Abstract
Background
Large-scale genome-wide association studies have identified multiple genetic variants that are associated with elevated body mass index (BMI) or the risk of obesity in Caucasian or Asian populations. We examined whether these variants are individually associated with obesity in Chinese children, and also assessed their cumulative effects and predictive value for obesity risk in Chinese children.
Methods
We genotyped 40 single nucleotide polymorphisms (SNPs) and conducted association analyses for 32/40 SNPs with an estimated minor allele frequency >1 % in 2 030 unrelated Chinese children, including 607 normal-weight, 718 overweight, and 705 obese individuals from two cross-sectional study groups. Logistic regression and linear regression under the additive model were used to examine associations, and the area under the receiver operating characteristic curve (AUCROC) was reported as prediction summary.
Results
We identified obesity association for 6 SNPs near SEC16B, RBJ, CDKAL1, TFAP2B, MAP2K5 and FTO (odds ratios (ORs) ranged from 1.19 to 1.41, nominal two-sided P-values < 0.05). Association (Bonferroni corrected) of rs543874 near SEC16B and rs2241423 near MAP2K5 had presumably stronger effects on obesity in Chinese children than in Caucasian populations. Their risk alleles were also associated with BMI standard deviation score (BMI-SDS) variability. We demonstrated the cumulative effects of the 32 SNPs on obesity risk (per risk allele: OR = 1.06, 95 % CI: 1.03-1.11, P = 4.84 × 10-4) and BMI-SDS (β = 0.04, 95 % CI: 0.02-0.06, P = 3.69 × 10-7). The difference in AUCROC for a model with covariates (age, age square, sex and study group) and the model including covariates and all 32 SNPs was 2.8 % (P = 0.0002).
Conclusion
While six SNPs were individually associated with obesity in Chinese children, the 32 common variants identified by recent GWA studies had cumulative effects and resulted in a limited increase in the AUCROC predictive value for childhood obesity.
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-016-0268-4) contains supplementary material, which is available to authorized users.
Keywords: Obesity, BMI, Gene, Variant, Children
Background
The rapid increase of obesity prevalence has been a major public health challenge in both developed and developing countries. Obesity is a major risk factor for many common chronic diseases such as type 2 diabetes mellitus, cardiovascular disease, and many forms of cancer [1]. Although the reason for the increase in obesity prevalence has been largely attributed to environmental factors, including changes in dietary patterns and lifestyle, genetic factors play an important role in obesity susceptibility [2]. The heritability of the variance of body mass index (BMI) ranged from 40 % to 70 % [3].
In 2010, a meta-analysis of genome-wide association (GWA) studies for BMI was conducted in 249,796 adult individuals of European ancestry by the Genetic Investigation of Anthropometric Traits (GIANT) consortium. They confirmed 14 known obesity susceptibility loci and identified 18 new loci associated with BMI at a genome-wide significance level (P < 5 × 10-8) [4].
Heritability estimations for BMI or obesity are higher in children, compared with adults [5]. Although the currently known major common variants related to obesity overlap to a substantial degree between children and adults, a GWA study in French and German populations identified 2 new loci for childhood obesity including rs121458332 in SDCCAG8 and rs13278851 near TNKS/MSRA. The latter locus had an effect in children and adolescents only [6]. In 2012, another meta-analysis of GWA studies identified 2 new loci for childhood obesity in populations of European ancestry (rs9568856 of OLFM4, rs9299 of HOXB5) [5].
Asians account for 60 % of the world’s population and have higher percentages of body fat and increased metabolic disease risk than individuals of European ancestry with the same BMI. Thus, a genetic study in an Asian population can not only facilitate the dissection of genetic architecture of obesity, but also identify genetic variants of particular importance in Asians [7]. Recently, two GWA studies in East Asian populations reported 4 new loci (rs2206734 of CDKAL1, rs11142387 of KLF9, rs261967 of PCSK1, rs12597579 of GP2) associated with BMI [7, 8].
Many replication studies for the 32 loci identified by GIANT have been performed in multiple ethnic populations, including Asians [9-14]. Studies on the SNPs near SDCCAG8 and TNKS/MSRA have led to mixed results [9,12,13]. Similarly, the 2 loci for childhood obesity (OLFM4 and HOXB5) and the 4 loci found in East Asians (CDKAL1, KLF9, PCSK1 and GP2) were not among the top hits of a large scale GWA studies meta-analysis which focused on the tails of the adult BMI distribution [13]. Recently, 28 SNPs from the 32 loci reported by GIANT and 4 additional loci identified in East Asians were studied in Chinese adults [15], but only 4 SNPs near TMEM18, PCSK1, BDNF and MAP2K5 were confirmed (nominal P-values < 0.05). The effects of these SNPs in Chinese children were unclear.
In the present study, we genotyped 40 single nucleotide polymorphism (SNPs) and conducted association analyses of the 32 variants that had an estimated minor allele frequency >1 % in 2 030 unrelated Chinese children, including 607 normal-weight, 718 overweight, and 705 obese individuals. The purpose of this case-control study was to (a) examine whether the common variants are individually associated with obesity in Chinese children and (b) assess the cumulative effects and predictive value for obesity in Chinese children.
Methods
Subjects
We conducted an association study in two independent study groups, recruited from the urban regions of Beijing, China. The first study group, including 386 obese, 400 overweight and 151 normal-weight individuals, came from the study on Adolescent Lipids, Insulin Resistance and Candidate Genes (ALIR) in nine middle schools of Dongcheng District of Beijing. The second study group, including 319 obese, 318 overweight and 456 normal-weight individuals, was from the Comprehensive Prevention Project for Overweight and Obese Adolescents (CPOOA) with physical exercise and healthy nutrition as instruments in five elementary and middle schools of the Haidian District of Beijing. The ascertainment strategies for the two study groups have been described in detail previously [16,17]. The two studies were approved by the ethics committee of Peking University Health Science Center. Written informed consent was provided by all participants and, in the case of minors, their parents.
