Summary
Cellular DNA replication origins direct the recruitment of replicative helicases via the action of initiator proteins belonging to the AAA+ superfamily of ATPases. Archaea have a simplified subset of the eukaryotic DNA replication machinery proteins and possess initiators that appear ancestral to both eukaryotic Orc1 and Cdc6. We have reconstituted origin-dependent recruitment of the homohexameric archaeal MCM in vitro with purified recombinant proteins. Using this system, we reveal that archaeal Orc1-1 fulfills both Orc1 and Cdc6 functions by binding to a replication origin and directly recruiting MCM helicase. We identify the interaction interface between these proteins and reveal how ATP binding by Orc1-1 modulates recruitment of MCM. Additionally, we provide evidence that an open-ring form of the archaeal MCM homohexamer is loaded at origins.
Introduction
In archaea and eukaryotes, the MCM replicative helicase is loaded by origin-bound initiator proteins (Bell, 2012; Yardimci and Walter, 2014). In eukaryotes, the initiator is the Origin Recognition Complex (ORC), comprised of Orc1-6. ORC interacts with the Orc1-related protein Cdc6 to recruit MCM(2-7) in complex with Cdt1 (Yardimci and Walter, 2014). Archaea possess a subset of the proteins found in eukaryotes, including one or more proteins related both to Orc1 and Cdc6 as well as a homo-hexameric MCM complex. Additionally, some archaea encode a distant homolog of Cdt1, called WhiP (Barry and Bell, 2006; Robinson and Bell, 2007). Mirroring the multiplicity of candidate initiator proteins, a number of archaeal species have been demonstrated to replicate their chromosomes from multiple replication origins; for review see (Samson and Bell, 2014). Members of the genus Sulfolobus have three replication origins (oriC1-3) in their 2.2 – 3 Mbp chromosomes and we have demonstrated that all three origins fire once per cell cycle (Duggin et al., 2008; Lundgren et al., 2004; Robinson and Bell, 2007; Robinson et al., 2007; Robinson et al., 2004; Samson et al., 2013).
Studies in S. islandicus revealed that each origin is defined by a distinct initiator protein. More specifically, replication initiation at oriC1 requires Orc1-1, oriC2 requires Orc1-3 and oriC3 is defined by WhiP (Samson et al., 2013). Significantly, it is possible to delete individual initiator proteins and retain cell viability. Previously, we demonstrated that we can complement a chromosomal deletion mutant of orc1-1 with a plasmid-borne copy and restore firing at its cognate chromosomal origin, oriC1. Studies with a mutated form of Orc1-1 revealed that ATP binding by Orc1-1 is required for origin firing in vivo. However, expression of a form of Orc1-1 that binds but fails to hydrolyze ATP, a “Walker B” mutant in which the glutamate residue that activates the water molecule for nucleophilic attack during ATP hydrolysis has been substituted by alanine (E147A), results in an over replication phenotype in vivo. In agreement with the in vivo data, a cell extract-based in vitro MCM loading assay revealed that the ATP-bound Walker B mutant form of Orc1-1 was proficient at recruiting MCM in vitro whereas the ADP-bound form of Orc1-1 was inactive (Samson et al., 2013). More recently, studies of an analogous Walker B mutant of Cdc6 in S. cerevisiae have also shown that nucleotide binding but not hydrolysis is required for Cdc6 function in vitro (Coster et al., 2014; Kang et al., 2014). Notably, the archaeal Orc1/Cdc6 proteins have been demonstrated to undergo a single round of ATP hydrolysis leaving ADP tightly bound in the active site (Singleton et al., 2004). This observation, coupled with the cell-cycle regulated transcription of the Orc1-1, support a model where newly synthesized Orc1-1 binds ATP just prior to the onset of S-phase allowing MCM recruitment; subsequent hydrolysis of ATP to ADP then inactivates the Orc1-1, thereby generating a permissive temporal window for MCM recruitment to origins (Samson et al., 2013).
In the well-understood E. coli system, the initiator protein DnaA recruits the DnaB helicase via the action of a dedicated helicase-loader, DnaC (Costa et al., 2013). Similarly, origin-bound ORC in eukaryotes requires Cdc6 and Cdt1 to recruit the MCM(2-7) heterohexamer. However, it has not been determined whether archaeal Orc1-1 contacts MCM directly or via a helicase-loader intermediary.
Another unresolved issue is how ATP-binding affects Orc1-1’s ability to recruit MCM. Further, it is not known how the archaeal MCM is loaded onto replication origins. To address these issues, we have established an in vitro MCM loading assay that employs recombinant Orc1-1 and MCM. Exploiting this assay, in parallel with in vivo studies, we show that Orc1-1 contacts MCM directly, without a helicase-loader intermediary. We map the interaction interface between the proteins and reveal a surprising parallel with an interaction mode observed between human single-stranded DNA binding protein and DNA repair factors. Our work also provides insight into how Orc1-1 responds to ATP to promote its ability to interact with MCM. Finally, we observe that an open-ring form of the archaeal homohexameric MCM is preferentially recruited to origins.
Results
A Defined in vitro System for Origin-Dependent MCM Recruitment
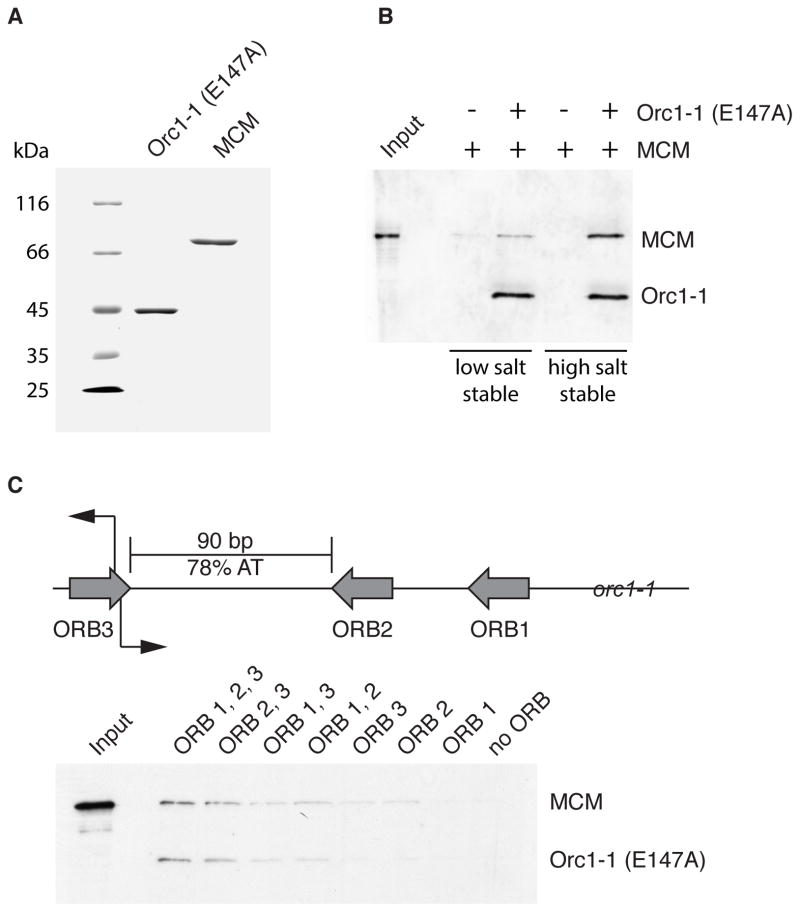
We previously described a cell extract-based system that mediates specific in vitro loading of MCM onto the oriC1 replication origin of S. islandicus (Samson et al., 2013). This origin requires the Orc1/Cdc6 protein Orc1-1 for function in vivo and in vitro and MCM loading is stimulated by use of a version of the Orc1-1 protein that binds to but fails to hydrolyze ATP. We have now refined this system and reconstituted origin-dependent recruitment of MCM into a salt resistant DNA-bound complex using recombinant Orc1-1 and MCM purified from E. coli (Figures 1A and 1B). Thus, we reveal that Orc1-1, in addition to possessing an ORC-like function in binding to replication origins, has a Cdc6-like function in directing MCM recruitment. Therefore, the archaeal Orc1/Cdc6 proteins represent the common ancestor of both present-day Orc1 and Cdc6.
