1. Introduction
The Clinical Genome Resource (ClinGen) is an NIH-funded collaborative program that brings together a variety of projects designed to provide high quality, curated information on clinically relevant genes and variants. ClinGen's EHR (Electronic Health Record) Workgroup aims to ensure that ClinGen is accessible to providers and patients through EHR and related systems. This paper describes the current scope of these efforts and progress to date. The ClinGen public portal can be accessed at www.clinicalgenome.org.
2. Overview of ClinGen
ClinGen is building a centralized and authoritative resource that defines the clinical relevance of genes and genomic variants for precision medicine and research. ClinGen began in 2013 as an NIH-funded collaboration between three funded projects. It has since grown to include over 350 researchers and clinicians from over 90 different institutions. ClinGen's diverse membership is organized into various Workgroups that collectively aim to “engage the genomics community in data sharing efforts, develop the infrastructure and standards to support curation activities, and incorporate machine-learning approaches to speed the identification of clinically relevant variants” (www.clinicalgenome.org)[1].
ClinGen's EHR Workgroup is tasked with ensuring that ClinGen is accessible to providers and patients through clinical information systems (i.e., EHR and other related systems). The Workgroup is responding by developing strategies for accessing ClinGen from clinical information systems.
This paper lays out the vision for the EHR Workgroup by:
Providing an overview of ClinGen, including articulation of its place within the EHR ecosystem.
Introducing the scope of its integration efforts to date.
Describing how ClinGen aims to serve as a resource for providing variant interpretations in healthcare settings.
3. ClinGen Resources
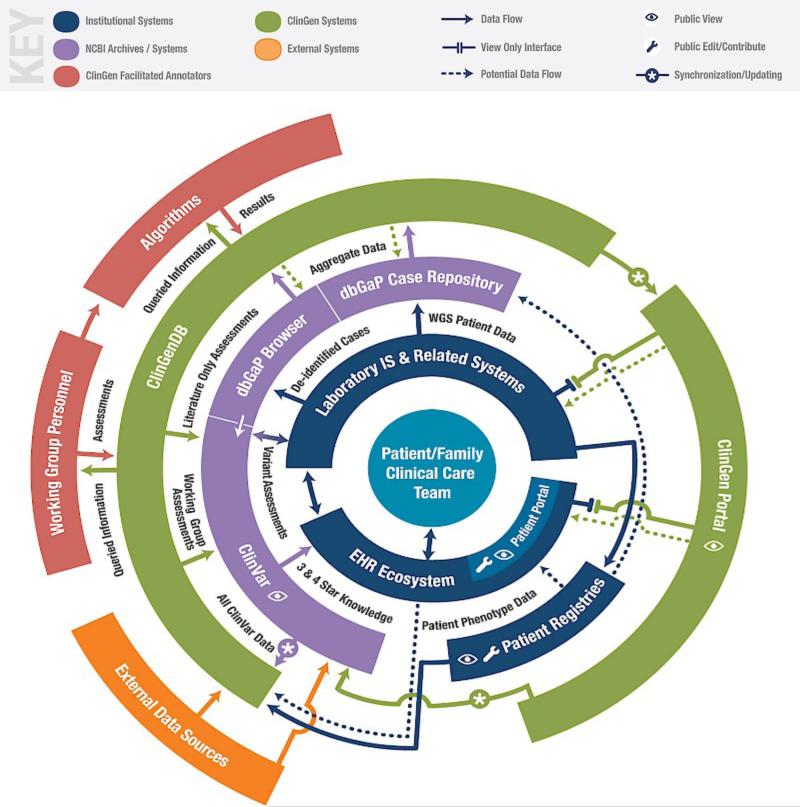
The ClinGen system map (Figure 1) illustrates a patient-centered conceptual framework for ClinGen and provides a vision for the interface of ClinGen resources with the EHR ecosystem and other institutional clinical systems. Table 1 describes many of the resources shown in Figure 1.
Figure 1.
ClinGen System Map (For a full description of the map see http://interactive.clinicalgenome.org/clingen-resource-system-map/)
Table 1.