Anthropometric measurements, including height and weight, were determined according to standard protocols. BMI was calculated as weight in kilograms divided by the squared height in meters. We used the BMI percentile criteria to define obesity, overweight and normal-weight in children and adolescents, which were determined in a representative Chinese population [18]. According to the criteria (Table 1), the children and adolescents with an age- and gender-specific BMI ≥ 95th percentile were defined as obese, while those with a BMI between 85th and 95th percentile were overweight and those with a BMI between 15th and 85th percentiles were normal-weight. Individuals with cardiovascular or metabolic diseases were excluded. The sex- and age-specific BMI standard deviation score (BMI-SDS) was calculated by using the growth reference data of the World Health Organization for children and adolescents aged 5–19 years [19].
Table 1.
Body mass index reference for screening overweight and obesity in Chinese school-age children [18]
| Age (years) | Boys | Girls | ||
|---|---|---|---|---|
| Overweight | Obesity | Overweight | Obesity | |
| 7- | 17.4 | 19.2 | 17.2 | 18.9 |
| 8- | 18.1 | 20.3 | 18.1 | 19.9 |
| 9- | 18.9 | 21.4 | 19.0 | 21.0 |
| 10- | 19.6 | 22.5 | 20.0 | 22.1 |
| 11- | 20.3 | 23.6 | 21.1 | 23.3 |
| 12- | 21.0 | 24.7 | 21.9 | 24.5 |
| 13- | 21.9 | 25.7 | 22.6 | 25.6 |
| 14- | 22.6 | 26.4 | 23.0 | 26.3 |
| 15- | 23.1 | 26.9 | 23.4 | 26.9 |
| 16- | 23.5 | 27.4 | 23.7 | 27.4 |
| 17- | 23.8 | 27.8 | 23.8 | 27.7 |
| 18 | 24.0 | 28.0 | 24.0 | 28.0 |
The general characteristics of the study samples are shown in Table 2. The study consisted of 2 030 Chinese children, including 607 normal-weight (mean age 12.55 ± 3.04 years, mean BMI 18.77 ± 2.50 kg/m2), 718 overweight (mean age 13.26 ± 2.36 years, mean BMI 23.86 ± 2.20 kg/m2) and 705 obese (mean age 12.85 ± 2.59 years, mean BMI 28.12 ± 3.94 kg/m2).
Table 2.
General characteristics of the study samples
| Normal-weight group | Overweight group | Obese group | |
|---|---|---|---|
| Number | 607 | 718 | 705 |
| Female (%) | 323(53.2) | 268(37.3) | 221(31.3) |
| Age | 12.55 ± 3.04 | 13.26 ± 2.36 | 12.85 ± 2.59 |
| Height(cm) | 156.20 ± 15.86 | 161.96 ± 12.97 | 162.74 ± 13.81 |
| Weight(kg) | 47.09 ± 13.52 | 63.65 ± 13.61 | 76.11 ± 19.96 |
| BMI (kg/m2) | 18.77 ± 2.50 | 23.86 ± 2.20 | 28.12 ± 3.94 |
| BMI-SDS | 0.003 ± 0.71 | 1.57 ± 0.27 | 2.49 ± 0.48 |
Data are expressed as mean ± standard deviation, if not indicated otherwise. BMI body mass index, SDS Standard deviation score
Selection of SNPs and genotyping
We selected 40 obesity-related loci identified by five recent GWA studies, with one representative SNP for each locus. Firstly, we selected the 32 SNPs reported by Speliotes et al [4]. Then we selected 4 SNPs (rs121458332 of SDCCAG8, rs13278851 of TNKS/MSRA, rs9568856 of OLFM4, rs9299 of HOXB5), which were identified by two GWA studies that focused on children [5,6]. Additionally, we selected 4 SNPs (rs2206734 of CDKAL1, rs11142387 of KLF9, rs261967 of PCSK1, rs12597579 of GP2), which were associated with BMI in two GWA studies of East Asian populations [7, 8].
Fasting venous blood samples were collected. Genomic DNA was extracted from blood leukocytes by the phenol/chloroform extraction method. Sequenom’s MassARRAY system (Sequenom, San Diego, CA, USA) was applied to genotype the 40 SNPs. Primers, including a pair of amplification primers and an extension primer for each SNP, were designed with SpectroDESIGNER software (Sequenom, San Diego, CA). A multiplex polymerase chain reaction was performed, and unincorporated double stranded nucleotide triphosphate bases were dephosphorylated with shrimp alkaline phosphatase followed by primer extension. The purified primer extension reaction was spotted onto a 384-element silicon chip (SpectroCHIP, Sequenom) and analyzed in the Matrix assisted laser desorption ionization time of flight mass Spectrometry (MALDI-TOF MS, Sequenom). The resulting spectra were processed with MassArray Typer (Sequenom, San Diego, CA).
Multiplex SNP assays designs failed for 6 out of 40 SNPs (rs10968576, rs4771122, rs10150332, rs12444979, rs29941 and rs261967); these were replaced by 6 proxy SNPs (rs16912921, rs9579083, rs10145154, rs6497416, rs29942, rs261966) with strong linkage disequilibrium (r2 > 0.80) in populations comparable to the discovery studies in HapMap data Release 24/phase II Nov08 (http://hapmap.ncbi.nlm.nih.gov; Han Chinese in Beijing, China (CHB) for rs261966, Utah residents with Northern and Western European ancestry (CEU) for the other 5 SNPs).
As shown in Additional file 1: Table S1, the call rates for 40 SNPs were above 95.0 %. We exclude one monomorphic (rs6497416 of GPRC5B), one triallelic (rs4836133 of ZNF608) and six rare variants with minor allele frequency below 1 % (in all genotyped individuals) from the subsequent analyses resulting in 32 SNPs. In the normal-weight group, 31 of the 32 SNPs showed no evidence for deviations from Hardy-Weinberg equilibrium (HWE; all P > 0.05). For one SNP (rs7138803) near FAIM2 we observed some evidence for a deviation from HWE (P = 0.01) but a double-checking of the genotype data revealed no obvious genotyping artifacts.
