Abstract
Background
The Parkinson’s Progression Marker Initiative is an international multi-center study whose main goal is investigating markers for Parkinson’s disease (PD) progression as part of a path to a treatment for the disease. This manuscript describes the baseline genetic architecture of this study, providing not only a catalog of disease linked variants and mutations, but also quantitative measures with which to adjust for population structure.
Methods
383 newly-diagnosed typical PD cases, 65 atypical PD and 178 healthy controls from the Parkinson’s Progression Marker Initiative study have been genotyped on the NeuroX and/or Immunochip arrays. This data is freely available to all researchers interested in pursuing PD research within the Parkinson’s Progression Marker Initiative.
Results
Parkinson’s Progression Marker Initiative represents a study population with low genetic heterogeneity. We recapitulate known PD associations from large-scale genome-wide association studies and refine genetic risk score models for PD predictability (area under the curve ~ 0.74). We show the presence of 6 LRRK2 p.G2019S and 9 GBA p.N370S mutation carriers.
Conclusions
The Parkinson’s Progression Marker Initiative study and its genetic data are useful in studies of PD biomarkers. The genetic architecture described here will be useful in the analysis of myriad biological and clinical traits within this study.
Keywords: Parkinson’s disease, genetics, genetic risk score, GWAS
INTRODUCTION
The Parkinson’s Progression Marker Initiative (PPMI, http://www.ppmi-info.org/) is a multi-center international collaborative effort sponsored by the Michael J. Fox Foundation for Parkinson’s Research (MJFF). At its core PPMI is a longitudinal observational study that aims to identify one or more markers of progression for Parkinson’s disease (PD), a critical step in the development and testing of new treatments.
PPMI uses a large variety of imaging methods, clinical measures and extensive biological sampling. Subjects for the study are collected from a number of sites across North America, Israel, Europe, and Australia. The core PPMI dataset centers on the acquisition of data from 400 newly diagnosed PD cases, 200 healthy controls, and 70 individuals who, while clinically diagnosed as PD cases, fail to show evidence of dopaminergic deficit by DAT scan. This latter group of patients are referred to as SWEDDs (subjects without evidence of dopaminergic deficit).
A critical part of the PPMI study is the deposition of samples and data in a repository that is readily accessible to the scientific and clinical community. In creating a relatively open data structure, the study sponsors aim to facilitate analysis, collaboration, and progress. As a part of this study all data generated from PPMI samples is required to be returned to the repository. We describe here the generation and analysis of the genetic data within PPMI. These data, which comprise more than 400,000 genotypes per subject, allow a genetic characterization of the PPMI samples; this includes the identification of known disease causing mutations and risk variants, in addition to metrics that can be readily used in the quality control and genetic classification of samples. We argue here that these data should be widely used to both understand the role of genetic influence in progression and biomarkers, and where appropriate to adjust for the effects of genetic background in analyses.
PATIENTS AND METHODS
Patients
PPMI actively recruited early-stage untreated PD subjects along with age appropriate controls. Currently, the PPMI cohort is divided into three subsets of participants: healthy controls (HC), etiologically typical PD cases, and an additional set of atypical PD subjects without dopaminergic deficit (SWEDD). The SWEDD subset are individuals presenting clinical features that meet criteria for a PD diagnosis but have normal dopamine transfer imaging scan data (DatScan).
The recruitment of PD, SWEDD and HC subjects has been previously described in detail elsewhere (1). PD subjects in PPMI are required to demonstrate at least asymmetric resting tremor or asymmetric bradykinesia or some combination of bradykinesia, resting tremor and/or rigidity within two years of diagnosis. They must be untreated for PD at the time of enrollment, as well as for the prior two years. SWEDD subjects include the PD criteria although DatScans show no dopaminergic deficit with high sensitivity and specificity after clinical diagnosis (2). HC subjects must be clinically ascertained as not having any neurologic dysfunction and Montreal Cognitive Assessment (MoCA) > 26 (3).
Genetic data
All available biobanked DNA specimens from all three participant subsets were genotyped using two genotyping arrays, ImmunoChip and NeuroX.