ClinGen System Map resources
| ClinVar (ClinGen's variant interpretation repository) | ClinVar is a database maintained by the National Center for Biotechnology Information (NCBI) that aggregates information about genomic sequence variation and its relationship to human health. From its public release in April 2013, ClinVar was designed to be a fully public and freely available resource.[2] (www.ncbi.nlm.nih.gov/clinvar/submitters). ClinVar archives assertions of clinical significance for variants related to specific conditions, and thus, relies on voluntary submissions from clinical testing labs, researchers, locus-specific databases, and other groups. |
| GenomeConnect (ClinGen's Patient Registry) | GenomeConnect is a patient registry that collects patient-entered phenotypic information and genetic testing reports. ClinGen created GenomeConnect in September 2014 to give individuals who have completed genetic testing the opportunity to share their genetic and phenotypic information with the clinical and research communities. Participants are also given the opportunity to connect with other families or individuals with the same genetic variant(s). The rich phenotypic and genotypic data in GenomeConnect will eventually serve as a resource for ClinGen curators to use in their interpretation of rare genetic variants and genes implicated in disease. |
| Curation interfaces (under development) | Included among ClinGen's tools and resources under development are the ClinGen curation interfaces. Through deployment of these interfaces, ClinGen aims to provide a standardized means for curating evidence (from ClinVar, published literature, and other sources) regarding both the clinical validity and actionability of genes and the pathogenicity of variants. Experts from an appropriate clinical domain will provide the final assertion for a gene or variant will review this evidence. |
| Other ClinGen Portal tools and resources | Other ClinGen portal tools and resources include: • A search of the structural variation database (formally ISCA), • A standard form to contact a lab to inquire more about a particular case/variant, • IRB Submission Guidance, • Guidance for clinical laboratories submitting to ClinVar, • The interactive ClinGen system map (Figure 1), • The ClinGen Web Resources page (See section 3), • ClinGen pathogenicity calculator, • A link to ClinGen Data Model Workgroup documents and, • Webinars covering: Interpreting Genomic Variants, Using NCBI Resources, Evaluating Copy Number Variants, Dosage Sensitivity Map, GenomeConnect, and more coming soon... |
One of the most critical resources that ClinGen relies on for evidence curation is ClinVar. [1][2] The variant assertions in ClinVar that have been reviewed by an Expert Panel will be available through the ClinGen portal, along with the complete set of evidence found to support each assertion. The resources available at the ClinGen portal, including the curation interface and Web Resources page, will interact with clinical systems and support programmatic access by those systems. Access will include the delivery of information obtained from the expert curation of evidence derived from ClinVar and other sources.
4. ClinGen's EHR Workgroup
The specific aims for this group are to: (1) design strategies for EHR access to ClinGen resources through the use of e-resource/infobutton standards proposed in meaningful use stage 3; (2) optimize the utility of ClinGen by engaging key clinical stakeholders in its design and organization; (3) implement strategies for accessing ClinGen through at least one EHR system and assess its general application to other clinical systems; and (4) coordinate development of a plan for sustainability that outlines resources and processes needed to execute strategies for accessing ClinGen through EHR systems. The Web Resources page is the first ClinGen product to interface with clinical systems.
5. Web Resources Page and OpenInfobutton
The ClinGen Web Resources page provides a “one stop shop” for high quality genomic medicine resources and for exploring the use of “infobuttons” to support access to these resources from EHR systems. Upon completion of the curation interface, the EHR Workgroup will work to specify the formats required to access ClinGen curation products using infobuttons. Our goal to support the information needs of clinicians, researchers and patients motivates these efforts.
The genomic medicine resources for the Web Resources page were selected and organized base on an informal survey of laboratory professionals, researchers, and clinicians attending the ClinGen reception at the 2015 American College of Medical Genetics and Genomics annual meeting. Selected resources were then organized into tabs by target audience and ordered according to the priorities outlined by survey participants. The resulting Web Resources page is available on ClinGen's public portal and is structured to ensure easy access based on the target audience.