Statistical analyses
The genotype data of the normal-weight group were tested for deviations from Hardy–Weinberg equilibrium using χ2 tests (see above). F-statistics (FST), a metric representing the effect of population subdivision, was calculated according to the following formula, FST = (P1-P2)2/ ((P1 + P2)*(2-(P1 + P2))), where P1 is the allele frequency estimate in the population of the discovery study and P2 is allele frequency estimate based on the total sample of our study [20,21]. A FST value ≥ 0.10 indicates large genetic differentiation [22].
Logistic regression was performed to examine the effect of each SNP on risk of obesity or overweight (categorical variable). Linear regression was performed to examine the effect of each SNP allele on BMI-SDS variability. Both logistic regression and linear regression were carried out under a (log)-additive genetic model with adjustment for age, age square, sex and study group (ALIR and CPOOA). For each of the 6 proxy SNPs, the allele which was correlated with the effect allele of the original SNP in the discovery study was defined as the effect allele, while the effect alleles of other SNPs were the same as the discovery studies, for comparing our results with the published data [4-8].
To identify cumulative effects of these SNPs, we created a genetic risk score (GRS) for each individual by summing up the number of effect alleles of the SNPs. We did not weight the risk alleles on the basis of their individual effect sizes because no well-accepted effect sizes were available for each of the SNPs, and it has been shown that weighting of risk alleles may have only limited effects [23]. Again logistic regression was used to calculate odds ratio (OR) of the GRS-32 from all 32 SNPs that met our minor-allele frequency cut-off (see above) for the risk of obesity or overweight. Linear regression was performed to examine the effect of GRS-32 on BMI-SDS variability.
SPSS 18.0 software was used for the above statistical analyses (SPSS, Chicago, IL). In addition to effect sizes estimates (i.e. per allele odds ratios (OR) and 95 % confidence intervals (95 % CI)), we reported nominal two-sided P-values. We applied a nominal significance level of α = 0.05 (two-sided). Adjustment was made for multiple testing using Bonferroni correction for 32 SNPs, i.e. resulting in αBF = 0.05/32 = 0.00156 (two-sided). Difference in effect size of each SNP between our study and the discovery study was examined by testing heterogeneity with MANTRA software, which was developed by Morris AP [24] for trans-ethnic meta-analysis of genome-wide association studies. P(heterogeneity) is the posterior probability of heterogeneity in allelic effects, which is derived from transethnic meta-analysis. If P(heterogeneity) > 50 %, there is the evidence of heterogeneity in allelic effects between the present and discovery studies [24]. The receiver operating characteristic (ROC) curves comparing normal-weight and obese children were produced by logistic regression, and the areas under the curve (AUCROC) from different models were compared by MedCalc software. Based on the published minor allele frequencies (see Table 3) and applying a (log)-additive genetic model, a sample size of 705 obese cases and 607 normal-weight controls has a comparisons-wise power ranging between 0.94-0.99 for a true allelic OR of 1.5 or 0.33-0.64 for a true allelic OR of 1.2 (α = 0.05; two-sided). Accounting for multiplicity these numbers changed to 0.63-0.98 or 0.05-0.20, respectively (αBF = 0.00156; two-sided). Similarly, analyzing BMI-SDS in a sample of 2 030 children, leads to a comparisons-wise power ranging between 0.51-0.89 for a true allelic β of 0.10 (in units of BMI-SDS) or 0.17-0.36 for a true allelic β of 0.05 (α = 0.05; two-sided). Accounting for multiplicity these numbers changed to 0.12-0.51 or 0.02-0.06, respectively (αBF = 0.00156; two-sided). These power calculations were performed using Quanto software (University of Southern California, Los Angeles, CA).
Table 3.
Hardy-Weinberg equilibrium (HWE) test and effect allele frequency (EAF) in Chinese children for32 GWA studies -derived SNPs
| SNP | Nearest gene | Chr | Position | Allele | HWE P-valuea | EAFb | EAFc | F ST | |
|---|---|---|---|---|---|---|---|---|---|
| Effect | Other | ||||||||
| Speliotes et al. 2010 (in children and adults of European ancestry) [4] | |||||||||
| rs1558902 | FTO | 16 | 52361075 | A | T | 0.58 | 0.13 | 0.42 | 0.105e |
| rs2867125 | TMEM18 | 2 | 612827 | C | T | 0.20 | 0.91 | 0.83 | 0.014 |
| rs571312 | MC4R | 18 | 55990749 | A | C | 0.67 | 0.23 | 0.24 | 0.000 |
| rs10938397 | GNPDA2 | 4 | 44877284 | G | A | 0.54 | 0.31 | 0.43 | 0.015 |
| rs10767664 | BDNF | 11 | 27682562 | A | T | 0.30 | 0.55 | 0.78 | 0.059 |
| rs2815752 | NEGR1 | 1 | 72585028 | A | G | 0.97 | 0.92 | 0.61 | 0.134e |
| rs7359397 | SH2B1 | 16 | 28793160 | T | C | 0.17 | 0.16 | 0.40 | 0.071 |
| rs9816226 | ETV5 | 3 | 187317193 | T | A | 0.36 | 0.97 | 0.82 | 0.060 |