Figure 1. A Defined System for Origin-Dependent Recruitment of MCM.
(A) Coomassie-stained 10% SDS-PAGE gel of 2 μg of purified recombinant Orc1-1 (E147A) and MCM. (B) Association of MCM with immobilized origin DNA in reactions with and without Orc1-1 following low and high salt washes. Proteins were detected by immuno-blotting after SDS-PAGE. Input shows the input MCM only. (C) Cartoon of the organization of oriC1. The grey arrows indicate the ORB consensus elements, the initiation sites mapped in vivo are indicated by black arrows. MCM loading assays were performed with oriC1 derivatives containing point mutations in the ORB elements (8). The remaining intact ORB elements are named. Proteins were detected by immuno-blotting after SDS-PAGE of high salt-stable recruited material. See also Figure S1 and S5.
Having established the assay, we sought to determine the origin-dependence of the reaction. We have shown previously that Orc1-1 binds non-cooperatively to three sites at oriC1, Origin Recognition Boxes (ORB) 1, 2 and 3 (Robinson et al., 2004). These elements contain a conserved dyad repeat flanked on one side by a G-rich sequence (Figure S1). This “G-string” confers a polarity to the ORB elements with ORB2 and ORB3 in opposite orientations relative to each other and ORB1 in the same orientation as ORB2. Structural studies have shown that an individual ORB element is bound by a monomer of Orc1-1 (Gaudier et al., 2007). In addition, we have shown that mutation of the conserved dyad symmetric element within an ORB abrogates recognition by Orc1-1 (Robinson et al., 2004). The inverted repeat ORB2 and ORB3 elements flank the 90 base-pairs of intervening DNA that is 78% AT-rich and a candidate duplex unwinding element. Studies in S. solfataricus revealed replication initiates in vivo at the inner boundary of ORB3 (Robinson et al., 2004).
To test the importance of the ORB elements, we introduced base substitutions into ORB1, 2 and 3 individually and in all possible combinations and assayed the abilities of these mutant DNA substrates to support MCM loading in vitro. As seen in Figure 1C, mutation of ORB2 and ORB3 abrogates the formation of salt-resistant MCM complexes on DNA (Figure 1C). Congruently, a construct lacking ORB1 loads as efficiently as wild-type, all other mutants showed reduced loading. Thus, we have established a highly specific, biochemically-defined assay for origin-dependent recruitment of archaeal MCM by the Orc1-1 protein.
The C-Terminal Winged-Helix Domain of MCM Mediates Recruitment
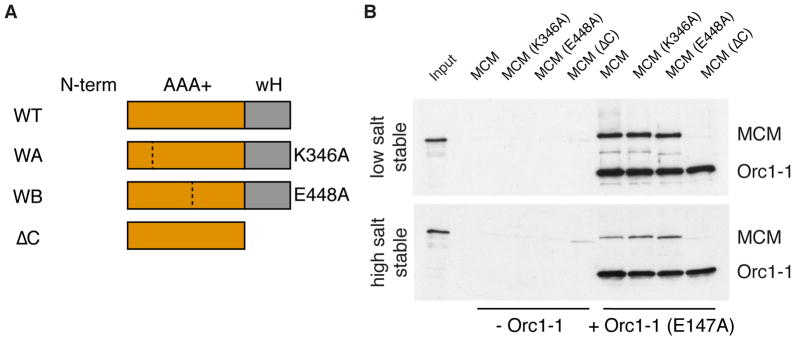
Next, we sought to identify the determinants within MCM that facilitate recruitment to origins. Studies in budding yeast have implicated distinct ATPase sites within the hetero-hexameric MCM(2-7) in helicase loading (Coster et al., 2014; Kang et al., 2014). Some of these effects appear to arise from reduced stability of the MCM(2-7) hexamer upon impairment of a given active site. Previously, we have shown that mutations to the conserved Walker A and Walker B sites of Sulfolobus MCM, while abolishing ATPase activity, do not impact on the stability of the hexamer (Moreau et al., 2007). Thus, we can separate ATP effects on hexamer stability from active site participation in recruitment. We tested the Walker A and Walker B mutant versions of MCM and found that neither ATP binding nor ATP hydrolysis by MCM are required for its recruitment to the replication origin by Orc1-1 (Figures 2A and 2B).
Figure 2. The Molecular Basis of MCM’s Interaction with Orc1-1.
(A) Cartoon of the MCM proteins used in the experiment shown in the panel to the right. Dotted lines indicate the site of amino acid substitutions. WT= wild-type, WA = Walker A mutation (K346A), WB = Walker B mutation (E448A), ΔC = truncation of C-terminal wH-domain. (B) Association of indicated MCM proteins with immobilized origin DNA in reactions with and without Orc1-1 following low and high salt washes. Proteins were detected by immuno-blotting after SDS-PAGE. Input shows the input WT MCM only See also Figure S5.
Archaeal and eukaryal MCM double-hexamerize via their N-terminal domains (Costa and Onesti, 2009; Fletcher et al., 2003; Remus et al., 2009). The dependence of MCM loading on the inverted repeats of ORB2 and ORB3, combined with our previous high resolution mapping of the start site of replication (Robinson et al., 2004), is compatible with a model in which a double hexamer of MCM is loaded on the intervening AT-rich DNA, with the C-terminal domains of MCM being oriented towards the ORB-bound Orc1-1 proteins. To test the importance of the C-terminal domain of MCM, we initially created a version lacking the C-terminal 68 amino acids. Although active as a helicase (Barry et al., 2007), this version of the protein was not recruited to oriC1 by Orc1-1, indicating that the C-terminus of the archaeal MCM is required for MCM’s recruitment to origins (Figures 2A and 2B).