Briefly, the Immunochip is an Illumina Infinium based array that interrogates 196,524 variants. The Immunochip was designed in 2009 by investigators interested in inflammatory and autoimmune disorders, however this content also included ~2000 variants that had been prioritized for follow up by PD genome wide association study (4). The content of the Immunochip is available on the PPMI site and this platform has been previously described (5).
Briefly, NeuroX was designed to include over 240,000 exonic variants, as well as over 24,000 variants specific to the study of neurodegenerative disease (6). These neurodegenerative disease-focused variants include loci derived from the largest completed meta-analyses of PD cases and controls, which identified many of the known PD mutations and additional rare/high-risk variants (7). The content for NeuroX is also available via the PPMI website.
For both NeuroX and ImmunoChip genotyping was performed as per the manufacturer's protocol (Illumina Inc, CA). Initial quality control centered on genotype success rate per individual using a cutoff of 95%, followed by concordance of reported and genotype sex. Subsequently the rates of heterozygosity and homozygosity were assessed to ensure that for each individual they were within an expected range (within 6 standard deviations of the population mean).
In addition to genotyping from these arrays, the APOE e2/e3/e4 genotypes (derived from SNPs rs7412 and rs429358) were generated for 301 subjects are available at PPMI; these variants were genotyped using TaqMan genotyping as previously described (8).
Statistical analyses
In order to assess for sample handling errors that would not be detected by gender checks we compared genotype concordance across polymorphisms common to both genotype platforms.
In an attempt to detect genetic outliers principal components analyses of common variants derived from NeuroX (minor allele frequency, MAF > 5%) were performed. The resulting metrics were used to compare PPMI subjects to HapMap Phase 3 samples and generate estimates of continental ancestry (9).
Two genetic risk scores (GRS) were generated for this study as a means of summarizing the cumulative effect of known genetic risk variants. The first can be referred to as the common variant GRS (cGRS), and is a composite of the 28 replicated risk loci identified in Nalls et al., 2014 (7). The second GRS can be referred to as the common and rare variant GRS (crGRS), and is an expansion of the previously described GRS to include two additional rare risk variants detected in PPMI that are well known to be associated with PD: p.N370S in GBA and p.G2019S in LRRK2 (Table S1) (10,11) . To create each GRS, the number of risk alleles per variant of interest are scaled by multiplying them by the natural log of the odds ratio published for that variant, then the scaled risk allele counts per variant are summed per sample. Risk estimates for the cGRS and crGRS were taken from Nalls et al., 2014, and odds ratios of rare alleles at p.N370S with 3.33 and p.G2019S with 9.620 were taken from the PDgene database and 23AndMe [www.pdgene.org and www.23andme.com] (12). The common and rare variant GRS were also recalculated from this GWAS replication series using publicly available data to facilitate more direct comparisons using identical methods. For each relatively normally distributed GRS, risk scores were Z transformed based on the mean and standard deviation of the HC subset (Figure S1, Panel A). Therefore, all regression models are modeling change in terms of one standard deviation from the HC subset mean. As part of this transformation, the HC population was used as a reference with a theoretical mean of 0 and standard deviation of 1 for each GRS.
Logistic regression was used to quantify a trend of risk associated with a one standard deviation change from the control population mean to assess risk associated with PD, SWEDD or combined PD and SWEDD status compared to the same set of pooled controls separately for both the rare and common GRS estimates. Stepwise modeling was used to generate the most parsimonious models. Covariates used include up to five eigenvectors from principal components analysis of PPMI sample genetic data after quality control and filtering, age at onset for cases or age at most recent follow-up for controls, and family history within 2nd degree or closer relatives. All analyses of PD and controls used age, family history and principal components to account for population substructure. Analyses of PD combined with SWEDD and controls used the same covariates as prior analyses, while age was dropped from the models incorporating only SWEDD and controls. Predictive power of each model was quantified by generation of receiver operator curves. Single variant analyses for all 3 subsets were carried out for all variants comprising both risk scores using identical logistic regression models with the same covariates. Although these single variant analyses possess relatively low power with estimates of 15.6% power after Bonferroni correction for 30 tests and and less than 1% power for genome-wide significance assuming modest effect sizes seen in GWAS (Cohen’s h = 0.2 with 367 cases and 165 controls).