| rs3817334 | MTCH2 | 11 | 47607569 | T | C | 0.16 | 0.33 | 0.41 | 0.007 |
| rs29942d | KCTD15 | 19 | 39001117 | C | T | 0.76 | 0.25 | 0.31 | 0.004 |
| rs543874 | SEC16B | 1 | 176156103 | G | A | 0.80 | 0.22 | 0.19 | 0.001 |
| rs987237 | TFAP2B | 6 | 50911009 | G | A | 0.52 | 0.18 | 0.18 | 0.000 |
| rs7138803 | FAIM2 | 12 | 48533735 | A | G | 0.01 | 0.28 | 0.38 | 0.011 |
| rs713586 | RBJ | 2 | 25011512 | C | T | 0.77 | 0.49 | 0.47 | 0.000 |
| rs2241423 | MAP2K5 | 15 | 65873892 | G | A | 0.67 | 0.41 | 0.78 | 0.142e |
| rs2287019 | QPCTL | 19 | 50894012 | C | T | 0.09 | 0.82 | 0.80 | 0.001 |
| rs1514175 | TNNI3K | 1 | 74764232 | A | G | 0.91 | 0.78 | 0.43 | 0.128e |
| rs2112347 | FLJ35779 | 5 | 75050998 | T | G | 0.88 | 0.44 | 0.63 | 0.036 |
| rs16912921d | LRRN6C | 9 | 28403461 | A | C | 0.19 | 0.30 | 0.34 | 0.002 |
| rs3810291 | TMEM160 | 19 | 52260843 | A | G | 0.58 | 0.29 | 0.67 | 0.145e |
| rs1555543 | PTBP2 | 1 | 96717385 | C | A | 0.81 | 0.87 | 0.59 | 0.099 |
| rs9579083d | MTIF3 | 13 | 26915270 | C | G | 0.87 | 0.15 | 0.23 | 0.010 |
| rs4929949 | RPL27A | 11 | 8561169 | C | T | 0.79 | 0.40 | 0.52 | 0.014 |
| rs206936 | NUDT3 | 6 | 34410847 | G | A | 0.30 | 0.51 | 0.21 | 0.098 |
| Scherag et al. 2010 (in children and adults of European ancestry) [6] | |||||||||
| rs12145833 | SDCCAG8 | 1 | 241550377 | T | G | 0.55 | 0.91 | 0.87 | 0.004 |
| rs13278851 | TNKS/MSRA | 8 | 9788282 | A | G | 0.28 | 0.14 | 0.11 | 0.002 |
| Bradfield et al. 2012 (in children of European ancestry) [5] | |||||||||
| rs9568856 | OLFM4 | 13 | 52962982 | A | G | 0.34 | 0.33 | 0.16 | 0.039 |
| rs9299 | HOXB5 | 17 | 44024429 | A | G | 0.44 | 0.53 | 0.65 | 0.015 |
| Okada et al. 2012 (in East Asians) [7] | |||||||||
| rs2206734 | CDKAL1 | 6 | 20802863 | G | A | 0.66 | 0.59 | 0.59 | 0.000 |
| rs11142387 | KLF9 | 9 | 72188152 | C | A | 0.96 | 0.33 | 0.46 | 0.018 |
| Wen et al. 2012 (in East Asians) [8] | |||||||||
| rs261966d | PCSK1 | 5 | 95875343 | G | A | 0.53 | 0.45 | 0.42 | 0.001 |
| rs12597579 | GP2 | 16 | 20165368 | C | T | 0.99 | 0.73 | 0.80 | 0.007 |
Chr Chromosome; Position: NCBI build 36.3 (NCBI, Bethesda, MD); EAF Effect allele frequency, HWE Hardy-Weinberg equilibrium
aHWE P-value in normal-weight Chinese children
bEffect allele frequency in all genotyped individuals of the present study
cEffect allele frequency of SNPs in the discovery studies (Ref. [4-8]), and that of each proxy SNP is from NCBI database among the same populations that in the discovery study
dProxy SNPs were used in the present study to replace the SNPs from the discovery studies
e F ST > 0.10 as indicator of large genetic differentiation between population in the present study and that in the discovery study
Results
Effect allele frequencies
The effect allele frequencies of 32 SNPs and FST values between the population in the present study and that in the discovery study are shown in Table 3. All effect allele frequencies in the present study were similar to those reported in the HapMap Han Chinese (http://hapmap.ncbi.nlm.nih.gov/). The FST values between the present study and the discovery study varied from 0 (rs987237 of TFAP2B, rs2206734 of CDKAL1) to 0.145 (rs3810291 of TMEM160). Based on the FST values, we found that 23/28 SNPs from three GWA studies of Europeans and 4/4 SNPs from two GWA studies of East Asians had similar effect allele frequencies in our study. Only 5 SNPs near FTO, MAP2K5, NEGR1, TNN13K, TMEM160 from the GIANT study on BMI variability showed large genetic differentiation between the Europeans and our study population (FST value ≥ 0.10).
Individual associations of 32 SNPs with obesity
Table 4 shows the results of the allelic association analyses of the 32 SNPs with obesity in Chinese children. We identified the nominally significant associations with obesity for effect alleles of 6 SNPs at FTO, SEC16B, TFAP2B, RBJ, MAP2K5 and CDKAL1 (ORs for the effect allele ranged between 1.19 and 1.41, nominal two-sided P < 0.05). SNP rs543874 near SEC16B and rs2241423 near MAP2K5 remained significant after Bonferroni correction for multiple testing (P < 0.00156, Bonferroni corrected for 32 SNPs). Fig. 1 shows ORs and 95 % CI for the association with obesity for the 32 SNPs in the present study and the published ORs for each SNP reported in the GWA studies. As shown in Fig. 1, overall 30 of the 32 SNPs yielded directionally consistent effects, i.e. the ORs of the SNPs in this study were comparable with those detected in the discovery studies. However, the effect sizes of two SNPs at SEC16B and MAP2K5 showed heterogeneity between the present and discovery studies (P(heterogeneity) > 50 %). The associations of obesity with SNP alleles at rs543874 of SEC16B (OR = 1.41, 95 % CI: 1.15-1.73) or at rs2241423 of MAP2K5 (OR = 1.34, 95 % CI: 1.12-1.59) seemed to be stronger in Chinese children than in Caucasians (OR = 1.10, 95 % CI: 1.06-1.14; OR = 1.07, 95 % CI: 1.04-1.10, respectively) [4], which was also indicated by non-overlapping 95 % confidence intervals in Fig. 1.