A Candidate Protein-Protein Interaction Site in the wH Domain of MCM
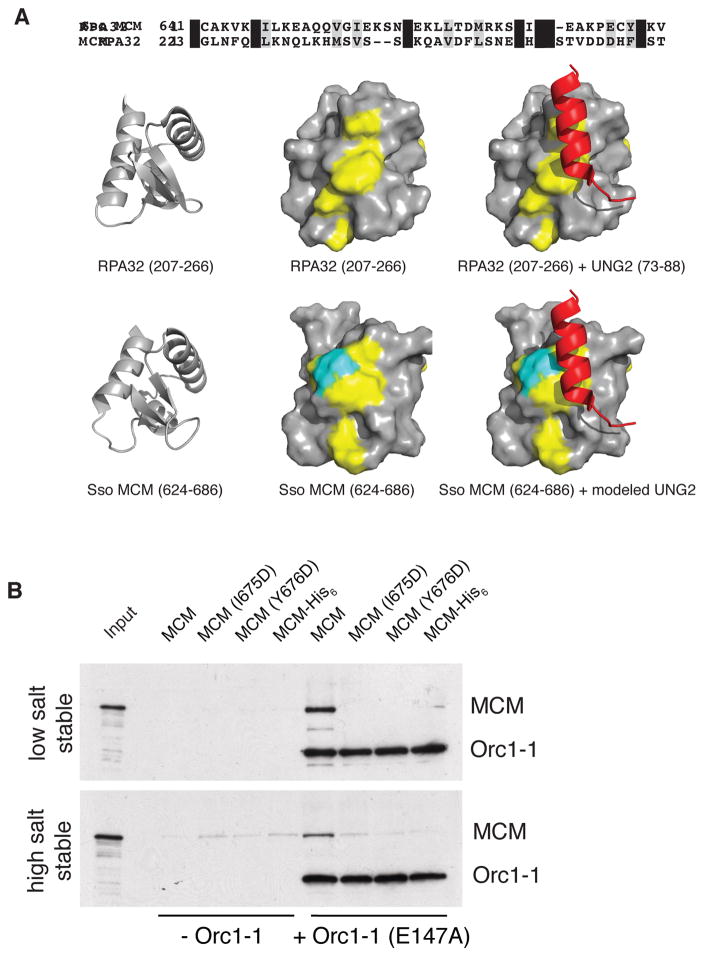
NMR studies have shown that the C-terminal domain of Sulfolobus MCM forms a winged-helix (wH) fold (Wiedemann et al., 2015). Initial analyses using the Phyre 2 server (Kelly et al., 2015) and subsequent analyses with DALI (Holm and Rosenstrom, 2010) revealed a hitherto undocumented similarity to a wH domain present in human RPA32 (see Figure 3A; RMSD 1.28 Å over 44 residues; 1.6 Å over 62 residues; see also Supplemental Information). This domain of RPA32, a component of the single-stranded DNA binding protein, is a known protein-protein interaction module that mediates interactions with a number of proteins, including the uracil N-glycosylase, UNG2 (Mer et al., 2000). Furthermore, key residues important for mediating contacts with RPA32’s partners were conserved in the MCM wH domain. Although the overall sequence identity is modest, we observed that the conserved residues formed a track along the surface of the protein that was coincident with the binding position of the UNG2 peptide on RPA32, colored in yellow in Figure 3A. Importantly, this conserved candidate protein-protein interaction surface is not conserved in other structurally-related wH domains that mediate protein-DNA interactions (Supplemental Experimental Procedures).
Figure 3. The wH Domain of MCM is a Conserved Protein-Protein Interaction Module.
(A) Sequence alignment of the wH domains of Sulfolobus MCM and human RPA32. Structures of the indicated regions of human RPA32 (PDB 1DPU) and Sulfolobus MCM (PDB 2M45) are shown below. The left-hand and middle images show cartoon and surface representations of the isolated wH-domains with identical amino acids colored in yellow. The C-terminal residue of MCM is shown in cyan. The right-hand images show the complex of RPA32 with a peptide from human UNG2; the lower image is a model of Sulfolobus MCM interacting with UNG2 generated by superimposing the wH domains of the two species. (B) Association of the indicated MCM proteins with immobilized origin DNA in reactions with and without Orc1-1 after low and high salt washes. Proteins were detected by immuno-blotting after SDS-PAGE. Input shows the input WT MCM only. See also Supplemental Experimental Procedures and Figure S2 and S5.
We speculated that a similar mode of interaction might exist with MCM. Accordingly, we generated a homology-based model of the wH of MCM interacting with the UNG2-derived peptide and used this to guide mutagenesis of MCM. We mutated Ile 675 and Tyr 676 separately to aspartate. Additionally, we observed that the C-terminal residue of MCM (Val 686) is positioned adjacent to the putative interaction site, so we generated a derivative in which we appended an eight amino acid extension (LEHHHHHH) to the protein’s C-terminus (Figure 3A). Circular dichroism confirmed that the resultant proteins were folded similarly to wild-type (Figure S2A). We further checked the integrity of the wH module by performing DNA binding and ATPase assays. As determined previously (Jenkinson and Chong, 2006; Barry et al., 2007; Weidemann et al., 2015), loss of the wH domain results in a marked activation of the ATPase activity of the MCM (Fig. S2C). Notably, the I675D, Y676D and C-terminally His6-tagged protein bound DNA and hydrolyzed ATP similarly to wild-type MCM (Figures S2B and S2C). We next tested the impact of the point mutations and the C-terminal extension on MCM’s ability to be recruited by Orc1-1. As can be seen in Figure 3B, introduction of either I675D or Y676D amino acid substitutions or introduction of the C-terminal His-tag abrogates the ability of MCM to be recruited to the replication origin.
Previous work has implicated the extreme C-terminus of MCM3 in Cdc6-dependent recruitment of MCM(2-7) to DNA-bound ORC CDC6 in budding yeast (Frigola et al., 2013). As the C-terminus of MCM3 is also predicted to form a wH domain, our findings suggest that this interaction module has been conserved between archaea and eukaryotes.
Mapping the MCM-Interaction Region of Orc1-1
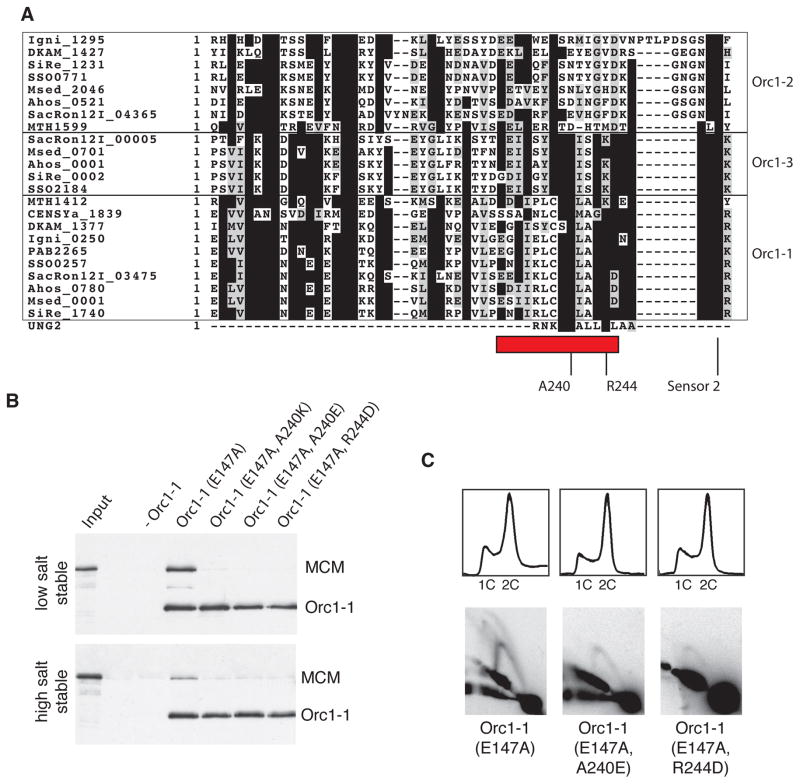
Next, we sought to identify the region of Orc1-1 responsible for recruitment of MCM. Remarkably, comparison of the sequence of the uracil N-glycosylase (UNG2) peptide that interacts with the human RPA32 with the sequence of archaeal Orc1/Cdc6 proteins reveals a related sequence in Orc1-1 (Figure 4A). As with the RPA32 and MCM wH domains, sequence conservation was very limited. Nevertheless, we were intrigued that this region mapped to a surface-exposed alpha-helix on the lid sub-domain of the Orc1-1 AAA+ ATPase domain (Figure S3A). Furthermore, an analysis of the sequence of a range of archaeal Orc1/Cdc6 sequences revealed that this motif was conserved in Orc1-1 and the oriC2-defining Orc1-3 proteins across a diverse range of archaeal species. Both of these proteins have demonstrated roles in promoting DNA replication at their cognate origins. However, the sequence motif was not conserved in Orc1-2 and its orthologs. In Sulfolobus, Orc1-2 is an Orc1/Cdc6 paralog that is not required for replication at any origin (Samson et al., 2013). Further, Orc1-2’s expression is induced in response to cellular stresses, such as UV-induced DNA damage and heat-shock (Tachdjian and Kelly, 2006; Frols et al., 2007; Gotz et al, 2007) and is thought to be a repressor of replication (Robinson et al., 2004). Thus, this candidate interaction motif is a signature of replication-promoting Orc1/Cdc6 proteins.
