or increases in portions of the fundamental niche that are not or are only rarely realized in homoploid populations (e.g. Peers et al., 2013). Due to our sampling design (on average 29 S. carniolicus individuals per location) it cannot be ruled out that rarely occupied habitats have remained under-sampled and that our estimates of niche breadth are therefore conservative.
A consequence of niche displacement is that upon secondary contact polyploids and their lower-ploid ancestors can form stable contact zones more easily than expected from the cytotypes’ ecological niches, especially if cytotypes still compete for resources or are prone to fitness loss due to the formation of inferior hybrids in heteroploid crosses, as probably is the case for S. carniolicus, where pollination barriers are lacking and polyploid cytotypes are interfertile (Sonnleitner et al., 2013). Hence, niche displacement may be (co-)responsible for secondary contact zones being common (Petit et al., 1999). An alternative yet not mutually exclusive consequence of niche displacement can be that overall ecological niches of polyploids and their lower-ploid ancestors converge after cytotypes have attained largely or completely non-overlapping distributions. This may contribute to the observed similarity of climatic niches of diploid plants and their polyploid derivatives (Glennon et al., 2014), or the perceived niche expansion of invasive polyploids in their non-native ranges (te Beest et al., 2011). Testing any of these hypotheses is currently not possible because studies on ecological differentiation among cytotypes usually do not discriminate between homoploid and heteroploid populations. As is evident from the present study, taking niche displacement into account will be essential for understanding ecological consequences of polyploidy, a major force generating biodiversity.
SUPPLEMENTARY INFORMATION
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Table S1: an overview of the sample sites and DNA ploidy level of sampled individuals. Figures S1–S4: detailed information concerning niche differentiation of the four groups of S. carniolicus and niche displacement within groups between homoploid and heteroploid sites.
ACKNOWLEDGEMENTS
We thank Daniela Stawik, Christian Gilli, Božo Frajman and Manfred Schmucker for help with fieldwork. This work was supported by the Austrian Science Fund (P20736-B16 to P.S. followed by G.M.S.).
LITERATURE CITED
- Arthur W. 1982. The evolutionary consequences of interspecific competition. Advances in Ecological Research 12: 127–187. [Google Scholar]
- Baack EJ. 2005. To succeed globally, disperse locally: effects of local pollen and seed dispersal on tetraploid establishment. Heredity 94: 538–546. [DOI] [PubMed] [Google Scholar]
- Balao F, Herrera J, Talavera S. 2011. Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: a multivariate morphological approach. New Phytologist 192: 256–265. [DOI] [PubMed] [Google Scholar]
- Beans CM. 2014. The case for character displacement in plants. Ecology and Evolution 4: 862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD. 1971. The duration of meiosis. Proceedings of the Royal Society of London, Series B: Biological Sciences 178: 277–299. [DOI] [PubMed] [Google Scholar]
- Bolker BM, Brooks ME, Clark CJ, et al. 2009. Generalized linear mixed models: a practical guide for ecology and evolution. Trends in Ecology & Evolution 24: 127–135. [DOI] [PubMed] [Google Scholar]
- Cooley JR. 2007. Decoding asymmetries in reproductive character displacement. Proceedings of the Academy of Natural Sciences of Philadelphia 156: 89–96. [Google Scholar]
- Dayan T, Simberloff D. 2005. Ecological and community-wide character displacement: the next generation. Ecology Letters 8: 875–894. [Google Scholar]
- Diekmann M. 2003. Species indicator values as an important tool in applied plant ecology – a review. Basic and Applied Ecology 4: 493–506. [Google Scholar]
- Dray S, Dufour AB. 2007. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software 22: 1–20. [Google Scholar]