Table 4.
Association of 32 GWA studies-derived SNP alleles with obesity in Chinese children
| SNP | Nearest gene | Chr. | Allele | Obesity VS Normal-weight | |||
|---|---|---|---|---|---|---|---|
| Effect | other | OR | 95 % CI | Two-sided | |||
| P-value | |||||||
| Speliotes et al. 2010 (in children and adults of European ancestry) [4] | |||||||
| rs1558902 | FTO | 16 | A | T | 1.31 | 1.01-1.69 | 0.039 |
| rs2867125 | TMEM18 | 2 | C | T | 1.06 | 0.79-1.42 | 0.703 |
| rs571312 | MC4R | 18 | A | C | 1.16 | 0.94-1.42 | 0.166 |
| rs10938397 | GNPDA2 | 4 | G | A | 1.19 | 0.98-1.43 | 0.077 |
| rs10767664 | BDNF | 11 | A | T | 1.10 | 0.93-1.30 | 0.273 |
| rs2815752 | NEGR1 | 1 | A | G | 1.07 | 0.79-1.46 | 0.645 |
| rs7359397 | SH2B1 | 16 | T | C | 1.06 | 0.85-1.34 | 0.597 |
| rs9816226 | ETV5 | 3 | T | A | 0.90 | 0.56-1.45 | 0.669 |
| rs3817334 | MTCH2 | 11 | T | C | 1.05 | 0.88-1.25 | 0.626 |
| rs29942 | KCTD15 | 19 | C | T | 1.01 | 0.83-1.23 | 0.949 |
| rs543874 | SEC16B | 1 | G | A | 1.41 | 1.15-1.73 | 0.001 |
| rs987237 | TFAP2B | 6 | G | A | 1.27 | 1.02-1.59 | 0.032 |
| rs7138803 | FAIM2 | 12 | A | G | 1.06 | 0.87-1.30 | 0.542 |
| rs713586 | RBJ | 2 | C | T | 1.19 | 1.00-1.42 | 0.048 |
| rs2241423 | MAP2K5 | 15 | G | A | 1.34 | 1.12-1.59 | 0.001 |
| rs2287019 | QPCTL | 19 | C | T | 0.89 | 0.72-1.10 | 0.283 |
| rs1514175 | TNNI3K | 1 | A | G | 1.11 | 0.90-1.36 | 0.332 |
| rs2112347 | FLJ35779 | 5 | T | G | 1.02 | 0.85-1.21 | 0.865 |
| rs16912921 | LRRN6C | 9 | A | C | 0.95 | 0.79-1.15 | 0.628 |
| rs3810291 | TMEM160 | 19 | A | G | 0.93 | 0.77-1.12 | 0.456 |
| rs1555543 | PTBP2 | 1 | C | A | 0.84 | 0.65-1.08 | 0.174 |
| rs9579083 | MTIF3 | 13 | C | G | 1.08 | 0.85-1.37 | 0.519 |
| rs4929949 | RPL27A | 11 | C | T | 0.90 | 0.75-1.07 | 0.215 |
| rs206936 | NUDT3 | 6 | G | A | 0.99 | 0.83-1.18 | 0.919 |
| Scherag et al. 2010 (in children and adults of European ancestry)[6] | |||||||
| rs12145833 | SDCCAG8 | 1 | T | G | 0.96 | 0.71-1.31 | 0.799 |
| rs13278851 | TNKS/MSRA | 8 | A | G | 0.90 | 0.71-1.14 | 0.375 |
| Bradfield et al. 2012 (in children of European ancestry) [5] | |||||||
| rs9568856 | OLFM4 | 13 | A | G | 1.08 | 0.89-1.30 | 0.438 |
| rs9299 | HOXB5 | 17 | A | G | 1.07 | 0.90-1.27 | 0.449 |
| Okada et al. 2012 (in East Asians) [8] | |||||||
| rs2206734 | CDKAL1 | 6 | G | A | 1.21 | 1.01-1.44 | 0.035 |
| rs11142387 | KLF9 | 9 | C | A | 0.90 | 0.75-1.08 | 0.268 |
| Wen et al. 2012 (in East Asians) [7] | |||||||
| rs261966 | PCSK1 | 5 | G | A | 1.03 | 0.87-1.22 | 0.764 |
| rs12597579 | GP2 | 16 | C | T | 1.02 | 0.84-1.24 | 0.871 |
SNP alleles which are associated with obesity risk in Chinese children are highlighted in bold. Chr. Chromosome
Fig. 1.
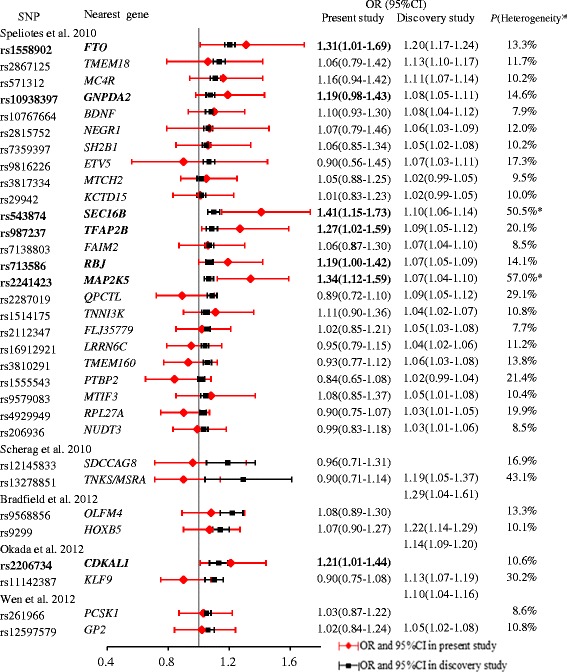
Forest plot showing the ORs (95 % CI) for associations between obesity and 32 SNPs in this study and the reported ORs in the discovery GWA studies. SNP alleles which are associated with obesity risk in Chinese children are highlighted in bold. # P(heterogeneity) is the posterior probability of heterogeneity in allelic effects, which is derived from transethnic meta-analysis [24] *P(heterogeneity) > 50 %, providing evidence of heterogeneity in allelic effects between the present and discovery studies
We also found directionally consistent associations of the 6 SNPs with risk of overweight (see Additional file 2: Table S2), but none was significant after Bonferroni correction for 32 loci.
