Abstract
The human KIR genes contain multiple promoters that control the process of gene activation and variegated expression of KIR on NK and T cells. Specific subfamilies of KIR genes have differences in the timing and tissue-specificity of expression: however, previous studies of the proximal KIR promoters have not shown significant differences in activity between differentially expressed KIR gene subsets. The recent identification of an intermediate KIR promoter (ProI) associated with KIR2DL1 expression suggested a central role for this element in KIR expression. The current study identifies ProI elements in all of the KIR genes, revealing four classes of ProI that correspond with four distinct expression phenotypes of KIR sub-groups: KIR2DL2/S2/L3 that are expressed early in reconstituting NK after transplant; KIR2DL4 that is expressed by CD56-bright NK in a non-variegated manner; KIR3DL3 that is not expressed by circulating NK cells; and the remaining KIR that are expressed by subsets of CD56-dim NK. The four classes of ProI are structurally diverse and display distinct functional properties. Altogether, these results indicate that KIR ProI elements contribute to the tissue/cell type specificity of KIR transcription, and cooperate with the probabilistic proximal promoter to control KIR expression.
Keywords: human, NK cells, KIR, transcription, AP1
Introduction
The receptors for class I MHC expressed by human and mouse NK cells have evolved from distinct gene families to perform identical functions1. The murine Ly49 class I receptors are related to c-type lectins2, whereas the human killer cell immunoglobulin-like receptors (KIR) are members of the Ig-superfamily3. In addition to their convergent functional evolution, the human and mouse gene families share a specialized pattern of variegated expression resulting in the majority (80%) of NK cells expressing from one to three receptors out of a repertoire of more than ten receptors in any given genotype4–7. This generates subsets of NK cells that are tuned to recognize altered expression of specific class I alleles. The KIR and Ly49 gene families make use of a similar molecular mechanism to generate stochastic expression on NK cells8. Bi-directional promoters are found upstream of each gene, and the relative affinity of binding sites for transcription factors involved in sense versus antisense promoter activity determines the probability of generating the sense transcript required for gene activation. In the Ly49 genes, the bi-directional promoter is only active in immature NK and it is located several kb upstream of the promoter associated with Ly49 expression in mature NK cells. There is a clear separation of probabilistic gene activation from the onset of protein expression in the mouse genes. In contrast, the bi-directional KIR promoter is adjacent to the first coding exon of the gene and it produces sense transcripts in mature, KIR-expressing NK cells, suggesting a role for this element in both gene activation and protein expression. A recent study of a weakly expressed KIR2DL1 allele demonstrated that transcription from an additional promoter (ProI) located ~300 bp upstream was required for KIR2DL1 expression9. ProI transcripts correlated with protein expression, whereas proximal transcripts did not. In addition, non-translatable splice variants of the proximal transcript were detected in cells with decreased ProI activity. These results suggested that the principal role of the proximal bi-directional promoter was to control variegated expression, similar to the distal Ly49 element, and that a separate upstream promoter was required for protein expression. A previous study of KIR2DL4 transcripts had revealed the presence of distal and intermediate promoter transcripts in this non-variegated KIR10. A distal promoter is located 11 kb upstream and an intermediate promoter is present approximately 900 bp upstream of the KIR2DL4 translation initiation site. Transcripts from the distal and intermediate KIR2DL4 promoters are spliced to a site 190 bp upstream of the KIR2DL4 start codon, allowing translation of these messages, thus bypassing the variegated expression associated with the bidirectional proximal promoter.
In order to determine if the presence of intermediate promoters was a general feature of KIR gene regulation, we searched for the presence of ProI elements in all members of the KIR gene family. The current study reveals that all KIR genes possess intermediate promoters, and the characteristics of ProI elements define KIR sub-groups that have distinct expression patterns. ProI therefore appears to represent the key element controlling KIR expression patterns, and the primary function of the proximal promoter is the control of variegated expression.
Results
Identification of KIR2DL1-related ProI elements
A comparison of the region upstream of the KIR2DL1 ProI transcription start site revealed a high degree of sequence conservation in many KIR genes, with the exception of KIR2DL4, KIR2DL2/L3/S2, KIR3DL2, and KIR3DL3 (Figure 1). Interestingly, the genes with low homology represent KIR with distinct expression characteristics. KIR2DL4 is expressed by CD56-bright and dim NK cells in a non-variegated manner11, 12, KIR2DL2/L3/S2 are the first receptors to be expressed by NK cells after transplant13, KIR3DL2 is more highly expressed by T cells than other KIR14, and KIR3DL3 is not expressed by circulating CD56 dim NK cells, but has been found in decidual CD56 bright NK cells15. A luciferase reporter assay system was used to test the promoter activity of the predicted promoter regions shown in Figure 1a. The genes with a high degree of homology to KIR2DL1 demonstrated varying levels of activity, but no ProI activity was observed for this region of the KIR2DL4, KIR2DL2/L3/S2, and KIR3DL2 genes (Figure 1b). The KIR3DL3 sequence present in this region had very little homology to KIR2DL1, but it was found to have a high level of promoter activity in uterine cancer lines.
Figure 1.