Figure 4. Identification of the MCM-Interacting Interface of Orc1-1.
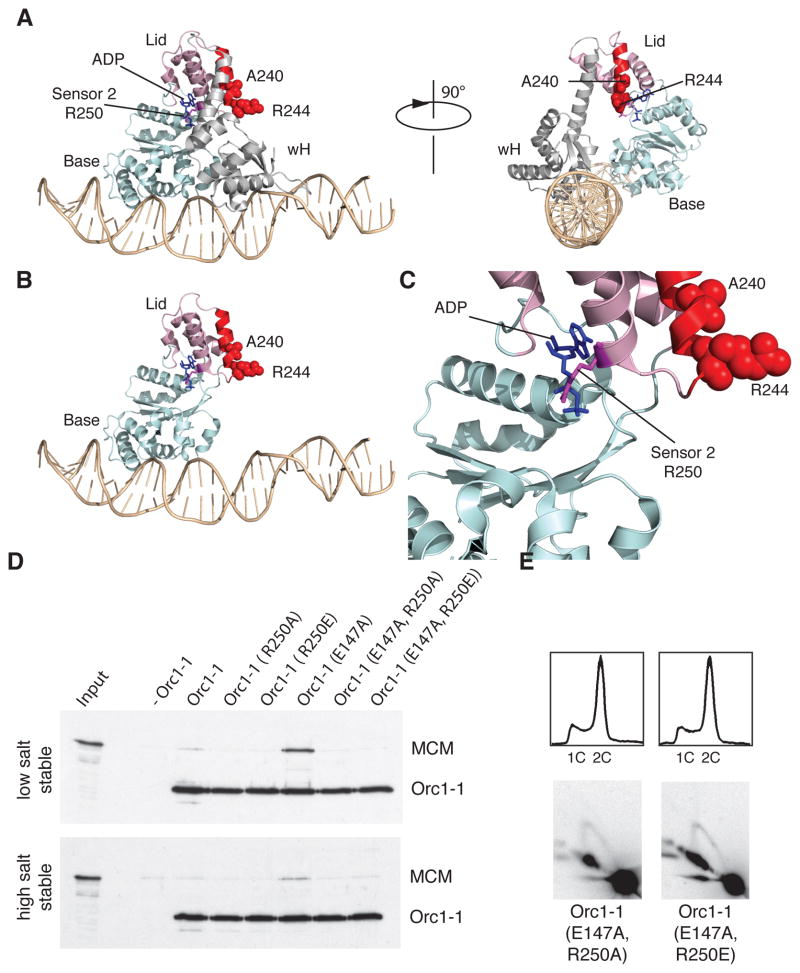
(A) Alignment of Orc1/Cdc6 proteins from a diverse range of archaeal species. Proteins are indicated by species abbreviations followed by annotated ORF number. Crenarchaea : SiRe – Sulfolobus islandicus, SSO – Sulfolobus solfataricus, SacRon12I – Sulfolobus acidiocaldarius, DKAM – Desulfurococcus kamchatkensis, Igni – Ingicoccus hospitalis, Msed – Metallosphaera sedula. Euryarchaea : PAB - Pyrococcus abyssi, MTH – Methanothermobacter thermautotrophicus. Thaumarchaea : CENSY – Cenarchaeum symbiosum. UNG2 is the sequence of the peptide derived from human ING2 that forms a complex with the RPA32 wH domain. The representatives of clades related to Sulfolobus Orc1-1, Orc1-2 and Orc1-3 are indicated and key conserved residues highlighted, including the Sensor 2 residue, R250 (S. islandicus Orc1-1 numbering). (B) Association of the MCM with immobilized origin DNA in reactions with and without the indicated Orc1-1 proteins following low and high salt washes. Proteins were detected by immuno-blotting after SDS-PAGE. (C) The upper panels show the results of flow cytometry of Δorc1-1 cells expressing the indicated Orc1-1 proteins from a plasmid (see ref. 5). The lower panels show the results of 2D gels to interrogate firing of oriC1 in vivo in the complemented strains. See also Figure S3, S5 and S6 and Table S1.
To test the importance of this candidate MCM-Recruitment Motif (MRM) we made amino acid substitutions (A240E, A240K and R244D) of the most conserved surface exposed residues (Figure 4A). These substitutions did not impact on origin DNA binding by the resultant proteins in vitro or in vivo following expression in a Δorc1-1 strain of S. islandicus (Figures S3B and S3C). However, they abolished interaction with MCM in the in vitro recruitment assay and impaired origin firing in vivo as determined by 2D neutral-neutral agarose gel electrophoresis (Figures 4B and 4C). Additionally, these proteins did not support the over-replication phenotype in vivo caused by the Walker B (E147A) mutant form of the Orc1-1, as adjudged by flow-cytometry (Figure 4C).
Basis of ATP Dependence of the Orc1-1 MCM Interaction
As mentioned above, the MCM-Recruitment Motif is located in the AAA+ ATPase module of Orc1-1 and forms a surface-exposed alpha-helix in the so-called lid sub-domain of the AAA+ domain (Figure 5A–C). Intriguingly, the MRM helix is four residues removed from the Sensor 2 residue R250. The Sensor 2 residue is a conserved feature of many AAA+ proteins and plays a role in coordinating the γ-phosphate of ATP (Wendler et al., 2012). Depending on the particular protein in question, mutation of Sensor 2 can impair the ability of the protein to hydrolyze ATP and/or transduce structural changes upon ATP binding, hydrolysis and release (Erzberger and Berger, 2006; Wendler et al., 2012). Interestingly, mutation of the Walker B and Sensor 2 residues of yeast Cdc6 both lead to impairment of ATP hydrolysis by Cdc6 (Takahashi et al., 2002; Coster et al., 2014; Kang et al., 2014). However, while the Walker B mutant, as in archaea, is active at MCM loading, the Sensor 2 mutation is inactive (Coster et al., 2014; Evrin et al., 2013; Kang et al., 2014; Samson et al., 2013). We speculated, therefore, that these apparently paradoxical results could be explained by Sensor 2 acting to reposition the MRM-containing lid domain upon ATP binding.
Figure 5. Basis of ATP-Dependence of the Orc1-1 MCM Interaction.