In order to facilitate EHR systems’ access to the Web Resources page, the Workgroup is exploring use of the Health Level Seven (HL7) Context-Aware Knowledge Retrieval standard, also known as the “Infobutton Standard.”(www.hl7.org/implement/standards/product_brief.cfm?product_id=208) This standard provides an information model for the semantics and syntax of requests from an EHR to knowledge resources that enables those resources to respond with context-specific information. We have found that many of the sites available on ClinGen's Web Resource page require custom configuration in order to facilitate access via infobutton requests. The Workgroup is using OpenInfobutton, a freely available, HL7-compliant system, to make infobutton implementation possible for EHR systems. It accepts context parameters from an EHR and, using a knowledge base of resources and information needs, generates a set of links that direct the user to relevant information.[3]
These efforts are part of the overall ClinGen public portal development efforts being undertaken by the larger GlinGen membership, and are targeting existing information resources. Activities of the EHR Workgroup converge with and respond/adapt to curation interface development and the products of curation to further specify formats required to access this ClinGen information in a context-specific way using infobuttons. These specifications will then be incorporated into the Web Resources page to interface with various clinical systems. Among Workgroup members are several liaisons to groups internal and external to ClinGen that provide the relevant expertise for such specifications. In defining specifications, the EHR Workgroup interfaces closely with the ClinGen Data Model Workgroup, which has been given the task of creating standards to ensure interoperability between systems within ClinGen.
6. Discussion
Our current efforts focus on the development of the ClinGen Web Resources page for which we can use “infobuttons” to provide passive decision support for the information needs of clinicians, researchers and patients, thus lowering the barriers to access in a healthcare setting. As we work to broaden access to ClinGen, there is the potential for more robust approaches to address those needs.
Genomic sequencing leads to great challenges in information management, education, and communication. For example, from a healthcare consumer/patient perspective, McBride and colleagues found that individuals who use direct-to-consumer (DTC) genetic testing are likely to search for information to help them to understand their results, and found they feel confident in applying this information in making healthcare decisions.[4] It is important, therefore, that high quality genomic information resources are provided in a form that is understandable by patients. Current work with GenomeConnect has the potential to provide patients with access to information. However, success with such a model comes with several challenges given the sensitivity of genomic data and the controversies around (de-)identifiability, potential privacy risks and harms originating from unintended uses of data.[5]
From a healthcare provider perspective, DTC genetic testing customers are also likely to bring their test results to physicians.[6] This highlights a need to provide clinicians with the resources to keep up-to-date on appropriate uses of genetic data, including warding off common misinformation or misconceptions. Though these challenges are true for genetic test results generally, they are particularly salient for genomic sequencing data given the additional uncertainties surrounding the interpretation and reporting of variants of unknown significance (VUS).
A survey study investigating the information needs of cancer genetic counselors indicated that most counselors desired additional information from laboratories on VUS results in order to help contextualize the result for patient preferences. Some professional society guidelines have indicated that the performing laboratory is responsible for interpretation of VUS results, thus concerns have been expressed regarding the potential for healthcare providers to utilize VUS results in guiding patient medical management.[7] It is likely that information preferences for using genomic sequencing results will differ for different patients and clinical settings. Our hope is that the efforts of the EHR Workgroup will help establish an infrastructure that will facilitate work by others to specify the amount and type of information that is accessible, including preferences for receiving updates, in a context-specific way.
Most of the work to understand information needs of stakeholders within ClinGen has been approached from a laboratory perspective.[1] As illustrated in Figure 1, the effective use of genetics in medicine will increasingly require data exchange between laboratory information systems and the EHR ecosystem. Existing models for the exchange of information between laboratory and clinical information systems show promise for providing updated genetic information in a form that clinicians will use.[8] Providing consistent access to ClinGen resources across both laboratory and clinical environments should increase coordination and consistency. For this reason, the Workgroup will consider how to integrate ClinGen into both laboratory and healthcare settings.