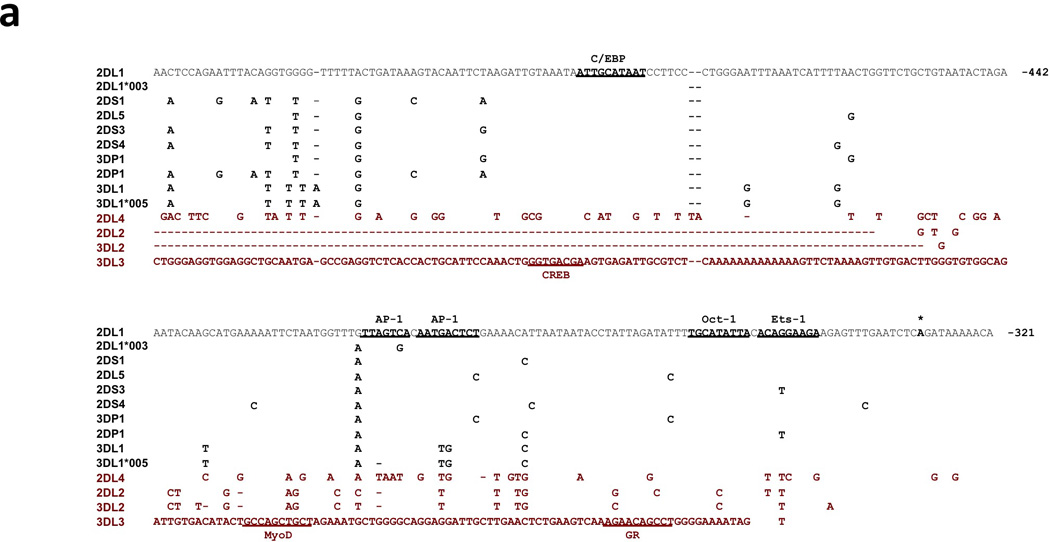
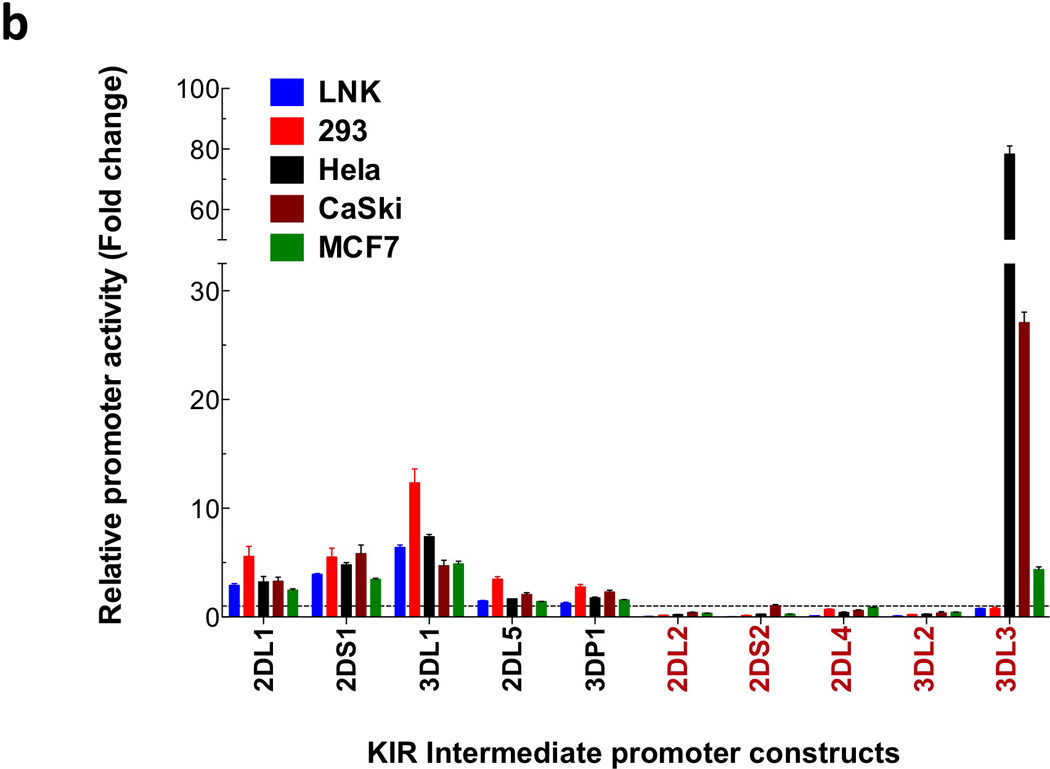
Comparison of the KIR2DL1 ProI region with other KIR genes. (a) The 240 bp ProI region of KIR2DL1 is shown, with only nucleotide differences displayed for other KIR genes. The location of the KIR2DL1 sequence relative to the start codon of the gene is indicated in bold at the right end of each line of sequence. Dashes indicate deletions relative to the KIR2DL1 sequence. Underlined bold sequence represents putative transcription factor-binding elements. The single bold A residue denoted by an asterisk indicates the transcription start site determined for KIR2DL1 ProI. KIR2DL1*003 represents the unique sequence found in the KIR2DL1*003 allele, and KIR3DL1*005 represent the sequence found in the KIR3DL1*001, *004, and *005 alleles. KIR listed in red print are genes with low homology to the KIR2DL1 ProI region. The complete sequence of the KIR3DL3 gene upstream of the Ets-1 site is shown due to a very low level of homology with KIR2DL1 in this region. (b) Luciferase activity of pGL3 constructs containing the KIR promoter regions depicted in (a). Constructs were transfected into the cell lines listed, and relative luciferase activity was determined 48 hours post-transfection. Values represent the mean, and error bars indicate the standard deviation of at least 3 independent experiments. KIR genes listed in red type correspond to the divergent KIR gene sequences identified in (a).
Allele and gene-specific polymorphisms in AP-1, Oct-1, and Ets-1 binding sites affect transcription factor binding
The KIR genes with a KIR2DL1-related ProI element were analyzed for the presence of putative transcription factor binding sites. Potential C/EBP, AP-1, Oct-1, and Ets-binding sites were identified. The C/EBP-binding site is conserved in all members of this class of ProI. However, gene-specific nucleotide changes are present in the AP-1 and Oct-1/Ets sites that are predicted to change the affinity for their respective transcription factors. In addition, the pair of adjacent AP-1 binding sites possess allele-specific polymorphisms in the KIR2DL1*003 and KIR3DL1*005 genes (Figure 1a). Gel-shift analysis was conducted with AP-1 or Oct-1/Ets site probes corresponding to individual KIR genes and alleles (Figure 2). The nucleotide changes present in the KIR2DL1*003 and KIR3DL1*005 probes resulted in a loss of AP-1 binding. Comparison of the tandem Oct-1 and Ets-1 binding sites revealed a loss of Ets-1 binding with probes from the KIR2DL2 and KIR2DS3 genes that have a G to T substitution in the core Ets-binding motif (Figure 1; Figure 2b). The KIR2DL2 gene has an additional T to C substitution in the Oct-1 site (Figure 1) that decreased observed binding to Oct-1 (Figure 2b).
Figure 2.