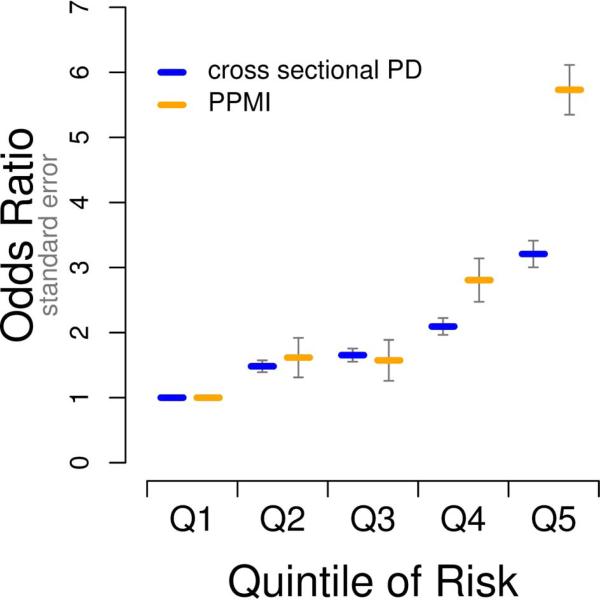
In addition, each of the 3 subsets used for analyses (PD, SWEDD, and SWEDD+PD) were split into quintiles of genetic risk based on each of the two GRS models. Risk associated with membership in each quintile of genetic risk was evaluated using identical regression models as those used for the trend test described above, but with the reference population being the lowest quintile of genetic risk. This model tests each increasing quintile of genetic risk as a possible predictor of case status across all 3 subsets compared to pooled controls; for each quintile subset the percentage of cases was also calculated.
RESULTS
Genotyping and Quality Control
For the Immunochip there is existing data on 524 successfully genotyped PPMI subjects (call rates > 95%, acceptable rates of heterozygosity and homozygosity, and concordance between self-reported and genetically ascertained sex). For NeuroX there is existing data on 619 successfully genotyped subjects (call rates > 95%, acceptable rates of heterozygosity and homozygosity (within +/− 6 SD of population mean), and concordance between self-reported and genetically ascertained sex). This data is readily available for analyses by qualified researchers after a brief application process that can be found at http://www.ppmi-info.org/access-data-specimens/download-data/. Once approved, any researcher with a valid login can activate the data access link below. The individual level genotype data for these arrays is available as plink binary files on the PPMI website (https://ida.loni.usc.edu/pages/access/geneticData.jsp).
The genotypes of variants typed both on Immunochip and NeuroX arrays (n=520) were compared and these showed an extremely high genotype concordance rate between the two platforms (99.94%). The rs7412 variant of APOE was also genotyped on NeuroX and this genotype showed ~99.3% concordance with the custom TaqMan genotyping across 5 redundant probes. Rs429358 could not be assayed accurately using currently available array technology, but using a proxy (rs4420638) we were able to achieve > 90% accuracy to tag the APOE epsilon-4 haplotype.
Population characterization
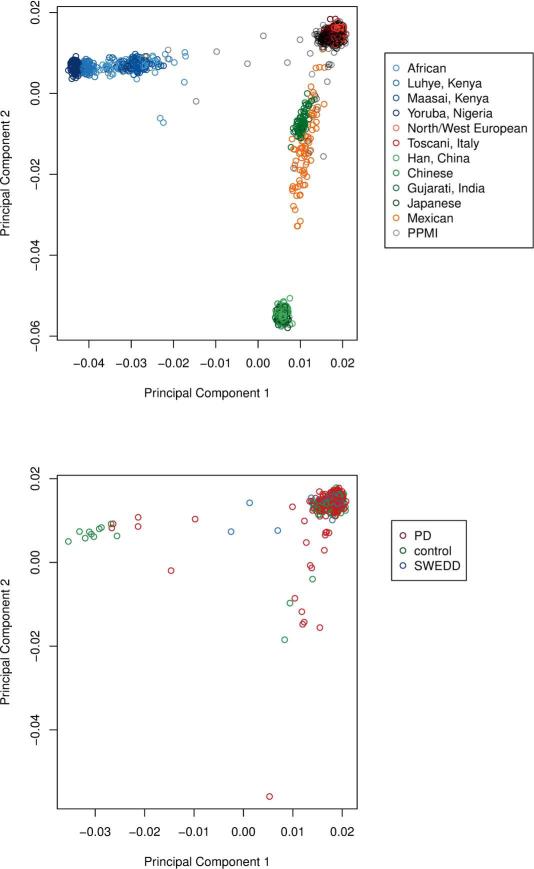
Principal component analysis revealed that from the 619 successfully genotyped subjects from the NeuroX array. Of these samples, 587 were of predominantly European ancestry and 32 subjects were of predominantly African or Asian continental ancestry. We have calculated principal components for all NeuroX genotyped subjects together (eigenvectors 1-20) and for the European ancestry subset separately. These eigenvectors are available for use in future studies to account for varying degrees of population substructure, as well as to give a more fine scale and accurate assessment of the continental ancestries of subjects included in PPMI (Figure 1).