(A) Structure of Orc1-1 ADP on DNA. The alpha-helix highlighted in red corresponds to the highlighted region of Orc1-1 in Figure S2A [Figure generated using Pymol (Schrödinger, LLC) from PDB file 2QBY]. The ADP moiety is shown in blue stick form and indicated. The Sensor 2 residue (R250) is in purple. Residues A240 and R244 are shown with atoms depicted as red spheres. The α/β “base” subdomain of the AAA+ domain is shown pale blue, the α-helical “lid” domain is in pink. The wH DNA binding domain of Orc1-1 is in gray. DNA is in wheat. Views from two perspectives are given, differing by a 90° rotation. (B) The same view as the left hand panel of A but with the wH domain removed for clarity. (C) A magnified view of the nucleotide binding pocket of Orc1-1 showing the incumbent ADP and the position of the Sensor 2 R250 residue. (D) Association of the MCM with immobilized origin DNA in reactions with and without the indicated Orc1-1 proteins following low and high salt washes. Proteins were detected by immuno-blotting after SDS-PAGE. Input shows the input WT MCM. (E) The upper panels show the results of flow cytometry of Δorc1-1 cells expressing the indicated Orc1-1 proteins from a plasmid. The lower panels show the results of 2D gels to interrogate firing of oriC1 in vivo in the complemented strains. See also Figure S3, S5 and S6.
Accordingly, we prepared versions of Orc1-1 with substitutions of the Sensor 2 R250 residue to either alanine or glutamate in both wild-type and Walker B mutant Orc1-1 (Figure 5D). We confirmed that the Sensor_2 mutants bound but failed to hydrolyze ATP (Figure S4A and data not shown). Importantly, these mutant proteins were still able to interact with origin DNA in vitro and in vivo (Figures S4B and S4C). However, neither single Sensor 2 nor double Sensor 2/Walker B mutant proteins could recruit MCM in vitro nor could the double mutants support replication initiation in vivo (Figure 5D and 5E). Furthermore, combining the Sensor 2 mutations with Walker B E147A abolished the over replication phenotype observed in vivo with the Walker B mutant alone.
Taken together, our data lead to a model for the molecular basis of ATP-dependent recruitment of MCM by the conserved Orc1/Cdc6 proteins. We propose that interaction of Orc1-1 with ATP leads to repositioning of the MRM-containing lid-domain due to coordination of the γ-phosphate of ATP by the Sensor 2 residue. Consequently, the MRM interacts with MCM, leading to MCM’s recruitment to the origin. Subsequent hydrolysis of ATP to ADP then repositions the MRM, preventing its interaction with MCM and thus inactivates the Orc1-1 protein. We note that as well as preventing extra rounds of MCM recruitment, this ATP hydrolysis dependent switch within Orc1-1 could facilitate release of MCM following recruitment. However, since the Walker B mutant supports replication initiation at oriC1 in vivo (Figure 4), this putative release is not a prerequisite for origin firing (Samson et al., 2013).
An Open-Ring Form of MCM is Preferentially Recruited by Orc1-1
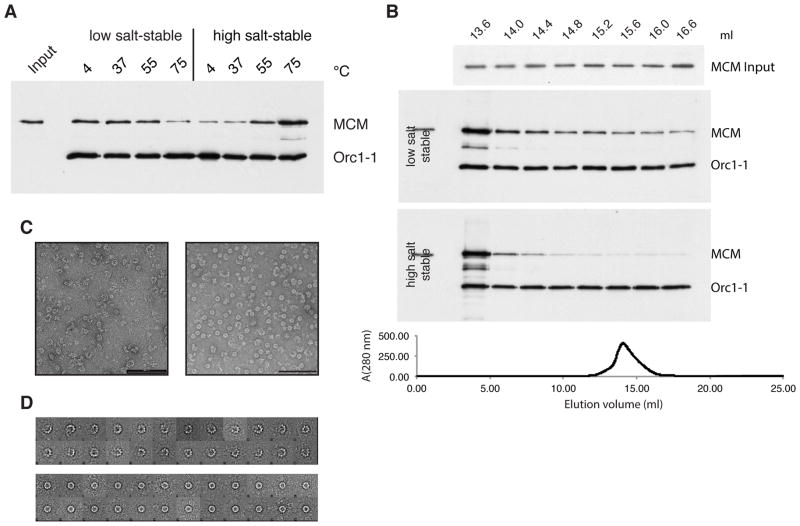
Our data indicate that there is no requirement for the energy released by ATP hydrolysis by either Orc1-1 or MCM to effect loading. As seen in Figure 2, a Walker B mutant Orc1-1 efficiently recruits and loads both Walker A and Walker B mutant MCM. Further, the Walker B mutant of Orc1-1 is proficient at MCM loading at oriC1 in vivo. This argues against a model in which the MCM ring is actively opened, loaded, resealed and released by Orc1-1 in a manner analogous to sliding clamp loading by the clamp loader: for a review of potential MCM loading mechanisms see (Yardimci and Walter, 2014). Eukaryotic MCM(2-7) has been observed to adopt both open and closed ring conformations with an open gate between MCM2 and MCM5 being of key importance for loading the hexamer (Bochman and Schwacha, 2008; Costa et al., 2011; Samel et al., 2014). Similarly, archaeal MCMs have been observed in a range of conformations (Costa and Onesti, 2009; Gomez-Llorente et al., 2005). We therefore sought to determine whether a particular conformer of MCM is recruited preferentially to origins. Previous work has shown that MCM from the thermophilic archaeon Methanothermobacter thermautotrophicus undergoes a temperature-dependent transition between open- and closed-ring forms, with treatment at the physiological growth temperature favoring the open-ring form (Gomez-Llorente et al., 2005). To determine if temperature might similarly affect Sulfolobus MCM, we incubated MCM at a range of temperatures prior to addition to loading assays (performed at 50°C). Pre-treatment of recombinant MCM at 75°C, the growth temperature of Sulfolobus, strongly favored generation of the high salt-resistant, loaded MCM on the origin (Figure 6A).
Figure 6. An Open-Ring Form of MCM is Preferentially Recruited to oriC1 by Orc1-1.
(A) MCM was treated at the indicated temperatures prior to addition to the loading assay. Following low and high salt washes, origin-associated proteins were detected by immuno-blotting after SDS-PAGE. Input shows the input WT MCM. (B) MCM was fractionated over a Superose 6 gel filtration column. Elution was monitored at 280 nm (lower panel) and 0.4 ml fractions collected. Fractions starting at the indicated volume were diluted to equal concentrations (see “input” panel) and used in recruitment assays. As previously, proteins were detected by immuno-blotting after SDS-PAGE. (C) Representative electron micrographs of negative stained MCM recovered from early eluting fractions (left panel) and later eluting (right panel) fractions following gel filtration; the scale bar indicates 100 nm. (D) 2D class averages of the particles in the early eluting fractions (upper panel) and later eluting fractions (lower panel).
Additionally, we fractionated Sulfolobus MCM over a gel filtration column and observed elution in a single peak compatible with a predominantly hexameric form of the protein. We then tested the ability of equal concentrations of MCM from different fractions to be recruited by Orc1-1 (Figure 6B). To do so, we quantified the amount of MCM in the various fractions and diluted the more concentrated fractions to the same concentration as the most dilute. We then used the same volume and concentration of MCM from all fractions in our recruitment assay. We observed a striking disparity across the elution peak, with MCM from the early eluting fractions being far more efficiently recruited by Orc1-1 than the later-eluting material. As gel filtration chromatography separates protein on basis of size and shape, we sought to address the conformations of the protein in the early and late fractions.