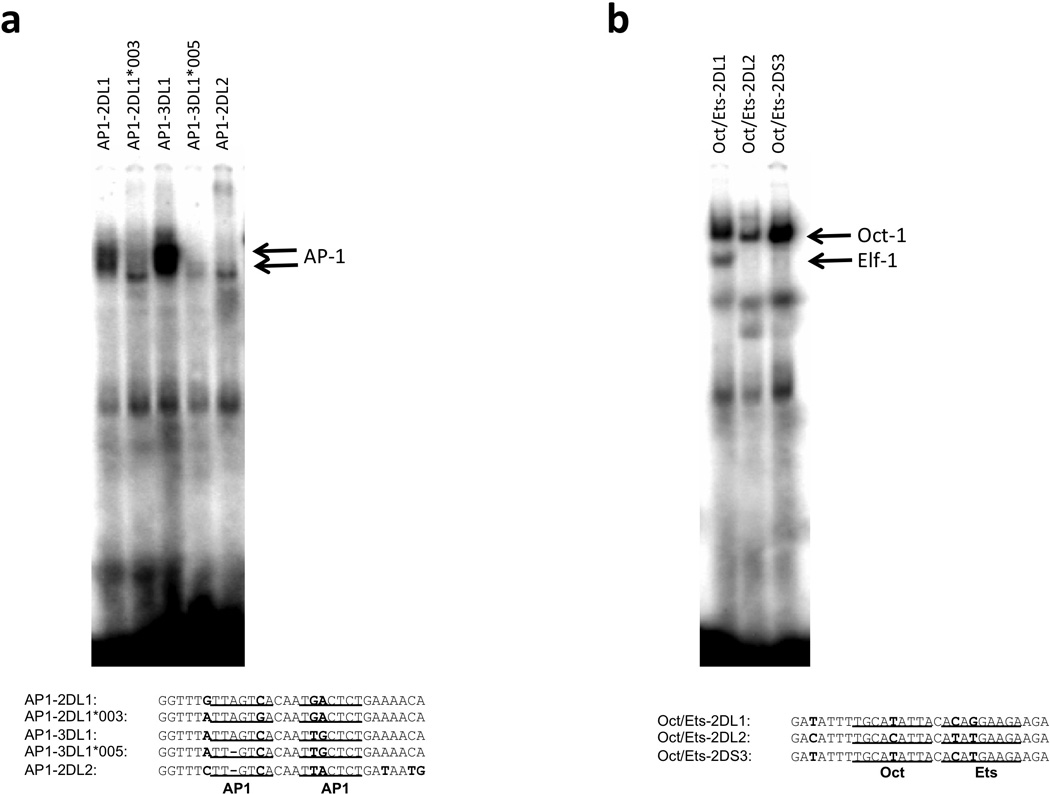
ProI SNPs in predicted binding sites alter transcription factor binding. (a) EMSA analysis of the predicted AP-1 binding region. Jurkat nuclear lysates were incubated with the oligonucleotides shown below. Predicted AP1-binding sites are underlined, and nucleotide differences are in bold type. The bands corresponding to an AP1 complex are indicated by arrows. (b) EMSA analysis of the tandem Oct and Ets sites in Jurkat nuclear lysates. Oligonucleotide probes are shown below with Oct and Ets sites underlined, and nucleotide differences in bold type. Oct-1 and Elf-1 complexes are indicated by arrows.
A recent study of expression patterns of KIR alleles confirmed the low expression intensity of KIR3DL1*005 relative to other KIR3DL1 alleles, and revealed that the average intensity of KIR2DL1*003 expression is lower than the KIR2DL1*002 allele16. Therefore, reduced intensity of KIR2DL1 expression may be associated with a reduced activity of the KIR2DL1*003 ProI element. Interestingly, the average frequency of expression of KIR2DL1*003 was slightly higher than KIR2DL1*002, suggesting that the intermediate promoter polymorphism does not affect the frequency of KIR positive cells.
Analysis of the enhanced activity of the KIR3DL3 intermediate promoter in cervical cancer lines
Although the sequence of the KIR3DL3 ProI fragment 5’ of the Ets-1 site had no significant homology to KIR2DL1, it possessed a high level of activity in two cervical cancer cell lines, Hela and CaSki (Figure 1b), suggesting that this gene might be expressed in uterine tissue. However, RT-PCR analysis of these cell lines did not detect any KIR3DL3 mRNA (data not shown), indicating that the increased transcriptional activity in Hela and CaSki may not be due to tissue specificity of the KIR3DL3 promoter per se, but instead it may be associated with changes in transcription activity due to transformation of these cervical cancer cell lines by papilloma virus, with the KIR3DL3 ProI being responsive to the virus-induced alterations.
The KIR3DL3 ProI region contains a putative CREB/AP1 element. EMSA revealed that the CREB/AP1 site produced a strong complex in Hela cells, with only faint bands appearing in the other nuclear lysates tested, suggesting that this region plays a role in the enhanced activity of the KIR3DL3 ProI element in these cells (Figure 3a). Antibody-inhibition of complex formation on the putative CREB-binding site revealed that numerous members of the Fos, Jun and CREB family of transcription factors were capable of binding to this element (Figure 3b). A comparison of the factors bound to the KIR3DL1 AP1 site in Hela nuclear lysate showed a similar pattern of Fos/Jun binding, but no detectable ATF/CREB binding to the KIR3DL1 AP1 site (Figure 3c). This indicates that the higher activity of the KIR3DL3 ProI element in Hela cells relative to other cell types may be due to higher expression of CREB family members in these cells, or perhaps a result of papilloma virus transformation and production of the viral E7 protein that is known to associate with the AP-1 family of transcription factors17.
Figure 3.
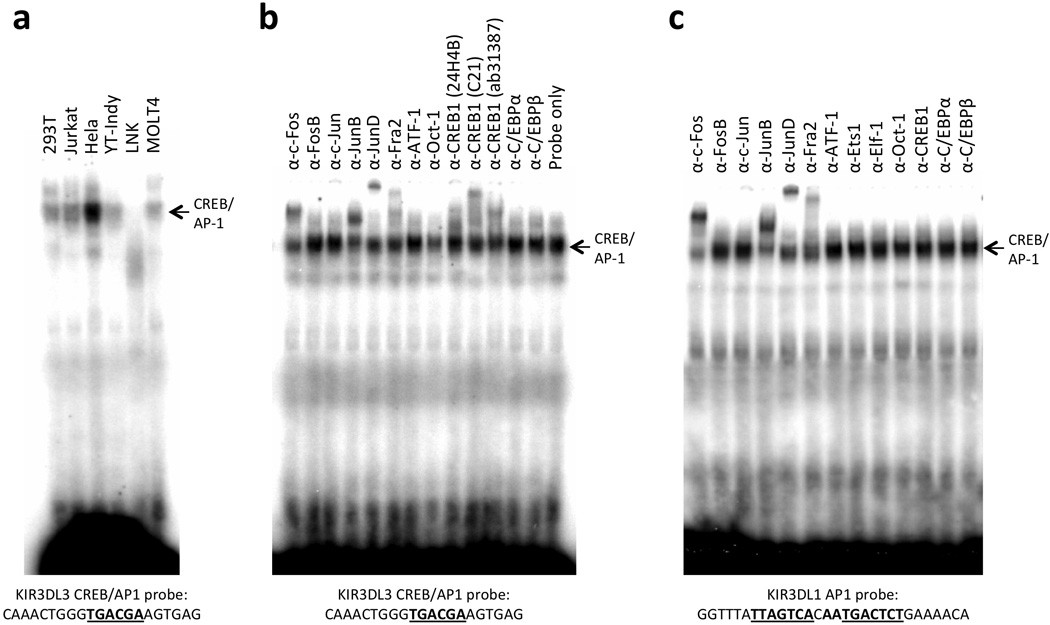
Enhanced formation of CREB/AP1 complexes in Hela cell nuclear lysates. (a) Comparison of complexes formed by a probe containing the KIR3DL3 CREB/AP1 site in nuclear extracts from various cell lines. Probe sequence is shown underneath the panel with the CREB site underlined. The position of the CREB/AP1 complex is indicated by the labeled arrow. (b) The ability of a panel of antibodies reacting with AP1, CREB or control transcription factors to inhibit the complex formed in Hela cells is shown. Probe and CREB/AP1 complex are indicated as in (a). The numbers in parentheses after each of the CREB1 antibodies indicate different commercial antibodies tested. (c) Similarity of AP1 binding but not CREB binding to the KIR3DL1 tandem AP1 sites. The sequence of the probe used is shown below, with the AP1 sites underlined. The position of the CREB/AP1 complex is indicated by the labeled arrow.