Figure 1. Population substructure within the PPMI genetics dataset.
Abbreviations for HapMap Phase 3 reference populations as follows: ASW: African ancestry in Southwest USA; CEU: Utah residents with Northern and Western European ancestry from the CEPH collection; CHB: Han Chinese in Beijing, China; CHD: Chinese in Metropolitan Denver, Colorado; GIH: Gujarati Indians in Houston, Texas; JPT: Japanese in Tokyo, Japan; LWK: Luhya in Webuye, Kenya; MEX: Mexican ancestry in Los Angeles, California; MKK: Maasai in Kinyawa, Kenya; TSI: Toscani in Italia; YRI: Yoruba in Ibadan, Nigeria.
GRS analyses in this report are restricted to the 587 confirmed European ancestry PPMI subjects whose estimates for eigenvectors 1 and 2 from principal components were within 9 standard deviations of the mean for the combined CEU and TSI HapMap populations (13). This was done to reduce population stratification and more closely recapitulate genetic risk models from our previous work, which centered on individuals of European ancestry (7). Descriptive statistics were calculated using this subset of 587 subjects (Table 1, Part A). One related pair of subjects was detected via identity by descent estimates (14). This sibling pair included one SWEDD and one PD sample, in analyses in which PD and SWEDD subjects were jointly compared to healthy controls, the PD sample from this pair was removed (Table 1, Part B).
Table 1.
Descriptive statistics and GRS regression summaries for PD, SWEDD and healthy controls.
| Part A. Descriptive statistics | |||||
| Subset | PD | SWEDD | Controls | ||
| Total in analyses (N) | 367 | 55 | 165 | ||
| Age in years (mean, SD) | 64.256, 9.598 |
63.018, 10.117 | 63.794, 1.588 | ||
| Females (%) | 33.242 | 34.545 | 33.333 | ||
| Family history of PD (%) | 25.068 | 30.909 | 5.454 | ||
| N370S carriers (%) | 1.912* | 1.818 | 0.606 | ||
| G2019S carriers (%) | 1.3624 | 0 | 0 | ||
| SNCA multiplication (%) | 0 | 1 | 0 | ||
| Common variant GRS (mean, SD in Z units) |
0.511, 1.034 | 0.323, 0.976 | 0, 1 | ||
| Common and rare variant GRS (mean, SD in Z units) |
0.596, 1.201 | 0.347, 1.065 | 0, 1 | ||
|
Part B. GRS logistic regression
models |
|||||
| Subset | Odds ratio of trend |
95% confidence interval |
P-value of trend |
AUC | AUC confidence interval |
| PD versus controls, common variant GRS |
1.691 | 1.372, 2.083 | 8.39E-07 | 0.741 | 0.698-0.784 |
| SWEDD versus controls, common variant GRS |
1.371 | 0.971, 1.934 | 0.073 | 0.696 | 0.608-0.784 |
| PD and SWEDD versus controls, common variant GRS |
1.617 | 1.328, 1.969 | 1.69E-06 | 0.716 | 0.674-0.759 |
| PD versus controls, common and rare variant GRS |
1.736 | 1.413, 2.134 | 1.59E-07 | 0.748 | 0.706-0.791 |
| SWEDD versus controls, common and rare variant GRS |
1.359 | 0.970, 1.904 | 0.075 | 0.694 | 0.606-0.782 |
| PD and SWEDD versus controls, common and rare variant GRS |
1.701 | 1.396, 2.074 | 1.40E-07 | 0.733 | 0.691-0.775 |
Star denotes one missing genotype. Odds ratios and other summary statistics are based on a single standard deviation (Z) change in the GRS distribution from the mean of the healthy controls. Age refers to onset in cases and age at last clinic visit in controls. Family history refers to any first or second degree relatives with PD from self-report.