To this end, we performed negative stain electron microscopy and observed that the leading edge of the peak contained a preponderance of an open-ring form of MCM with 78% of particles (n=1454) in the open ring conformation; in contrast, the later-eluting material, which was recruited to a far lesser extent, contained 90% closed-ring form (n=1564; Figures 6C and 6D). 2D class averages of open-ring forms from the early eluting fractions revealed a variety of open-ring conformations with some variability in the number of protomers visible. Taken together, these data reveal that an open-ring conformation of archaeal MCM facilitates its recruitment and thus loading on replication origins.
Discussion
While high resolution structures of archaeal replication initiator proteins have been available for several years, comparatively little is known about the function and regulation of these proteins in vitro and in vivo. Our current work provides answers to a number of unresolved issues and yields insight into the function and evolution of the eukaryotic replication machinery. The genome sequences of archaea have revealed the presence of candidate initiator proteins with sequences that appeared ancestral to both Orc1 and Cdc6 in eukaryotes. While a considerable body of functional and structural information has been accrued to document the Orc1-like functions of these proteins in origin definition, it has been unclear whether these proteins additionally directly recruit the MCM helicase. In our current work we provide direct evidence that Sulfolobus Orc1-1 both binds the replication origin oriC1 and recruits MCM. This is a departure from the paradigm of both bacterial replication and higher eukaryotic replication systems that employ dedicated helicase loader proteins (DnaC in E. coli and the helicase co-loaders Cdc6 and Cdt1 in higher eukaryotes) which act as adaptors between origin-bound initiators and incoming helicases. Intriguingly, trypanosomes, which are very early branching eukaryotes, encode a single Orc1/Cdc6 ortholog, reminiscent of that found in archaea. Furthermore, trypanosomes lack a detectable homolog of Cdt1 (Tiengwe et al., 2012). This suggests that the duplication and specialization of function of the ancestral orc1/cdc6 took place within the eukaryotic lineage. Presumably this transition imparts greater regulatory potential to the eukaryotic preRC, concomitant with increasing genome size and number of replication origins during eukaryotic evolution.
In Sulfolobus islandicus, we observe a simple binary relationship of one initiator protein per replication origin. Our previous work has revealed that the ATP-bound form of the Orc1-1 protein is active in MCM recruitment while the ADP-bound form is not. Importantly, the cell-cycle dependent transcription of the orc1-1 gene coupled with the observation that these proteins undergo a single turnover ATP hydrolysis event provides a simple model for regulation of origin firing. However, while we could detect subtle differences between the protease sensitivities of ATP and ADP-bound forms of Orc1-1, the basis of the ATP-dependent activation of the protein was unresolved. By identifying the interaction interface between Orc1-1 and MCM our current data suggest a mechanism for the ATP-dependent regulation of Orc1-1 activity. More specifically, the Sensor 2 arginine residue is crucial for Orc1-1 function and we propose that, upon coordinating the gamma phosphate of ATP, it repositions the immediately adjacent MCM-recruitment helix, thereby facilitating MCM’s recruitment. As alluded to above, Sensor 2 also plays a key role in Cdc6’s function in budding yeast (Takahashi et al., 2002; Evrin et al., 2013) and we suggest this residue may play an analogous role in sensing nucleotide status of the Cdc6 protein. Significantly, as in archaeal Orc1-1, ATP hydrolysis by Cdc6 is not required for its function, rather the period of ATP occupancy of the active site confers a temporal window during which MCM can be loaded (Samson et al., 2013; Coster et al., 2014; Kang et al., 2014).
We observe that an open ring form of MCM is preferentially recruited and loaded by Orc1-1. While it possible that additional cellular factors, not present in our recombinant-protein based system, may modulate MCM conformation in vivo, we note that treatment of MCM at the physiological growth temperature of Sulfolobus, favors the generation of the open ring form. Direct parallels can be drawn with eukaryotic MCM(2-7) which contains a gate between MCM2 and MCM5, the function of which is essential for MCM(2-7) loading in vitro (Samel et al., 2014).
Thus, despite the organizational simplicity of the archaeal replication machinery, our data reveal clear and fundamental mechanistic parallels with the eukaryotic apparatus. We note that recent crystal structures of Drosophila ORC (Bleichert et al., 2015) and electron microscopy models of yeast ORC Cdc6 MCM (Sun et al., 2013) suggest an orientation of Cdc6 that is compatible with the interaction interface that we have determined between archaeal Orc1/Cdc6 and MCM being utilized in the eukaryotic pre-Replication Complex.
Experimental Procedures
Protein Purification
S. islandicus Orc1-1 and MCM proteins were expressed in and purified from E. coli Rosetta cells (Figures S5A–C). Wild-type and mutant Orc1-1 proteins possessed C-terminal His6-tags and were purified as described previously (Dueber et al., 2007; Samson et al., 2013). In order to express MCM proteins, cultures were grown in LB at 37 °C to an OD600 = 0.6-0.8 and induced with 1 mM IPTG overnight at 25 °C. Cells were lysed in 1X TBS, 1 mM DTT containing Roche Mini-Complete Protease Inhibitors using a French press. The soluble lysate was heat treated for 20 minutes at 68 °C, and the heat-stable fraction was purified over a HiTrap Heparin column (GE Healthcare). Proteins were further purified over a HiLoad 26/600 Superdex 200 column (GE Healthcare) in 2X TBS, 1 mM DTT. Alternatively, following purification over a Heparin column, wild-type MCM was fractionated over a Superose 6 10/300 column (GE Healthcare) in 2X TBS, 1 mM DTT. Peak fractions were diluted with column buffer to 10 ng/μl and immediately used in MCM recruitment assays (see below).
MCM Recruitment Assay
Substrate DNA was prepared as described (Samson et al., 2013). Mutated substrates were prepared by site directed mutagenesis. Binding reactions were assembled in Protein LoBind tubes (Eppendorf) and contained 200 pM DNA in 50 μl of binding buffer (20 mM Tris acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9) plus 20 ng/μl poly dG:dC. Reactions were pre-heated to 50 °C in a water bath and either Orc1-1 protein buffer or Orc1-1 protein (1 μM final) were added and supplemented with 2 mM ATP. Following a 10 minute incubation at 50 °C, 50 μl of recombinant MCM [12.9 nM] plus competitor DNA and 2 mM ATP were added to the Orc1-1 reactions and incubated for 30 or 45 minutes at 50 °C. The reactions were then washed twice by magnet-mediated bead pelleting and resuspension in 100 μl binding buffer, followed by one wash in 100 μl binding buffer containing 500 mM potassium acetate. Following this final wash, beads were boiled in 1X SDS-PAGE loading buffer, run on SDS-PAGE gels, transferred to PVDF membranes and subjected to western analyses. All wash steps employed pre-warmed buffers and were performed in the 50°C water bath. In the recruitment assays performed with the mutant ORB DNA substrates, recombinant Orc1-1 was added into the reaction at 25 nM with the MCM mixture. Reactions were then incubated for 45 minutes at 50 °C and wash and elution steps were performed as described above.
In the assays analyzing the effect of temperature on subsequent MCM recruitment, MCM was pre-incubated at 4 °C, 37 °C, 55 °C, or 75 °C for 10 minutes before adding to the Orc1-1 binding reaction.