Individual association of 32 SNPs with BMI-SDS
We additionally examined the association between the 32 common variants and BMI-SDS variability in all 2 030 children (see Additional file 3: Table S3). There were 5 SNPs that showed significant evidence for allelic association with BMI-SDS variability (nominal P <0.05); except for rs2206734 (P = 0.133), 5 of the 6 SNPs associated with obesity were also associated with BMI-SDS. Again the allelic association of rs543874 near SEC16B and rs2241423 near MAP2K5 remained significant after Bonferroni correction for multiple testing (P < 0.00156, Bonferroni corrected for 32 SNPs).
Cumulative effects of these SNP alleles
Figure 2 shows a shift towards an increased number of risk alleles in the obese group compared to the controls (χ2 tests overall P = 0.001). To assess the cumulative effects of 32 SNPs, we created a GRS-32 by summing up the number of effect alleles of the SNPs. Multiple logistic regression revealed that on average, each additional effect allele of GRS-32 was associated with an adjusted 1.06-fold increased odds for obesity (95 % CI: 1.03-1.11, P = 4.84 × 10-4). Similarly, the average per allele increase for BMI-SDS was 0.04 (95 % CI: 0.02-0.06, P = 3.69 × 10-7) in the overall sample. The 32 variants explained 3.30 % of the variance in BMI-SDS.
Fig. 2.
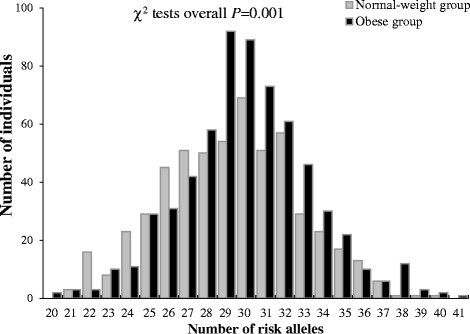
Distributions (histograms) of the number of risk alleles at 32 loci from GWA studies in obese (black) and normal-weight (gray) Chinese children.
The area under the ROC curve (AUCROC) for prediction of risk of obesity using a model including only covariates (age, age square, sex and study group) was 0.735 (95 % CI: 0.709-0.760), which increased to 0.763 (95 % CI: 0.738-0.787) when additionally including the GRS-32 (Difference: 2.8 %, P = 0.0002 for difference between the two models).
Discussion
In this cross-sectional study, we investigated the association of 32 common variants identified by five recent GWA studies in Chinese children. Except for a recent study of 28 SNPs in Chinese adults [15], only 12 loci identified in European population prior to 2010 have been investigated for their effects on BMI or obesity in Chinese populations so far [14, 25-29].
We found nominally significant associations to obesity risk in Chinese children for effect alleles of 6 SNPs near FTO, SEC16B, TFAP2B, RBJ, MAP2K5 and CDKAL1. We compared our findings to the results of a recent study among Chinese adults aged 50-70 years [15]. Although the 6 nominally significant SNP alleles of our study were not significant in that study, the 95 % CIs of ORs overlapped. Similarly, some SNP alleles identified by recent GWA studies [4-8] also confer susceptibility to obesity to Chinese though not meeting a formal significance level. Further exploration of our data revealed that the SNP alleles near SEC16B and MAP2K5 had presumably even stronger effects on obesity in Chinese children than in Caucasian populations (as based on the non-overlap of confidence intervals) and remained significant after correcting for multiple testing. These findings imply possible ethnic differences for effect sizes which have not been reported previously. Large-scaled studies or meta-analyses are required to clarify the ethnic difference of effect size.
In search of a better understanding of the genetic etiology of obesity and given the small individual effect sizes for loci identified in GWA studies that are likely missed applying formal significance testing, many researchers have aggregated information across loci to calculate a genetic risk score from the sum of risk alleles accumulated in an individual [30]. We calculated a genetic risk score for the cumulative effects of all 32 common variants from five GWA studies in Chinese children, including 4 SNPs associated with childhood obesity and 4 SNPs identified in East Asians. We showed that these variants had cumulative effects but a limited predictive value for obesity, which is consistent with previous studies in different populations [2, 14, 31-34].
There are several possible explanations for confirmation of only 6 SNPs in our sample. Firstly, the true effects of the 26 SNPs without formal statistical significance might be smaller than in original populations – a phenomenon called the winners curse. Consequently, our study would be underpowered to detect the effects. We noted only one of the four SNPs (rs2206734 of CDKAL1) that were initially associated with in East Asians achieved significance in this study. However, all these 26 SNPs without statistical significance (including 3 SNPs of East Asians) had directionally consistent effects on obesity compared to the original studies (Fig. 1). Moreover, the cumulative effect analysis of all 32 SNPs demonstrated a clear dosage effect, suggesting polygenic contribution of the alleles at these loci with smaller effect sizes. Secondly, our data suggest that there is a possible ethnic differentiation between Chinese and other ethnic groups. Among 26 loci without a formal significant association, 3 loci (NEGR1, TNN13K, TMEM160) had different effect allele frequencies between Europeans and our Chinese individuals (FST value ≥ 0.10). Thirdly, none of the 6 proxy SNPs showed significant association, which awaits further studies in difference of linkage disequilibrium between Chinese and Caucasian populations.