A well-known challenge for using genomic sequence data clinically is that while genomic data can be persistent, our scientific understanding and subsequent clinical interpretations evolve over time. Currently, genomic sequence variants and the data interpretations tied to them are often not distributed to healthcare providers in structured form, which limits reuse and reinterpretation of these data.[9] A key recommendation from both a policy and research perspective to address this challenge is to determine the optimal way to transmit variant information and clinical interpretations in a standardized, unambiguous format.[10] As such, our evaluation of genomic medicine resources and configuration for use by infobuttons includes an assessment of existing ontologies to support standardized searches.
ClinGen is engaged in efforts to set internal data standards for the incorporation of genetic testing results into the EHR ecosystem. As a consequence, ClinGen Workgroups are engaged with standards organizations, including HL7, the Global Alliance for Genomics and Health, Logical Observation Identifiers Names and Codes, and the Displaying and Integrating Genetic Information Through the EHR Action Collaborative. The goals are to both inform and be informed by emerging standards, to ensure the rapid adoption of standards appropriate for medical genetics, and to ensure the compliance of ClinGen with those standards.
6. Conclusion
Current ClinGen EHR Workgroup efforts focus on the development of the Web Resources page and infobutton implementation. These efforts are part of ClinGen public portal development, with plans to support programmatic access by clinical systems.
Acknowledgments
The ClinGen Consortium is funded by The National Human Genome Research Institute (NHGRI), in conjunction with additional funding from The Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) and The National Cancer Institute (NCI), through the following grants and contracts: 1U41HG006834-01A1 (Rehm), 1U01HG007437-01 (Berg), 1U01HG007436-01 (Bustamante), and HHSN261200800001E.
Footnotes
Conflicts of Interest Statement
CLO, BH, JMC, SD, AM, TN, AN, EMR, MSW declare no conflicts of interest. SA works for Partners HealthCare which owns equity in GeneInsight, Inc., a company which distributes software to facilitate variant updates.
References
- 1.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, et al. ClinGen--the Clinical Genome Resource. N Engl J Med. 2015;372(23):2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cimino JJ, Jing X, Del Fiol G. Meeting the electronic health record “meaningful use” criterion for the HL7 infobutton standard using OpenInfobutton and the Librarian Infobutton Tailoring Environment (LITE). AMIA Annu Symp Proc. 2012;2012:112–120. [PMC free article] [PubMed] [Google Scholar]
- 4.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Characteristics of users of online personalized genomic risk assessments: implications for physician-patient interactions. Genet Med. 2009;11(8):582–587. doi: 10.1097/GIM.0b013e3181b22c3a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shabani M, Borry P. Challenges of web-based personal genomic data sharing. Life Sci Soc Policy. 2015;11(1):22. doi: 10.1186/s40504-014-0022-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldsmith L, Jackson L, O'Connor A, Skirton H. Direct-to-consumer genomic testing: systematic review of the literature on user perspectives. Eur J Hum Genet. 2012;20(8):811–816. doi: 10.1038/ejhg.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scherr CL, Lindor NM, Malo TL, Couch FJ, Vadaparampil ST. A preliminary investigation of genetic counselors' information needs when receiving a variant of uncertain significance result: a mixed methods study. Genetics in Medicine. 2015 doi: 10.1038/gim.2014.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilcox AR, Neri PM, Volk LA, Newmark LP, Clark EH, Babb LJ, Varugheese M, Aronson SJ, Rehm HL, Bates DW. A novel clinician interface to improve clinician access to up-to-date genetic results. J Am Med Inform Assoc. 2014;21(e1):e117–121. doi: 10.1136/amiajnl-2013-001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirts BH, Salama JS, Aronson SJ, Chung WK, Gray SW, Hindorff LA, Jarvik GP, Plon SE, Stoffel EM, Tarczy-Hornoch PZ, et al. CSER and eMERGE: current and potential state of the display of genetic information in the electronic health record. J Am Med Inform Assoc. 2015 doi: 10.1093/jamia/ocv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiley LK, Tarczy-Hornoch P, Denny JC, Freimuth RR, Overby CL, Shah N, Martin RD, Sarkar IN. Harnessing next-generation informatics for personalizing medicine: a report from AMIA's 2014 Health Policy Invitational Meeting. Journal of the American Medical Informatics Association. 2015 doi: 10.1093/jamia/ocv111. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]