The KIR2DL2/L3/S2 intermediate promoter is located further upstream
A previous examination of KIR2DL4 distal transcripts revealed the presence of an intermediate promoter approximately 900 bp upstream of the translation initiation site10, indicating that a ProI-like element might exist elsewhere for the other non-homologous genes. Due to the lack of detectable promoter activity in the KIR2DL2/L3/S2, and KIR3DL2 region homologous to KIR2DL1 ProI, a search for a distinct KIR2DL2 ProI was initiated. 5’-RACE was performed for the KIR2DL2 gene using primers upstream of the proximal promoter transcription start site, and a series of upstream gene fragments were tested for promoter activity. Figure 4a shows the KIR2DL2 region found to contain promoter activity. Mapping of promoter activity through the analysis of a series of PCR fragments localized optimal transcription activity to the region from −864 to −438 relative to the translation start site (Figure 4b). This region contains putative binding sites for YY1, Oct-1, IRF-1, C/EBP, and TBP. The transcriptional start site mapped by 5’ RACE was located within the nucleotide sequence of the 3’ KIR2DL2 primer associated with the highest promoter activity (−438; Figure 4b). There is a TATA-like element located 24 bp upstream of this start site, and a C/EBP site is located 59 bp upstream, indicating the structure of a typical TATA- based promoter. A construct lacking the transcription initiation site (−953 to −566 fragment; Fig 4c) has reduced promoter activity, and a construct lacking the TATA element had no detectable activity (−953 to −589 fragment; Fig 4c). The 5’ boundary of the effective promoter region is defined by a repetitive element insertion containing distal promoter elements that is present only in the KIR2DL2/L3/S2, KIR3DL2 genes. The decrease in promoter activity seen in constructs containing additional 5’ sequence (−1320, −1240, −1141) may be due to the generation of transcripts from the distal region that inhibit ProI function.
Figure 4.
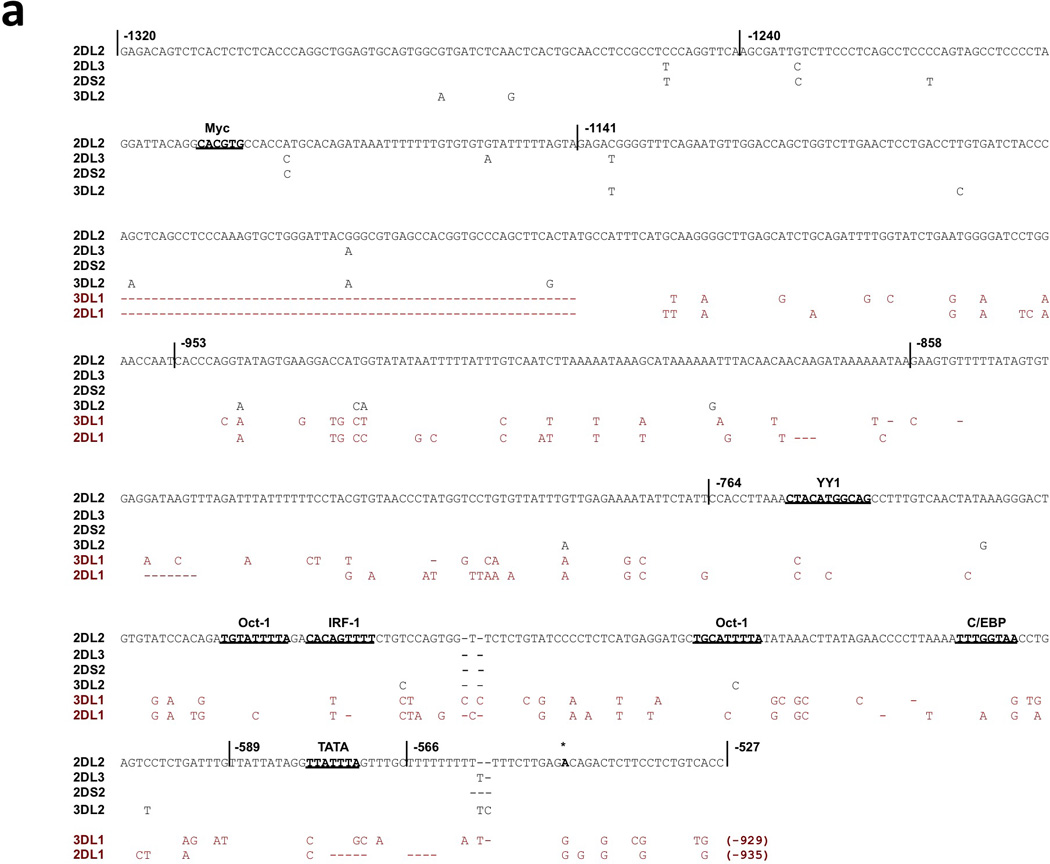
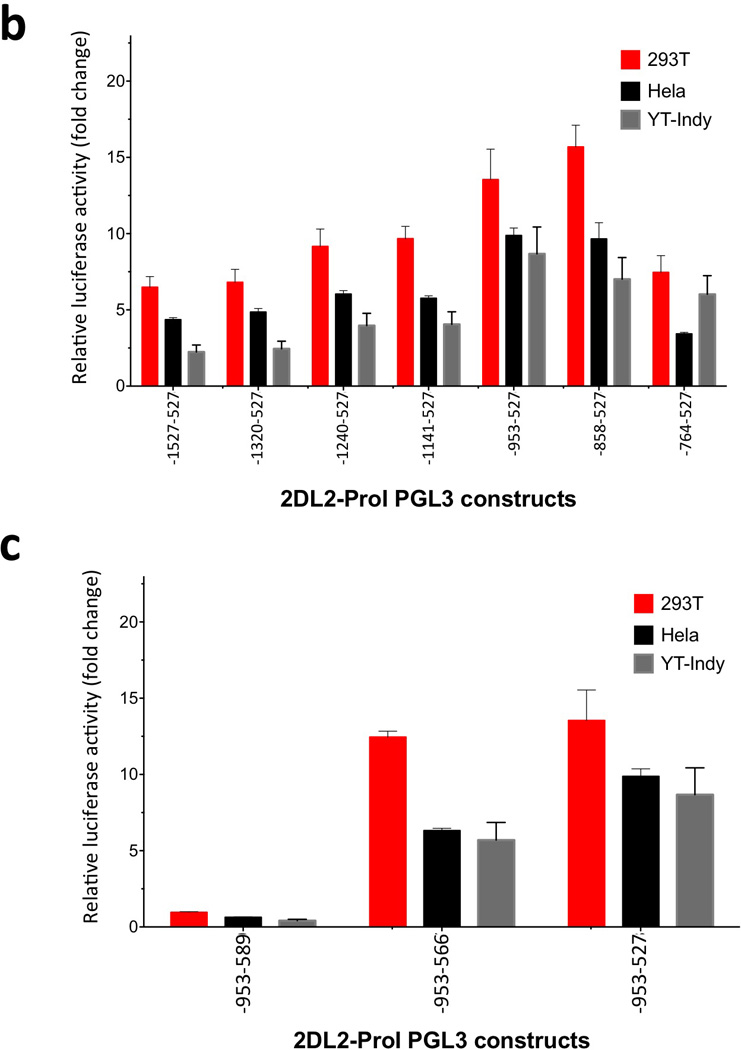
Identification of the KIR2DL2/L3/S2 and KIR3DL2 ProI region. (a) The sequence of the 793 bp KIR2DL2 region tested for ProI activity is shown with only nucleotide differences shown for the other KIR genes. The vertical lines through the sequence indicate the ends of PCR fragments tested for promoter activity as shown in (b), and the numbers shown indicate the position relative to the start codon of the KIR2DL2 gene. Predicted transcription factor binding sites are underlined in bold type. The non-conserved KIR2DL1 and KIR3DL1 sequences are shown in red type. The dashed lines show the location of a region that is deleted in the KIR2DL1 and KIR3DL1 genes. The single bold A residue denoted by an asterisk indicates the transcription start site determine for KIR2DL2 ProI. (b) Mapping of the 5’ boundary of KIR2DL2 ProI activity. PCR fragments were generated with a series of 5’ primers together with a common 3’ primer containing the transcription start site and cloned into the pGL3 reporter vector. Constructs were transfected into the cell lines listed, and relative luciferase activity was determined 48 hours post-transfection. Values represent the mean, and error bars indicate the standard deviation of at least 3 independent experiments. (c) Mapping of the 3’ boundary of KIR2DL2 ProI. Two additional pGL3 constructs were generated by PCR using the −864 primer found to mark the 5’ end of the active promoter region and 3’ primers deleting either the transcription initiation site (−477) or the putative TATA element (−500). The activity of the −864 to −438 construct is shown again for comparison.