- Duchoslav M, Šafářová L, Krahulec F. 2010. Complex distribution patterns, ecology and coexistence of ploidy levels of Allium oleraceum (Alliaceae) in the Czech Republic. Annals of Botany 105: 719–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dullinger S, Hülber K. 2011. Experimental evaluation of seed limitation in Alpine snowbed plants. PLoS ONE 6: e21537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar García P, Winkler M, Flatscher R, et al. 2012. Extensive range persistence in peripheral and interior refugia characterizes Pleistocene range dynamics in a widespread Alpine plant species (Senecio carniolicus, Asteraceae). Molecular Ecology 21: 1255–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber-Girard M, Felber F, Buttler A. 1996. Habitat differentiation in a narrow hybrid zone between diploid and tetraploid Anthoxanthum alpinum. New Phytologist 133: 531–540. [Google Scholar]
- Flatscher R, Escobar García P, Hülber K, et al. 2015. Underestimated diversity in one of the world’s best studied mountain ranges: the polyploid complex of Senecio carniolicus (Asteraceae) contains four species in the European Alps. Phytotaxa 213: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler NL, Levin DA. 1984. Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. American Naturalist 124: 703–711. [Google Scholar]
- Glennon KL, Ritchie ME, Segraves KA. 2014. Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecology Letters 17: 574–582. [DOI] [PubMed] [Google Scholar]
- Grace JB, Wetzel RG. 1981. Habitat partitioning and competitive displacement in cattails (Typha): experimental field studies. The American Naturalist 118: 463–474. [Google Scholar]
- Grant PR. 1975. The classical case of character displacement. Evolutionary Biology 8: 875–894. [Google Scholar]
- Hülber K, Sonnleitner M, Flatscher R, et al. 2009. Ecological segregation drives fine-scale cytotype distribution of Senecio carniolicus in the Eastern Alps. Preslia 81: 309–319. [PMC free article] [PubMed] [Google Scholar]
- Hülber K, Sonnleitner M, Suda J, et al. 2015. Ecological differentiation, lack of hybrids involving diploids, and asymmetric gene flow between polyploids in narrow contact zones of Senecio carniolicus (syn. Jacobaea carniolica, Asteraceae). Ecology and Evolution 5: 1224–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MTJ, Husband BC, Burton TL. 2003. Habitat differentiation between diploid and tetraploid Galax urceolata (Diapensiaceae). International Journal of Plant Sciences 164: 703–710. [Google Scholar]
- Laird NM, Ware JH. 1982. Random-effects models for longitudinal data. Biometrics 38: 963–974. [PubMed] [Google Scholar]
- Landolt E. 1977. Ökologische Zeigerwerte zur Schweizer Flora. Veröffentlichungen des Geobotanischen Institutes der ETH, Stiftung Rübel 64: 1–208. [Google Scholar]
- Landolt E, Bäumler B, Erhardt A, et al. 2010. Flora Indicativa: ecological indicator values and biological attributes of the Flora of Switzerland and the Alps . Bern: Haupt. [Google Scholar]
- Levin DA. 1983. Polyploidy and novelty in flowering plants. American Naturalist 122: 1–25. [Google Scholar]
- Levin DA. 2002. The role of chromosomal change in plant evolution . New York: Oxford University Press. [Google Scholar]
- Levin DA. 2003. The ecological transition in speciation. New Phytologist 161: 91–96. [Google Scholar]
- Lumaret R, Guillerm JL, Delay J, Loutfi AAL, Izco J, Jay M. 1987. Polyploidy and habitat differentiation in Dactylis glomerata L. from Galicia (Spain). Oecologia 73: 436–446. [DOI] [PubMed] [Google Scholar]
- McIntyre PJ. 2012. Polyploidy associated with altered and broader ecological niches in the Claytonia perfoliata (Portulacaceae) species complex. American Journal of Botany 99: 655–662. [DOI] [PubMed] [Google Scholar]
- Oswald BP, Nuismer SL. 2011. A unified model of autopolyploid establishment and evolution. The American Naturalist 178: 687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto SP, Whitton J. 2000. Polyploid incidence and evolution. Annual Review of Genetics 34: 401–437. [DOI] [PubMed] [Google Scholar]
- Palmer MW. 1993. Putting things in even better order: the advantages of Canonical Correspondence Analysis. Ecology 74: 2215–2230. [Google Scholar]