PD related variants
We identified 6 carriers of the LRRK2 p.G2019S variant (rs34637584; Table S1), all of whom were PD cases. In addition we identified 9 subjects who carried the GBA p.N370S PD risk variant (rs76763715; also called p.N409S; Table S1), including 7 PD patients, 1 SWEDD, and 1 control. No carriers of LRRK2 p.R1441H, p.Y1699C, or p.I2020T were identified.
The variants used for the generation of the GRS models were abstracted from NeuroX. In addition to posting all genotype data for NeuroX and ImmunoChip to the PPMI site, we have also posted these variants individually along with the individual genetic risk scores calculated here. In the GRS analysis, we recapitulate the associations between cGRS and crGRS and PD as seen in previous publications (7). Using similar data, we observed a highly significant association between PD and the crGRS (P-value = 1.59E-07). This shows that a one standard deviation increase in the crGRS above the HC mean is associated with an odds ratio of 1.74 (95% CI 1.413-2.134). When the SWEDD subset was compared to the same controls no significant GRS association was observed, although there was a suggestive trend that mirrored the PD association’s directionality (Table 1, Part B). Although we see no association between the GRS models and SWEDD status, when comparing the AUCs of the models that include the crGRS or the cGRS (as well as appropriate covariates such as family history, age and principal components), DeLong’s test showed no significant difference between the PD and SWEDD ROC curves (15). This could be due to low power to detect a difference because of the small number of SWEDD in PPMI; thus we cannot discount the possibility of a significant differences between these groups due to divergent genetic etiologies.
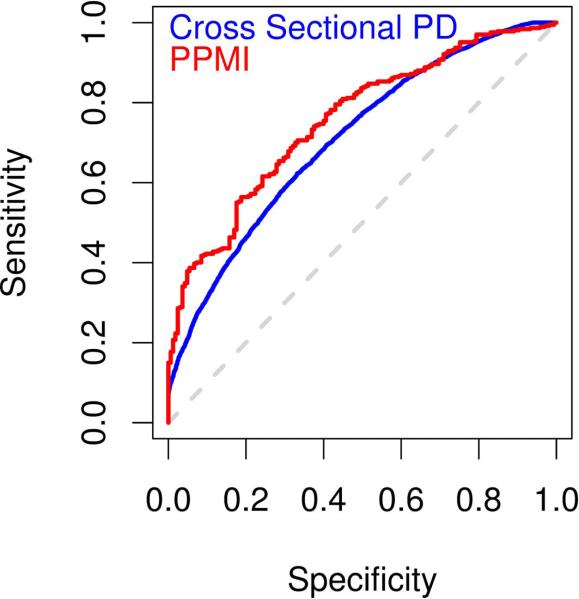
Our previous efforts in risk prediction analysis in large-scale, primarily cross-sectional studies of GWAS data have yielded an AUC of 0.633 based on a common GRS model (7). When comparing only the cGRS between the cross-sectional GWAS and longitudinal PPMI data (excluding all other factors from the model), the common variant GRS are 0.607 (95% CI 0.597-0.617) for the former and 0.631 (95% CI 0.581-0.681) for the latter, while the common and rare variant scores are 0.613 (95% CI 0.603-0.623) and 0.639 (95% CI 0.589-0.688) respectively. These differences in AUCs are not statistically significant when comparing GWAS data to PPMI; again this is likely due to the comparatively small sample size in PPMI. Notably, the depth of available data in PPMI has allowed us to include the variants p.G2019S and p.N370S as well as family history of PD in our model, further improving on previous efforts in which this data was not available for all participants. This leads to an increase in our ability to predict disease with an AUC of 0.748 for the crGRS in PD (Table 1, Part B and Figures 2 and S1).
Figure 2. Comparison of PPMI and large-scale cross-sectional meta-analysis results.
Panel A, areas under the curve from receiver operator curve analyses of crGRS; Panel B, comparison of odds ratios by quintile of genetic risk. The data for cross sectional analysis was taken from our previously published work (7).