Transcription factors bound by the KIR2DL2 Pro-I region
Oligonucleotide probes corresponding to putative YY1, Oct, IRF, and C/EBP sites from the KIR2DL2 ProI region were tested on nuclear lysates from Jurkat, and YT cells (Figure 5). Strong complexes were observed in Jurkat lysates with Oct and YY1 probes, with a weaker complex observed for C/EBP, and no significant binding to the Oct-IRF probe (Figure 5b). To confirm the identity of factors bound to each of the three probes that produced complexes in Jurkat nuclear lysates, inhibition was performed with a panel of antibodies. For each of the three predicted transcription factor-binding sites, complex inhibition or super-shift was observed with antibodies reactive with the transcription factor predicted to interact with the probe sequence (Figure 5c), indicating a potential role for YY1, Oct-1, and C/EBP in KIR2DL2 ProI activity.
Figure 5.

Identification of transcription factors binding to the KIR2DL2 ProI region. (a) Probes containing predicted binding sites are listed with the putative binding region underlined. (b) EMSA of the probes listed in (a) performed with either Jurkat (left panel) or YT-Indy (right panel) nuclear extracts. (c) Antibody inhibition of complexes formed in Jurkat nuclear lysates with Oct (left panel), YY1 (center panel), or C/EBP (right panel) probes. Antibodies used are listed above each lane, and those shown in red type produced complex inhibition and/or supershift.
Discussion
The initial characterization of the KIR promoter elements immediately upstream of the coding region18, 19 revealed a high degree of sequence conservation (91.1–99.6%), with only KIR2DL4 possessing a significantly divergent promoter structure (67% homology to KIR3DL1), consistent with its non-variegated expression at the earlier CD56-bright stage of NK cell development. The KIR promoters were divided into four groups based on polymorphisms found in YY1 and Sp1-transcription factor-binding sites: group 1= most variegated KIR genes; group 2= KIR2DL1/S1 and KIR2DL5/S5; group 3= KIR3DL3; group 4=KIR2DL4. Although these groupings provided a potential explanation for the distinct expression patterns of KIR3DL3 and KIR2DL4, they could not account for the observation that KIR2DL2/S2 and KIR2DL3 were the first KIR to be expressed by NK cells developing in vitro20, or after hematopoietic stem cell transplantation12. The subsequent discovery of the bi-directional nature of the KIR proximal promoter revealed that the polymorphisms found in the YY1 and Sp1 sites influenced the relative strength of competing sense and antisense promoters, and the loss of Sp1- binding correlated with increased sense promoter activity and a higher frequency of expression by NK cells21, 22.
The KIR gene groupings generated as a result of identifying the ProI elements upstream of the bi-directional proximal KIR promoter may provide an explanation for the more rapid expression of the KIR2DL2/S2/L3 genes in reconstituting NK. The inclusion of the framework KIR3DL2 gene in this group indicates that it should also be expressed early, however previous studies of reconstituting NK have not tested KIR3DL2 expression. The higher frequency of T cells expressing KIR3DL2 than other KIR may reflect an expanded window of opportunity for KIR3DL2 expression due to ProI activation earlier in T cell development.