The strengths of our study include: (a) Anthropometric measurements were taken by trained interviewers according to a standard protocol which minimized measurement errors; (b) Our study groups were relatively homogeneous, both coming from the urban area of Beijing; (c) Our study was conducted in Chinese children. Compared with adults, children have higher BMI or obesity heritability and most obese children have simple obesity without complications, which help to identify the effects of common variants on obesity.
The main limitation of the present study is the relatively small sample size and consequently, reduced statistical power. Moreover, the study is limited by the number of loci tested, which is growing as new genome-wide meta-analyses are conducted [35].
Conclusion
In conclusion, 6/32 tested SNPs were individually associated with obesity in Chinese children. Particularly the effect alleles at two SNPs near SEC16B and MAP2K5 had presumably stronger effects in Chinese children than in Caucasian populations. The 32 common variants identified by recent GWA studies had cumulative effects and predictive value for obesity in Chinese children. The study demonstrated the value of conducting genetic studies in different ethnic populations. Assuming that the high heritability estimates for obesity are correct, these results suggest that more common variants, along with rare variants, copy number variants, epigenetic effects or gene-gene and gene-environment interaction remain to be identified.
Acknowledgements
The study was supported by grants from National Natural Science Foundation of China (81172683, 81573170) and by grants from the German Bundesministerium für Bildung und Forschung (01KU0903, NGFN-Plus 01GS0820 and 01E01002 to AS) and the Deutsche Forschungsgemeinschaft (HI865/2-1). We thank all the children and adolescents and their parents for their participation.
Abbreviations
- GWA
Genome-wide association
- BMI
Body mass index
- SNPs
Single nucleotide polymorphisms
- AUCROC
Area under the receiver operating characteristic curve
- ORs
Odds ratios
- BMI-SDS
BMI standard deviation score
- GIANT
Genetic Investigation of Anthropometric Traits consortium
- ALIR
Study on Adolescent Lipids, Insulin Resistance and Candidate Genes
- CPOOA
The Comprehensive Prevention Project for Overweight and Obese Adolescents
- FST
F-statistics
- GRS
Genetic risk score
- ROC
Receiver operating characteristic curves.
Additional files
Basic genotyping information of 40 GWA studies-derived SNPs in Chinese children. (XLSX 16 kb)
Association of 32 GWA studies-derived SNP alleles with overweight in Chinese children. (XLSX 13 kb)
Association of 32 GWA studies-derived SNP alleles with BMI standard deviation score (BMI-SDS) variability in Chinese children. (XLSX 13 kb)
Footnotes
Competing interests
The authors all declare that they have no competing financial interests.
Authors’ contributions
HJW, AH, JH, YW and JM designed and supervised the project. HJW, JYS, AS and XRM performed data analyses. HG and TI conducted genotyping. All authors were involved in writing the paper and had final approval.
Contributor Information
Hai-Jun Wang, Email: whjun1@bjmu.edu.cn.
Anke Hinney, Email: anke.hinney@uni-due.de.
Jie-Yun Song, Email: songjieyun1983@126.com.
André Scherag, Email: andre.scherag@med.uni-jena.de.
Xiang-Rui Meng, Email: meng_xiangrui@163.com.
Harald Grallert, Email: harald.grallert@helmholtz-muenchen.de.
Thomas Illig, Email: Illig.Thomas@mh-hannover.de.
Johannes Hebebrand, Email: johannes.hebebrand@uni-due.de.
Yan Wang, Email: wangyan@bjmu.edu.cn.
Jun Ma, Phone: +86 10 82801624, Email: majunt@bjmu.edu.cn.
References
- 1.Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–1209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Li S, Zhao JH, Luan J, Luben RN, Rodwell SA, Khaw KT, et al. Cumulative effects and predictive value of common obesity-susceptibility variants identified by genome-wide association studies. Am J Clin Nutr. 2010;91(1):184–190. doi: 10.3945/ajcn.2009.28403. [DOI] [PubMed] [Google Scholar]
- 3.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27(4):325–351. doi: 10.1023/A:1025635913927. [DOI] [PubMed] [Google Scholar]
- 4.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradfield JP, Taal HR, Timpson NJ, Scherag A, Lecoeur C, Warrington NM, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44(5):526–531. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scherag A, Dina C, Hinney A, Vatin V, Scherag S, Vogel CI, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. Plos Genet. 2010;6(4):e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wen W, Cho YS, Zheng W, Dorajoo R, Kato N, Qi L, et al. Meta-analysis identifies common variants associated with body mass index in east Asians. Nat Genet. 2012;44(3):307–311. doi: 10.1038/ng.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okada Y, Kubo M, Ohmiya H, Takahashi A, Kumasaka N, Hosono N, et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat Genet. 2012;44(3):302–306. doi: 10.1038/ng.1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Bradfield JP, Zhang H, Sleiman PM, Kim CE, Glessner JT, et al. Role of BMI-associated loci identified in GWAS meta-analyses in the context of common childhood obesity in European Americans. Obesity (Silver Spring) 2011;19(12):2436–2439. doi: 10.1038/oby.2011.237. [DOI] [PubMed] [Google Scholar]
- 10.Dorajoo R, Blakemore AI, Sim X, Ong RT, Ng DP, Seielstad M, et al. Replication of 13 obesity loci among Singaporean Chinese, Malay and Asian-Indian populations. Int J Obes (Lond) 2012;36(1):159–163. doi: 10.1038/ijo.2011.86. [DOI] [PubMed] [Google Scholar]