The AUC seen in PPMI surpasses previous estimates reported by large scale meta-analyses, despite its current application in PPMI being a much smaller dataset. When examining covariates themselves including eigenvectors accounting for population substructure as well as family history and age, the AUC for PD is 0.702 (0.657-0.747) in PPMI; examining classification based solely on age and family history of PD in PPMI, the AUC is 0.605 (0.556-0.654). As previously described, the AUC including all nominated factors and covariates in the common and rare GRS model is 0.748 (0.706-0.790).
Based on DeLong’s test the inclusion of genetic information in the model is a significant improvement over the model using just family history and age (p = 1.55e-07) or the model that includes population structure, family history, and age (p-value = 0.0026).
DISCUSSION
In this report, we describe baseline associations in the PPMI dataset and provide metrics that afford adjustment for known associations, common mutations, and genetic background. We also show the utility of a genetic risk score approach, even in cohorts that are of insufficient size to detect individual variant associations.
Notably our efforts showed a low degree of genetic heterogeneity in the PPMI cohort. We identified a small number of non-European ancestry individuals based on PCA of informative genotypes. Of note, we also identified a pair of cryptically related individuals, being members of the SWEDD and PD groups. The principal components calculated as a part of this study should be considered for use as a quality control or adjustment parameter in much of the work intended for PPMI. For example, it is conceivable that biomarkers may behave differently based on ethnic background, and this could be accounted for by adjustment using the PCA metrics or by excluding genetic outliers.
We identified 6 carriers of the LRRK2 p.G2019S mutations; as expected all of the carriers were PD cases. We also identified 9 subjects who carried the GBA p.N370S PD risk variant , including 7 PD patients, 1 SWEDD, and 1 control. In both instances the frequency of these more common GBA and LRRK2 variants was close to that expected from previous work in European ancestry cohorts (10,16,17).
This dataset illustrates the use of GRS models in complex disease; there are two observations that are of particular note. First, the GRS performs marginally better in the longitudinal and deeply phenotyped PPMI study compared to typical cross sectional studies. If genetic risk across populations is assumed to be uniform, this increase in AUC associated with the cGRS in PPMI compared to cross sectional series of PD could likely be due to the low misdiagnosis rate afforded by the deep and longitudinal phenotyping in PPMI, particularly in comparison to that of a cross sectional study that does not include an imaging component. The observation that the PD GRS models performed poorly in the SWEDD cohort would suggest that as a whole the SWEDD group represents a disorder(s) with an etiology that is distinct from typical PD, or a mix of typical PD and atypical parkinsonism cases.
There are of limitations relating to the genotyping data described here; first, not all known PD mutations or neurological disease related variants are reliably assayed using NeuroX or ImmunoChip. For example both the GBA p.L444P and APOE e4 allele are not directly genotyped using these arrays. While the APOE e2/3/4 polymorphism has been directly genotyped using taqman in PPMI (data for which are available through the PPMI site), the GBA p.L444P variant remains untyped in this series at the time of writing. In addition both NeuroX and ImmunoChip are focused arrays, and while they contain a large number of putatively PD-associated variants, they do not assay common variability in a genome wide context. This may be limiting in the discovery of genetic variants associated with traits tested in the PPMI individuals.
In summary, here we have performed a baseline genetic characterization of the PPMI dataset. This work shows that genetically these cases appear to be consistent with typical PD, and that the genetic information included in this study will likely add significant value to future analyses.
Supplementary Material
Supplemental Figure S1: Summaries of analyses relating to estimates of risk using the step-wise regression modeling of the GRS based on both common and rare variation. Panel A, density distributions of the estimated genetic risk based on the common and rare variant GRS; Panel B, AUC estimate of PD and controls; Panel C, AUC estimate of SWEDD and controls; Panel D, AUC estimate of combined PD-SWEDD and controls; Panel E, distribution of odds ratios and 95% confidence intervals for each subset of analyses using the common and rare variant GRS by quintile of estimated genetic risk; Panel F, case frequency per quintile of estimated genetic risk based on the common and rare variant GRS.
Supplemental Text: PPMI investigator list.