A comparison of the intergenic regions of KIR family members (Figure 6) reveals that the distinct ProI elements have arisen as a result of differential insertion/deletion of repetitive elements in the region upstream of the conserved bi-directional proximal promoter. The KIR2DL1-related ProI elements are in a region of non-repetitive DNA, however the three other classes of ProI are all associated with repetitive elements. The KIR2DL2 and KIR2DL4 intermediate promoters are contained within LINE elements, whereas KIR3DL3 ProI contains a unique Alu element insertion. It is of interest to note that the previously characterized KIR3DL1 distal promoter (Pro-D)8, 10 is located within the same L1M5 LINE element that contains KIR2DL2 ProI. It appears that the KIR2DL2-related genes have replaced the KIR2DL1-associated Pro-I with a distal promoter element due to a 406 bp deletion of the ProI region. This event may account for the expression of KIR2DL2/L3/S2 earlier in NK development than other variegated KIR, since transcription from the distal promoter was detected at the earliest stages of NK cell differentiation23.
Figure 6.
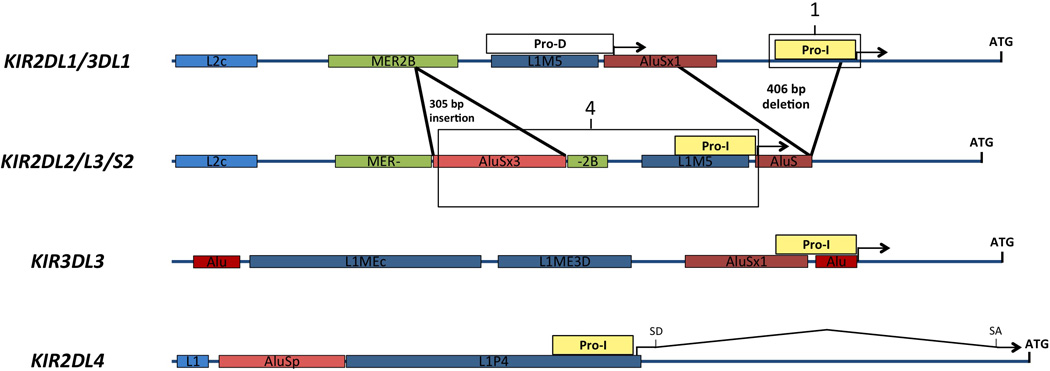
Generation of distinct ProI elements as a result of repetitive element insertion and deletion. A schematic of the four classes of KIR genes based on ProI analysis is shown. The start codon of each gene is indicated by (ATG) over a vertical line. Yellow rectangles labeled “ProI” indicate the intermediate promoters characterized. The boxes labeled (1) and (4) represent the promoter regions depicted in Figures 1a and 4a respectively. The white rectangle labeled “Pro-D” represents the previously characterized distal KIR3DL1 promoter. Insertions and deletions in KIR2DL2 relative to KIR2DL1 are indicated by heavy black lines marking locations of the events. Repetitive elements of different classes are indicated by different colored rectangles labeled with the name of the element. For the KIR2DL4 gene, (SD) and (SA) indicate the location of the splice donor and splice acceptors used to produce a spliced intermediate transcript.
Most KIR genes contain an approximately 2 kb intergenic region separating the start codon of one KIR gene from the polyA-addition site of the previous gene. The majority of studies examining factors that control KIR transcription have concentrated on the 300 bp immediately upstream of the start codon. The current study of intermediate promoters together with previous analyses of distal promoter elements indicates that the majority of the intergenic region plays a direct role in the regulation of transcription. The recent discovery of a direct correlation between intermediate promoter activity and protein expression of KIR2DL19, raises the possibility that ProI elements control KIR expression, and the proximal promoter region exists solely to generate variegated expression of the KIR genes via the stochastic production of antisense RNA, leading to gene silencing8. However, methylation of the proximal KIR promoter has been shown to control KIR expression24,25, indicating that, at a minimum, the proximal promoter cooperates with the upstream promoter to enable expression, since no KIR transcripts are generated if the proximal promoter region is methylated. A role for methylation of ProI in gene silencing is excluded by the observation that there are no CpG sites in the KIR2DL1 intermediate promoter region, while 18 CpG sites are present in the proximal promoter region. In addition, a RUNX-binding site located in the proximal promoter has been shown to be required for KIR expression, as non-expressed variants of the KIR2DL5 gene lack RUNX binding to this region26. Furthermore, demethylation of the silent KIR2DL5 allele gene leads to its expression, revealing a role for RUNX in gene activation. A possible collaboration between ProI and the proximal promoter in gene demethylation and activation is suggested by observations made on the KIR3DL3 gene. Trompeter et al.27 demonstrated that although KIR3DL3 contains an intact RUNX-binding site, it is methylated and not expressed in peripheral blood NK cells. However, expression can be induced by demethylating the proximal promoter region, indicating the presence of a fully functional promoter. Perhaps the lack of detectable KIR3DL3 ProI activity in NK cells revealed by the current study represents the missing element required for KIR3DL3 proximal promoter demethylation. Previous studies of distal KIR promoters revealed that the KIR3DL3 distal element is inverted and transcribes only in the antisense direction10. This observation, coupled with a lack of ProI transcription in NK cells, would result in a lack of distal transcripts traversing the proximal promoter region that may be required for access of RUNX to the DNA and subsequent demethylation of the region.
Whether or not the ProI elements of all KIR genes are absolutely necessary to drive protein expression is not known but can be tested by performing quantitative RT-PCR experiments comparing proximal and distal transcripts across all KIR genes. It will be important therefore, to assay levels of ProI transcripts in future studies examining the relationship between levels of KIR transcription and the intensity of receptor expression on the surface of NK cells.
Materials and Methods
Cell lines
HEK293 cells were cultured in DMEM media containing 10% fetal bovine serum (FBS), 100 U/ml each of penicillin and streptomycin (P/S), sodium pyruvate, and L-glutamine. YT-Indy and Jurkat cells were cultured in RPMI 1640 media containing 10% FBS and P/S. The LNK cell line was cultured in RPMI 1640 containing beta-mercaptoethanol, nonessential amino acids, 10% FBS, 100 U/ml P/S, sodium pyruvate, L-glutamine, Hepes, and 1000 U/ml of recombinant human IL-2. The cancer cell lines, Hela, CaSki and MCF7, were obtained from American Type Culture Collection (ATCC). Cells were maintained according to the supplier’s instructions.