- Parisod C, Holderegger R, Brochmann C. 2010. Evolutionary consequences of autopolyploidy. New Phytologist 186: 5–17. [DOI] [PubMed] [Google Scholar]
- Peers MJL, Thornton DH, Murray DL. 2013. Evidence for large-scale effects of competition: niche displacement in Canada lynx and bobcat. Proceedings of the Royal Society of London, Series B: Biological Sciences 280: 20132495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Bretagnolle F, Felber F. 1999. Evolutionary consequences of diploid–polyploid hybrid zones in wild species. Trends in Ecology & Evolution 14: 306–311. [DOI] [PubMed] [Google Scholar]
- Pfennig KS, Pfennig DW. 2009. Character displacement: ecological and reproductive responses to a common evolutionary problem. Quarterly Review of Biology 84: 253–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Ramsey TS. 2014. Ecological studies of polyploidy in the 100 years following its discovery. Philosophical Transactions of the Royal Society B 369: 20130352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönswetter P, Lachmayer M, Lettner C, et al. 2007. Sympatric diploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps are separated along an altitudinal gradient. Journal of Plant Research 120: 721–725. [DOI] [PubMed] [Google Scholar]
- Sonnleitner M, Flatscher R, Escobar García P, et al. 2010. Distribution and habitat segregation on different spatial scales among diploid, tetraploid and hexaploid cytotypes of Senecio carniolicus (Asteraceae) in the Eastern Alps. Annals of Botany 106: 967–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnleitner M, Weis B, Flatscher R, et al. 2013. Parental ploidy strongly affects offspring fitness in heteroploid crosses among three cytotypes of autopolyploid Jacobaea carniolica (Asteraceae). PLoS ONE 8: e78959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg D, Hedrén M. 2009. Habitat differentiation, hybridization and gene flow patterns in mixed populations of diploid and autotetraploid Dactylorhiza maculata s.l. (Orchidaceae). Evolutionary Ecology 23: 295–328. [Google Scholar]
- Suda J, Weiss-Schneeweiss H, Tribsch A, Schneeweiss GM, Trávníček P, Schönswetter P. 2007. Complex distribution patterns of di-, tetra-, and hexaploid cytotypes in the European high mountain plant Senecio carniolicus (Asteraceae). American Journal of Botany 94: 1391–1401. [DOI] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, et al. 2011. The more the better? The role of polyploidy in facilitating plant invasions. Annals of Botany 109: 19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak CJF. 1986. Canonical Correspondence Analysis: a new eigenvector technique for multivariate direct gradient analysis. Ecology 67: 1167–1179. [Google Scholar]
- Thompson JD, Lumaret R. 1992. The evolutionary dynamics of polyploid plants – origins, establishment and persistence. Trends in Ecology & Evolution 7: 302–307. [DOI] [PubMed] [Google Scholar]
- Thompson KA, Husband BC, Maherali H. 2014. Climatic niche differences between diploid and tetraploid cytotypes of Chamerion angustifolium (Onagraceae). American Journal of Botany 101: 1868–1875. [DOI] [PubMed] [Google Scholar]
- Treier UA, Broennimann O, Normand S, et al. 2009. Shift in cytotype frequency and niche space in the invasive plant Centaurea maculosa. Ecology 90: 1366–1377. [DOI] [PubMed] [Google Scholar]
- Van Dijk P, Bijlsma R. 1994. Simulations of flowering time displacement between two cytotypes that form inviable hybrids. Heredity 72: 522–535. [Google Scholar]
- Van Dijk P, Hartog M, Van Delden W. 1992. Single cytotype areas in autopolyploid Plantago media L. Biological Journal of the Linnean Society 46: 315–331. [Google Scholar]
- Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM. 2013. Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenetic and Genome Research 140: 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TE, Takebayashi N, Barker MS, Mayrose I, Greenspoon PB, Rieseberg LH. 2009. The frequency of polyploid speciation in vascular plants. Proceedings of the National Academy of Sciences, USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.