ATP Binding and ATPase Assays
100 μg of C-terminally hexahistidine tagged Orc1-1 proteins were immobilized on 50 μl bead volume of Ni-NTA agarose resin by incubation in 500 μl of Buffer A (10 % glycerol, 50 mM HEPES pH 7.9, 750 mM NaCl, 5 mM beta-mercaptoethanol) for 30 minutes at room temperature. Beads were recovered by brief centrifugation and resuspended in 1 ml Buffer A containing 6M guanidinium hydrochloride. After 30 minutes, beads were harvested and sequentially washed with 2 fold-decreasing concentrations of guanidinium hydrochloride (GuHCl) in Buffer A to a final concentration of 93.8 mM GuCl in Buffer A. Finally, beads were washed with 1 ml Buffer A before Orc1-1 proteins were eluted with Buffer A containing 250 mM imidazole. ATP binding was assessed by incubation of 10 μg of the refolded protein with buffer A supplemented with 0.375 MBq of α-32P ATP (111 TBq/mmol) in the presence of 2 mM MgCl2 for 15 minutes at room temperature. Reactions were then split in two and one half analyzed by native gel electrophoresis in Tris-glycine, before phosphorimaging of the wet gel; the other half was subjected to SDS PAGE and protein detected by staining with Comassie brilliant blue.
MCM ATPase activity was assessed using the colorimetric assay developed by (Chen et al, 1956). Reactions were performed at 50 °C for 30 minutes in 100 μl volumes in 20 mM Tris acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9 with 1μM MCM (monomer) and 1 mM ATP. Phosphate release was measured as described (Chen et al., 1956) and quantified by reference to a standard curve generated with known concentrations of potassium phosphate.
Construction of pSSREF
The pyrEF genes and promoter from Sulfolobus solfataricus P2 were cloned into the NotI and XmaI sites of pSSR to generate pSSREF, a vector selectable by both uracil auxotrophy and simvastatin resistance. The promoter and gene for S. islandicus orc1-1 were cloned into the SphI and SalI sites of pSSREF. Mutations to the orc1-1 gene were produced by site-directed mutagenesis.
Genetic Manipulation of Sulfolobus
S. islandicus REY15A (ΔpyrEF, ΔlacS, Δorc1-1) - a kind gift from Dr Qunxin She, Copenhagen University - was grown to early log phase and pelleted at room temperature (Samson et al., 2013). The cells were washed three times with 20 mM sucrose at room temperature and were then resuspended in 20 mM sucrose to an OD600 = 10–20. 200–800 ng of plasmid DNA were added to 50 μl of these cells. The cells were then electroporated at 1.2 kV, 25 μF, 600 Ω. 900 μl of pre-warmed high salt buffer (22.7 mM ammonium sulfate, 2.9 mM potassium sulfate, 1.3 mM potassium chloride, 9.3 mM glycine) were added and the cells were transferred to sterile tubes and incubated at 78 °C for 1–2 hours before plating on TSY (0.1 % tryptone, 0.2 % sucrose, 0.05 % yeast extract) plates + uracil [20 μg/ml] + 18 μM simvastatin. After incubation at 78 °C for 10–14 days, colonies were picked and restreaked onto fresh SCVY (0.2 % sucrose, 0.2 % casamino acids, 1X vitamin solution, 0.004 % yeast extract) plates lacking uracil and simvastatin. Isolated colonies on the SCVY plates were used to inoculate 1.5-ml TSY liquid cultures (lacking uracil and simvastatin). Transformants were confirmed by colony PCR and western blot detection of the Orc1-1 protein (Figure S6). Plasmids were recovered from S. islandicus and re-sequenced to confirm that no additional mutations had arisen in the orc1-1 gene. No additional mutations were detected.
Flow Cytometry
S. islandicus cells were fixed with 72% ethanol, stained with SYTOX Green (Invitrogen) and DNA contents were analyzed with an LSRII flow cytometer.
Neutral-Neutral 2D Gel Electrophoresis
S. islandicus cells were collected by centrifugation and washed three times with TEN buffer (50 mM Tris-Cl, pH 8; 50 mM EDTA, pH 8; 100 mM NaCl). Cell suspensions were then mixed with low melting point agarose [0.4 % final concentration] and dispensed into CHEF plug molds (Bio-Rad). Genomic DNA preparation and 2D gel electrophoresis were performed as described previously (Robinson et al., 2004; Samson et al., 2013). To analyze the oriC1 locus, DNA was digested with NdeI. DNA from the 2D gels was transferred to Hybond-XL membranes (GE Healthcare) by overnight capillary transfer and 32P-labelled DNA probes were hybridized to the membranes in Church-Gilbert buffer at 65 °C. Washes were performed as described in the Hybond-XL manual and membranes were exposed to phosphorimager screens that were scanned on a Typhoon 9210 phosphorimager (GE Healthcare).
Chromatin Immunoprecipitation and qPCR
Chromatin immunoprecipitation (ChIP) was performed as described (Samson et al., 2013). Specifically, formaldehyde was added to S. islandicus cultures in early log phase. After crosslinking for 20 minutes at room temperature, the reaction was quenched with 125 mM glycine, the cells were pelleted and washed with 1X PBS. To generate cell lysate, the cells were resuspended in TBS-TT (20 mM Tris, 150 mM NaCl, 0.1 % Tween-20, 0.1 % Triton X-100, pH 7.5) and lysed in a French press. After sonicating the lysate to generate DNA fragments ranging from 200–1000 bp, the extract was clarified by centrifugation. Based on the protein concentration, 10 μg of total protein were used in a 100-μl ChIP reaction. Samples were incubated for 2 hours at 4 °C with 3 μl of αOrc1-1 antibody and an additional hour after the addition of Protein A Sepharose. After washing with TBS-TT, TBS-TT containing 500 mM NaCl, and TBS-TT containing 0.5 % Tween-20 and 0.5 % Triton X-100, the protein-DNA complexes were eluted in 20 mM Tris, 10 mM EDTA, 0.5 % SDS, pH 7.8 at 65 °C for 30 minutes. The samples were then incubated with 10 ng/μl Proteinase K for 6 hours at 65 °C followed by 10 hours at 37 °C. The samples were extracted with phenol and chloroform and the DNA was precipitated with 100 % ethanol containing 20 μg of glycogen. After washing with 70 % ethanol and air-drying, the DNA was resuspended in 50 μl TE buffer.
QPCR was performed using a master mix from the Brilliant III SYBR Green QPCR kit (Agilent Technologies, Inc.), primers at 125 nM, and 1 μl of ChIP DNA. All reactions were performed in triplicate and analyzed using a Mastercycler ep realplex2 machine and analysis software (Eppendorf).
Fluorescence Polarization
Fluorescence polarization measurements were performed with a range of concentrations of Orc1-1 protein in 100 μl reaction volumes containing (20 mM Tris-acetate, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM DTT, pH 7.9) and 20 nM 6-FAM-labeled double-stranded oligonucleotides corresponding to ORB2. Binding was performed for 10 minutes at room temperature before analysis in a Synergy H1 plate reader. While the assays reported in this work were performed at room temperature, we also examined the effect of performing the reactions at 45°C. No significant changes in binding were observed at the higher temperature (data not shown). Data were plotted using Kaleidagraph version 4.5 (Synergy Software) and data were fit to a single-site binding model [y=Bmax*X/(Kd + X)] where Bmax is the maximum specific binding signal and X is the concentration of Orc1-1.
Circular dichroism
Circular dichroism (CD) measurements were performed with a Jasco J-715 spectrometer using a quartz cuvette with a 1.0 mm path length. The various MCM proteins were buffer exchanged into deionized water using Zebra Spin desalting columns (7000 MWCO) and diluted to 0.33 mg/ml [2.45 μM] with deionized water. UV wavelength scans were recorded at 25 °C from 300 to 190 nm. Averages for five CD spectra were presented. Ellipticity results were expressed as mean residue ellipticity in millidegrees (mdeg).