5’ RACE Analysis of KIR2DL2 ProI Transcripts
5’ RACE was performed on KIR2DL2-expressing NK92 cell RNA using the Invitrogen 5’ RACE System (Invitrogen, Carlsbad, CA, USA) following the manufacturers directions. Gene-specific cDNA was generated using a KIR2DL2/L3/S2-specific antisense primer located 147 bp upstream from the proximal promoter transcription start site (TSS): (5’-TTGACACCTTGCGTCCTTCACTAC-3’). A poly-dC tail was added to the cDNA and first round PCR amplification was performed using a poly-dG-anchor primer 5’ and a KIR2DL2 antisense primer located 294 bp upstream of the proximal TSS: (5’-GTAATGTGCAAAATGTCTAACAGG-3’) at an annealing temperature of 59° C for 35 cycles with Platinum PCR Supermix (Invitrogen). Excess primers from first round PCR reactions were removed with the Charge-Switch PCR Clean-Up Kit (Invitrogen). Nested PCR was subsequently performed with the anchor primer and a KIR2DL2 antisense primer located 366 bp upstream of the proximal TSS: (5’-TCCTGCTTGAGTTTCTAGTACTAA-3’) at an annealing temperature of 60° C for 30 cycles. PCR products were cloned into pCR2.1 Topo (Invitrogen) and sequences analyzed to map the 5’ start sites.
RT-PCR of KIR transcripts
Total RNA was purified from 1×107 cells with the RNeasy kit (Qiagen, Valencia, CA, USA) and cDNA synthesis was carried out using Random Hexamer primer, Taqman Reverse Transcription Reagents kit (Applied Biosystems Foster City, CA, USA) according to the manufacturer's instructions. Reactions were carried out using FastStart SYBR Green Master kit (Roche Diagnostics, Indianapolis, IN, USA) on the 7300 Real-Time PCR System (Applied Biosystems). The qRT-PCR was performed in duplicate and was repeated in at least three separate experiments9. Melting curve analyses were performed to verify the amplification specificity. Relative quantification of gene expression was performed according to the ΔΔ-CT method using StepOne Software 2.0 (Applied Biosystems).
Electrophoretic mobility shift assays (EMSA)
Nuclear extracts were prepared from the various cell lines using the CellLytic NuCLEAR extraction kit (Sigma-Aldrich, St. Louis, MO). Protein concentration was measured with a Bio-Rad protein assay (Hercules, CA), and samples were stored at −70°C until use. Double-stranded DNA oligonucleotide probes corresponding to the predicted TF binding sites of KIR ProI region were synthesized (Figures 2, 3, 5). Labeling and DNA-protein binding reactions were performed as previous described9. For antibody supershift experiments, nuclear extracts were incubated with 2 µL of antibody for 1 h on ice before the addition of 32P-labeled DNA probe. After the addition of labeled DNA probe, the binding reaction was incubated for an additional 20 min at room temperature. The antibodies used were cFos (6-2H-2F), FosB (102), cJun (H-79), JunB (C-11), JunD (329), Fra-2 (Q-20), ATF-1 (FI-1), Ets-1 (C-4), Elf-1 (C-20), Oct-1 (E-8, C-21 and 12F11), Oct-3/4 (C-10), Oct-2 (C-20), CREB-1 (24H4B), C/EBPα (D-5), C/EBPβ (H-7), C/EBPγ (H-50), SP-1 (E-3), IRF-1 (C-20), IRF-3 (SL-12), ICSBP (C-19), PU.1 (A-7), E4BP4 (B-1), and MyoD (C-20) from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) at gel shift grade (200 µg/0.1µl)
Generation of luciferase reporter plasmids
Promoter fragments were generated by PCR using the primers listed in Table 1 and cloned into the TOPO-TA vector (Invitrogen). Inserts were excised with XhoI/HindIII and cloned into pGL3 (Promega, Madison, WI, USA) to generate constructs in forward orientation. All constructs were verified by sequencing. Sequence analysis was carried out with the SeqWeb package at the NCI-Frederick supercomputing center and the Molecular Evolutionary Genetics Analysis (MEGA) software version 4.5.
Table 1.
Sequences and locations of primers used in this study
| Primer name & amplication | Sequence (5′ to 3′) | Construct length (bp) | |
|---|---|---|---|
| ProI promoter assay | |||
| Forward primer | |||
| 2DL1-For | AAACTCCAGAATTTACAGGTGGGG | ||
| 2DS1-For | AAAATCCAGAGTTTAAATGTGTGG | ||
| 3DL1-For | AAAATCCAGAATTTACATGTTGTG | ||
| 2DL5-For | AAACTCCAGAATTTACAGGTGTGG | ||
| 2DL2/3DL2-For | CTTGAGACAGACTCTTCCTCTGTCAC | ||
| 2DL4-For | CAGACCTTCAATTGACATATTGTG | ||
| 3DL3-For | CTGGGAGGTGGAGGCTGCAATGAG | ||
| Reverse primer | |||
| 2DL1/3DL1-Rev | GAGATTCAAACTCTTCTTCCTGTG | ||
| 2DL2/2DS2-Rev | GAGATTCAAACTCTTCTTCATATG | ||
| 2DL4-Rev | GAGATTCAAACTCTCCTTGATATG | ||
| 3DL2-Rev | GAGATTCAAACTTTTCTTCATGTG | ||
| 3DL3-Rev | GAGATTCAAACTCTTCTTCATGTG | ||
| 2DL2 5'-deletion assay | |||
| 2DL2-1527-For | ACAGGTGTTTATTGAACCAAC | 1,001 | |
| 2DL2-1320-For | GAGACAGTCTCACTCTCTCACC | 794 | |
| 2DL2-1240-For | AGCGATTGTCTTCCCTCAGCCTCC | 714 | |
| 2DL2-1141-For | GAGACGGGGTTTCAGAATGTTGG | 615 | |
| 2DL2-953-For | CACCCAGGTATAGTGAAGGACC | 427 | |
| 2DL2-815-For | CCTACGTGTAACCCTATGGTCC | 289 | |
| 2DL2-764-For | CCACCTTAAACTACATGGCAGCC | 234 | |
| 2DL2-527-Rev | GGTGACAGAGGAAGAGTCTGTCTC | ||
| 2DL2 3'-deletion assay | |||
| 2DL2-953-For | CACCCAGGTATAGTGAAGGACC | ||
| 2DL2-589-Rev | CAAATCAGAGGACTCAGGTTACC | 365 | |
| 2DL2-566-Rev | GCAAACTAAATAACCTATAATAACAAATCAGAGG | 388 | |
| 2DL2-527-Rev | GGTGACAGAGGAAGAGTCTGTCTC | 427 | |
Cell transfection and luciferase assays
The cell lines, including breast cancer cell line MCF7, cervical cancer cell lines Hela and CaSki, mouse NK cell line LNK, and human embryonic kidney 293T cells, were plated at 1×105 cells per well in a 24-well plate the day before transfection, and incubated overnight at 37°C in 5% CO2. For each well, 2–5 µl of HilyMax transfection reagent (Dojindo, Rockville, MD, USA) was diluted in 30 µl of growth medium without serum and incubated at room temperature for 5 min. The DNA mixture containing 1 µg of the specific reporter construct plus 5–25 ng of Renilla luciferase pRL-SV40 control DNA was then added to each well, and incubated at room temperature for 20 min. YT-Indy cells were transfected by electroporation with a BTX ECM 830 (Gentronics, San Diego, CA, USA) set at 500mV, with 5 pulses of 400 µs at an interval of 1 s. A total of 5×106 cells in 0.5 ml of serum free RPMI medium were transfected with 10µg of the specific reporter construct plus 0.1µg of the Renilla luciferase pRL-SV40 vector. Luciferase activity was assayed at 48 hr using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Measurement of the firefly luciferase activity of the KIR-ProI promoter constructs was normalized relative to the activity of the Renilla luciferase produced by the pRLSV40 control vector and each construct was tested in triplicate in at least three independent experiments.