ACKNOWLEDGEMENTS AND FUNDING
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov), and DNA panels, samples and clinical data from the National Institute of Neurological Disorders and Stroke Human Genetics Resource Center DNA and Cell Line Repository. This work was supported by the Intramural Research Program of the National Institute on Aging, (project numbers Z01-AG000949-02), human subjects protocol 2003-077. PPMI is supported by the Michael J. Fox Foundation for Parkinson’s Research (MJFF) and is co-funded by MJFF, Abbvie, Avid Radiopharmaceuticals, Biogen Idec, Bristol-Myers Squibb, Covance, Eli Lilly & Co., F. Hoffman-La Roche, Ltd., GE Healthcare, Genentech, GlaxoSmithKline, Lundbeck, Merck, MesoScale, Piramal, Pfizer and UCB.
Footnotes
PPMI investigators listed in the supplemental text.
REFERENCES
- 1.Parkinson Progression Marker Initiative The Parkinson Progression Marker Initiative (PPMI) Prog Neurobiol. 2011 Dec;95(4):629–35. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De la Fuente-Fernández R. Role of DaTSCAN and clinical diagnosis in Parkinson disease. Neurology. 2012 Mar 6;78(10):696–701. doi: 10.1212/WNL.0b013e318248e520. [DOI] [PubMed] [Google Scholar]
- 3.Dalrymple-Alford JC, MacAskill MR, Nakas CT, Livingston L, Graham C, Crucian GP, et al. The MoCA: well-suited screen for cognitive impairment in Parkinson disease. Neurology. 2010 Nov 9;75(19):1717–25. doi: 10.1212/WNL.0b013e3181fc29c9. [DOI] [PubMed] [Google Scholar]
- 4.International Parkinson’s Disease Genomics Consortium (IPDGC), Wellcome Trust Case Control Consortium 2 (WTCCC2) A two-stage meta-analysis identifies several new loci for Parkinson’s disease. PLoS Genet. 2011 Jun;7(6):e1002142. doi: 10.1371/journal.pgen.1002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013 Sep;14(9):661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 6.Nalls MA, Bras J, Hernandez DG, Keller MF, Majounie E, Renton AE, et al. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiol Aging. 2015 Mar;36(3):1605.e7–12. doi: 10.1016/j.neurobiolaging.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat Genet. 2014 Sep;46(9):989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Federoff M, Jimenez-Rolando B, Nalls MA, Singleton AB. A large study reveals no association between APOE and Parkinson’s disease. Neurobiol Dis. 2012 May;46(2):389–92. doi: 10.1016/j.nbd.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010 Sep 2;467(7311):52–8. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sidransky E, Nalls MA, Aasly JO, Aharon-Peretz J, Annesi G, Barbosa ER, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009 Oct 22;361(17):1651–61. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez DG, Paisán-Ruíz C, McInerney-Leo A, Jain S, Meyer-Lindenberg A, Evans EW, et al. Clinical and positron emission tomography of Parkinson’s disease caused by LRRK2. Ann Neurol. 2005 Mar;57(3):453–6. doi: 10.1002/ana.20401. [DOI] [PubMed] [Google Scholar]
- 12.Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide B-MM, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson’s disease genetics: The PDGene database. PLoS Genet. 2012 Mar 15;8(3):e1002548. doi: 10.1371/journal.pgen.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International HapMap Consortium A haplotype map of the human genome. Nature. 2005 Oct 27;437(7063):1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011 Jan 7;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988 Sep;44(3):837–45. [PubMed] [Google Scholar]
- 16.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, et al. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365(9457):415–6. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 17.Nichols WC, Pankratz N, Hernandez D, Paisán-Ruíz C, Jain S, Halter CA, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson’s disease. Lancet. 2005 Jan 29;365(9457):410–2. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1: Summaries of analyses relating to estimates of risk using the step-wise regression modeling of the GRS based on both common and rare variation. Panel A, density distributions of the estimated genetic risk based on the common and rare variant GRS; Panel B, AUC estimate of PD and controls; Panel C, AUC estimate of SWEDD and controls; Panel D, AUC estimate of combined PD-SWEDD and controls; Panel E, distribution of odds ratios and 95% confidence intervals for each subset of analyses using the common and rare variant GRS by quintile of estimated genetic risk; Panel F, case frequency per quintile of estimated genetic risk based on the common and rare variant GRS.
Supplemental Text: PPMI investigator list.