- 11.Hong KW, Oh B. Recapitulation of genome-wide association studies on body mass index in the Korean population. Int J Obes (Lond) 2012;36(8):1127–1130. doi: 10.1038/ijo.2011.202. [DOI] [PubMed] [Google Scholar]
- 12.Paternoster L, Evans DM, Nohr EA, Holst C, Gaborieau V, Brennan P, et al. Genome-wide population-based association study of extremely overweight young adults--the GOYA study. PLoS One. 2011;6(9):e24303. doi: 10.1371/journal.pone.0024303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berndt SI, Gustafsson S, Magi R, Ganna A, Wheeler E, Feitosa MF, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet. 2013;45(5):501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong J, Shi J, Qi L, Cui B, Gu W, Zhang Y, et al. Genetic susceptibility, birth weight and obesity risk in young Chinese. Int J Obes (Lond) 2013;37(5):673–677. doi: 10.1038/ijo.2012.87. [DOI] [PubMed] [Google Scholar]
- 15.Zhu J, Loos RJ, Lu L, Zong G, Gan W, Ye X, et al. Associations of genetic risk score with obesity and related traits and the modifying effect of physical activity in a Chinese Han population. PLoS One. 2014;9(3):e91442. doi: 10.1371/journal.pone.0091442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang D, Ma J, Zhang S, Hinney A, Hebebrand J, Wang Y, et al. Association of the MC4R V103I polymorphism with obesity: a Chinese case-control study and meta-analysis in 55,195 individuals. Obesity (Silver Spring) 2010;18(3):573–579. doi: 10.1038/oby.2009.268. [DOI] [PubMed] [Google Scholar]
- 17.Wang HJ, Zhang H, Zhang SW, Pan YP, Ma J. Association of the common genetic variant upstream of INSIG2 gene with obesity related phenotypes in Chinese children and adolescents. Biomed Environ Sci. 2008;21(6):528–536. doi: 10.1016/S0895-3988(09)60013-1. [DOI] [PubMed] [Google Scholar]
- 18.Ji CY. Report on childhood obesity in China (1)--body mass index reference for screening overweight and obesity in Chinese school-age children. Biomed Environ Sci. 2005;18(6):390–400. [PubMed] [Google Scholar]
- 19.WHO. Growth reference data for 5–19 years. Available at http://www.who.int/growthref/tools/en/.
- 20.Duan S, Zhang W, Cox NJ, Dolan ME. FstSNP-HapMap3: a database of SNPs with high population differentiation for HapMap3. Bioinformation. 2008;3(3):139–141. doi: 10.6026/97320630003139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weir BS, Cockerham CC. Estimating F-Statistics for the Analysis of Population Structure. 1984. pp. 1358–1370. [DOI] [PubMed] [Google Scholar]
- 22.Okada Y, Wu D, Trynka G, Raj T, Terao C, Ikari K, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssens AC, Moonesinghe R, Yang Q, Steyerberg EW, van Duijn CM, Khoury MJ. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genet Med. 2007;9(8):528–535. doi: 10.1097/GIM.0b013e31812eece0. [DOI] [PubMed] [Google Scholar]
- 24.Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol. 2011;35(8):809–822. doi: 10.1002/gepi.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng MC, Tam CH, So WY, Ho JS, Chan AW, Lee HM, et al. Implication of genetic variants near NEGR1, SEC16B, TMEM18, ETV5/DGKG, GNPDA2, LIN7C/BDNF, MTCH2, BCDIN3D/FAIM2, SH2B1, FTO, MC4R, and KCTD15 with obesity and type 2 diabetes in 7705 Chinese. J Clin Endocrinol Metab. 2010;95(5):2418–2425. doi: 10.1210/jc.2009-2077. [DOI] [PubMed] [Google Scholar]
- 26.Shi J, Long J, Gao YT, Lu W, Cai Q, Wen W, et al. Evaluation of genetic susceptibility loci for obesity in Chinese women. Am J Epidemiol. 2010;172(3):244–254. doi: 10.1093/aje/kwq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu L, Xi B, Zhang M, Shen Y, Zhao X, Cheng H, et al. Associations of six single nucleotide polymorphisms in obesity-related genes with BMI and risk of obesity in Chinese children. Diabetes. 2010;59(12):3085–3089. doi: 10.2337/db10-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xi B, Shen Y, Reilly KH, Zhao X, Cheng H, Hou D, et al. Sex-dependent associations of genetic variants identified by GWAS with indices of adiposity and obesity risk in a Chinese children population. Clin Endocrinol (Oxf) 2013;79(4):523–528. doi: 10.1111/cen.12091. [DOI] [PubMed] [Google Scholar]
- 29.Wang J, Mei H, Chen W, Jiang Y, Sun W, Li F, et al. Study of eight GWAS-identified common variants for association with obesity-related indices in Chinese children at puberty. Int J Obes (Lond) 2012;36(4):542–547. doi: 10.1038/ijo.2011.218. [DOI] [PubMed] [Google Scholar]
- 30.Klimentidis YC, Chen GB, Lopez-Alarcon M, Harris JJ, Duarte CW, Fernandez JR. Associations of obesity genes with obesity-related outcomes in multiethnic children. Arch Med Res. 2011;42(6):509–514. doi: 10.1016/j.arcmed.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.den Hoed M, Ekelund U, Brage S, Grontved A, Zhao JH, Sharp SJ, et al. Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59(11):2980–2988. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheung CY, Tso AW, Cheung BM, Xu A, Ong KL, Fong CH, et al. Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a chinese population. J Clin Endocrinol Metab. 2010;95(3):1395–1403. doi: 10.1210/jc.2009-1465. [DOI] [PubMed] [Google Scholar]
- 33.Sandholt CH, Sparso T, Grarup N, Albrechtsen A, Almind K, Hansen L, et al. Combined analyses of 20 common obesity susceptibility variants. Diabetes. 2010;59(7):1667–1673. doi: 10.2337/db09-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J, Bradfield JP, Li M, Wang K, Zhang H, Kim CE, et al. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity (Silver Spring) 2009;17(12):2254–2257. doi: 10.1038/oby.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen W, Zheng W, Okada Y, Takeuchi F, Tabara Y, Hwang JY, et al. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23(20):5492–5504. doi: 10.1093/hmg/ddu248. [DOI] [PMC free article] [PubMed] [Google Scholar]