Statistical analysis
Mann-Whitney-U and two-tailed t-test were performed using GraphPad Prism version 5 for Windows; p<0.05 was regarded to be statistically significant.
Acknowledgements
This project has been funded in whole or in part with federal funds from the National Cancer Institute (NCI), NIH, under contract HHSN261200800001E. This research was supported in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government.
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Anderson SK, Ortaldo JR, McVicar DW. The ever-expanding Ly49 gene family: repertoire and signaling. Immunol Rev. 2001;181:79–89. doi: 10.1034/j.1600-065x.2001.1810106.x. [DOI] [PubMed] [Google Scholar]
- 3.Carrington M, Norman P. The KIR Gene Cluster. Bethesda, MD: National Library of Medicine (US), NCBI; 2001. [Google Scholar]
- 4.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 5.Held W, Kunz B. An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire. Eur J Immunol. 1998;28:2407–2416. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 6.Kubota A, Kubota S, Lohwasser S, Mager DL, Takei F. Diversity of NK cell receptor repertoire in adult and neonatal mice. J Immunol. 1999;163:212–216. [PubMed] [Google Scholar]
- 7.Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson SK. Probabilistic bidirectional promoter switches: noncoding RNA takes control. Mol Ther Nucleic Acids. 2014;3:e191. doi: 10.1038/mtna.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright PW, Li H, Huehn A, O'Connor GM, Cooley S, Miller JS, Anderson SK. Characterization of a weakly expressed KIR2DL1 variant reveals a novel upstream promoter that controls KIR expression. Genes Immun. 2014;15:440–448. doi: 10.1038/gene.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 11.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D'Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7:739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 12.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer JC, Ottinger H, Ferencik S, Sribar M, Punzel M, Beelen DW, et al. Relevance of C1 and C2 epitopes for hemopoietic stem cell transplantation: role for sequential acquisition of HLA-C-specific inhibitory killer Ig-like receptor. J Immunol. 2007;178:3918–3923. doi: 10.4049/jimmunol.178.6.3918. [DOI] [PubMed] [Google Scholar]
- 14.Chan AT, Kollnberger SD, Wedderburn LR, Bowness P. Expansion and enhanced survival of natural killer cells expressing the killer immunoglobulin-like receptor KIR3DL2 in spondylarthritis. Arthritis Rheum. 2005;52:3586–3895. doi: 10.1002/art.21395. [DOI] [PubMed] [Google Scholar]
- 15.Trundley AE, Hiby SE, Chang C, Sharkey AM, Santourlidis S, Uhrberg M, et al. Molecular characterization of KIR3DL3. Immunogenetics. 2006;57:904–916. doi: 10.1007/s00251-005-0060-7. [DOI] [PubMed] [Google Scholar]
- 16.Dunphy SE, Guinan KJ, Chorcora CN, Jayaraman J, Traherne JA, Trowsdale J, et al. 2DL1, 2DL2 and 2DL3 all contribute to KIR phenotype variability on human NK cells. Genes Immun. 2015;16:301–310. doi: 10.1038/gene.2015.15. [DOI] [PubMed] [Google Scholar]
- 17.Antinore MJ, Birrer MJ, Patel D, Nader L, McCance DJ. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J. 1996;15:1950–1960. [PMC free article] [PubMed] [Google Scholar]
- 18.Stewart CA, van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 19.van Bergen J, Stewart CA, van den Elsen PJ, Trowsdale J. Structural and functional differences between the promoters of independently expressed killer cell Ig-like receptors. Eur J Immunol. 2005;35:2191–2199. doi: 10.1002/eji.200526201. [DOI] [PubMed] [Google Scholar]
- 20.Miller JS, McCullar V. Human natural killer cells with polyclonal lectin and immunoglobulin-like receptors develop from single hematopoietic stem cells with preferential expression of NKG2A and KIR2DL2/L3/S2. Blood. 2001;98:705–713. doi: 10.1182/blood.v98.3.705. [DOI] [PubMed] [Google Scholar]
- 21.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, et al. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 22.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4:e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cichocki F, Hanson RJ, Lenvik T, Pitt M, McCullar V, Li H, et al. The transcription factor c-Myc enhances KIR gene transcription through direct binding to an upstream distal promoter element. Blood. 2009;113:3245–3253. doi: 10.1182/blood-2008-07-166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, et al. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 25.Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, et al. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomez-Lozano N, Trompeter HI, de Pablo R, Estefania E, Uhrberg M, Vilches C. Epigenetic silencing of potentially functional KIR2DL5 alleles: implications for the acquisition of KIR repertoires by NK cells. Eur J Immunol. 2007;37:1954–1965. doi: 10.1002/eji.200737277. [DOI] [PubMed] [Google Scholar]
- 27.Trompeter HI, Gomez-Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C, et al. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4 and of KIR3DL3. J Immunol. 2005;174:4335–4343. doi: 10.4049/jimmunol.174.7.4135. [DOI] [PubMed] [Google Scholar]