Negative Stain Electron Microscopy
Negative stain grids were prepared by applying 3 μl of protein solution (~ 50 ng/μl) onto a carbon-coated copper grid. The grid was then stained with 2% uranyl acetate. Electron micrographs were recorded on a Philips CM10 equipped with a LaB6 filament operated at an acceleration voltage of 80 kV. Micrographs were recorded at a nominal magnification of 130,000X on a Veleta CCD camera resulting in an Å/pixel value of 3.7 at the sample level. 2D class averages of S. solfataricus MCM particles were obtained using IMAGIC (van Heel et al., 1996).
Supplementary Material
Acknowledgments
We thank James Berger for discussing data prior to publication. RYS is funded by NIH Training Grant T32GM007757. Work in SDB’s lab is also funded by the College of Arts and Sciences, Indiana University.
Footnotes
Supplemental Information includes six figures and Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barry ER, Bell SD. DNA replication in the archaea. Micro Mol Biol Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, McGeoch AT, Kelman Z, Bell SD. Archaeal MCM has separable processivity, substrate choice and helicase domains. Nucl Acid Res. 2007;35:988–998. doi: 10.1093/nar/gkl1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SD. Archaeal orc1/cdc6 proteins. Sub-cellular Biochem. 2012;62:59–69. doi: 10.1007/978-94-007-4572-8_4. [DOI] [PubMed] [Google Scholar]
- Bleichert F, Botchan MR, Berger JM. Crystal structure of the eukaryotic origin recognition complex. Nature. 2015;519:321–326. doi: 10.1038/nature14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, Schwacha A. The mcm2-7 complex has in vitro helicase activity. Mol Cell. 2008;31:287–293. doi: 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Chen PS, Tobibara TY, Warner H. Microdetermination of phosphorous. Anal Chem. 1956;28:1756–1758. [Google Scholar]
- Costa A, Hood IV, Berger JM. Mechanisms for initiating cellular DNA replication. Ann Rev Biochem. 2013;82:25–54. doi: 10.1146/annurev-biochem-052610-094414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18:471–U110. doi: 10.1038/nsmb.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A, Onesti S. Structural biology of MCM helicases. Criti Rev Biochem Mol Biol. 2009;44:326–342. doi: 10.1080/10409230903186012. [DOI] [PubMed] [Google Scholar]
- Coster G, Frigola J, Beuron F, Morris EP, Diffley JF. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell. 2014;55:666–677. doi: 10.1016/j.molcel.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber ELC, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci USA. 2008;105:16737–16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA plus proteins. Ann Rev Bioph Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- Evrin C, Fernandez-Cid A, Zech J, Herrera MC, Riera A, Clarke P, Brill S, Lurz R, Speck C. In the absence of ATPase activity, pre-RC formation is blocked prior to MCM2-7 hexamer dimerization. Nucl Acid Res. 2013;41:3162–3172. doi: 10.1093/nar/gkt043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XJS. The structure and function of MCM from archaeal M-thermoautotrophicum. Nat Struct Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- Frigola J, Remus D, Mehanna A, Diffley JFX. ATPase-dependent quality control of DNA replication origin licensing. Nature. 2013;495:339–343. doi: 10.1038/nature11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frols S, Gordon PML, Panlilio MA, Duggin IG, Bell SD, Sensen CW, Schleper C. Response of the hyperthermophilic archaeon Sulfolobus acidocaldarius to UV damage. J Bact. 2007;189:8708–8718. doi: 10.1128/JB.01016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–1216. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- Gomez-Llorente Y, Fletcher RJ, Chen XS, Carazo JM, San Martin C. Polymorphism and double hexamer structure in the archaeal minichromosome maintenance (MCM) helicase from Methanobacterium thermoautotrophicum. J Biol Chem. 2005;280:40909–40915. doi: 10.1074/jbc.M509760200. [DOI] [PubMed] [Google Scholar]
- Gotz D, Paytubi S, Munro S, Lundgren M, Bernander R, White MF. Responses of hyperthermophilic crenarchaea to UV irradiation. Genome Biol. 2007;8:R220. doi: 10.1186/gb-2007-8-10-r220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: conservation mapping in 3D. Nucl Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Warner MD, Bell SP. Multiple functions for Mcm2-7 ATPase motifs during replication initiation. Mol Cell. 2014;55:655–665. doi: 10.1016/j.molcel.2014.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Prot. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M, Andersson A, Chen LM, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: Synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci USA. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted Loading of Mcm2-7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci USA. 2007;104:5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Blood KA, McCallum SA, Edwards PAW, Bell SD. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD. Identification of two origins of replication in the single chromosome of the Archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- Samel SA, Fernandez-Cid A, Sun J, Riera A, Tognetti S, Herrera MC, Li H, Speck C. A unique DNA entry gate serves for regulated loading of the eukaryotic replicative helicase MCM2-7 onto DNA. Gene Dev. 2014;28:1653–1666. doi: 10.1101/gad.242404.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Bell SD. Archaeal chromosome biology. J Mol Micro BIotech. 2014;24:420–427. doi: 10.1159/000368854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Xu Y, Gadelha C, Stone TA, Faqiri JN, Li D, Qin N, Pu F, Liang YX, She Q, et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 2013;3:485–496. doi: 10.1016/j.celrep.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MR, Morales R, Grainge I, Cook N, Isupov MN, Wigley DB. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J Mol Biol. 2004;343:547–557. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- Sun J, Evrin C, Samel SA, Fernandez-Cid A, Riera A, Kawakami H, Stillman B, Speck C, Li H. Cryo-EM structure of a helicase loading intermediate containing ORC-Cdc6-Cdt1-MCM2-7 bound to DNA. Nat Struct Mol Biol. 2013;20:944–951. doi: 10.1038/nsmb.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachdjian S, Kelly RM. Dynamic metabolic adjustments and genome plasticity are implicated in the heat shock response of the extremely thermoacidophilic archaeon Sulfolobus solfataricus. J Bact. 2006;188:4553–4559. doi: 10.1128/JB.00080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Tsutsumi S, Tsuchiya T, Stillman B, Mizushima T. Functions of Sensor 1 and Sensor 2 regions of Saccharomyces cerevisiae Cdc6p in vivo and in vitro. J Biol Chem. 2002;277:16033–16040. doi: 10.1074/jbc.M108615200. [DOI] [PubMed] [Google Scholar]
- Tiengwe C, Marcello L, Farr H, Gadelha C, Burchmore R, Barry JD, Bell SD, McCulloch R. Identification of ORC1/CDC6-Interacting Factors in Trypanosoma brucei Reveals Critical Features of Origin Recognition Complex Architecture. PLoS ONE. 2012;7:e32674. doi: 10.1371/journal.pone.0032674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Wendler P, Ciniawsky S, Kock M, Kube S. Structure and function of the AAA+ nucleotide binding pocket. Bioch Bioph ACTA. 2012;1823:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Wiedemann C, Szambowska A, Hafner S, Ohlenschlager O, Guhrs KH, Gorlach M. Structure and regulatory role of the C-terminal winged helix domain of the archaeal minichromosome maintenance complex. Nucl Acid Res. 2015;43:2958–2967. doi: 10.1093/nar/gkv120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardimci H, Walter JC. Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol. 2014;21:20–25. doi: 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.