Abstract
Mitochondrial metabolism is necessary for the maintenance of oxidative TCA cycle function and mitochondrial membrane potential. Previous attempts to decipher whether mitochondria are necessary for biological outcomes have been hampered by genetic and pharmacologic methods that simultaneously disrupt multiple functions linked to mitochondrial metabolism. Here, we report that inducible depletion of mitochondrial DNA (ρ° cells) diminished respiration, oxidative TCA cycle function and the mitochondrial membrane potential resulting in diminished cell proliferation, hypoxic activation of HIF-1, and specific histone acetylation marks. Genetic reconstitution only of the oxidative TCA cycle function specifically in these inducible ρ° cells restored metabolites resulting in reestablishment of histone acetylation. In contrast, genetic reconstitution of the mitochondrial membrane potential restored ROS, which were necessary for hypoxic activation of HIF-1 and cell proliferation. These results indicate that distinct mitochondrial functions associated with respiration are necessary for cell proliferation, epigenetics, and HIF-1 activation.
Introduction
Mitochondria are well appreciated as biosynthetic and bioenergetic organelles for their role in producing metabolites and ATP, which are byproducts of the tricarboxylic acid (TCA) cycle and the mitochondrial membrane potential, respectively. The TCA cycle metabolites such as oxaloacetate and citrate generate cytosolic aspartate and acetyl-CoA that are required for pyrimidine and fatty acid synthesis, respectively (Boroughs and DeBerardinis, 2015). The TCA cycle produces reducing equivalents NADH and FADH2, which deliver their electrons to the electron transport chain (ETC) that ultimately utilizes oxygen as the final acceptor (respiration). Importantly, a consequence of electron flux through an intact ETC is to establish a mitochondrial membrane potential required for generation of ATP and biogenesis of iron-sulfur clusters (ISC), ancient protein cofactors that associate with proteins in the mitochondria, cytosol, and nucleus to perform diverse functions including respiration, protein translation, and genome maintenance (Veatch et al., 2009). Mitochondrial membrane potential is also utilized for protein import of nuclear DNA encoded proteins into the mitochondria.
An emerging idea is that mitochondria also function as signaling organelles (Chandel, 2015). Two notable mitochondrial-dependent signaling mechanisms involve the release of reactive oxygen species (ROS) for protein thiol oxidation and the release of citrate, which generates acetyl-CoA used for protein lysine acetylation. Mitochondrial ROS-dependent signaling controls numerous biological responses including proliferation, differentiation, and adaptation to stress as well as physiological and pathological outcomes such as immunity, cancer, and aging (Schieber and Chandel, 2014; West et al., 2015; Yun and Finkel, 2014). For example, mitochondrial-derived ROS have been implicated in hypoxic signal transduction through the activation of hypoxia-inducible transcription factor 1 (HIF-1), an important component of the oxygen-sensing pathway. Cytosolic acetyl-coA derived from mitochondrial citrate functions as a substrate for histone acetyltransferases to regulate epigenetics (Wellen et al., 2009).
Understanding the mechanisms by which respiration regulates diverse biological outcomes such as cell proliferation, epigenetics, and oxygen sensing has been a challenge because pharmacologic or genetic ablation of the ETC can simultaneously impair mitochondrial membrane potential to diminish ATP generation (bioenergetic), the production of ROS (signaling) and regeneration of NAD+ and FAD thus diminishing oxidative TCA cycle function (biosynthetic). Moreover, previous studies utilized respiratory deficient cells that have undergone metabolic rewiring, allowing them to proliferate and conduct ROS dependent signaling (Mullen et al., 2012; Sullivan et al., 2013; Weinberg et al., 2010). Additionally, restricting the production of TCA cycle intermediates in the cytosol rather than directly impairing respiration has been utilized to infer the role of mitochondria in regulation of epigenetics (Carey et al., 2015; Wellen et al., 2009). It is not known whether impairment of respiration would restrict production of the TCA cycle intermediates sufficient to regulate epigenetics. Hence, in the current study, we genetically ablated the ETC in an inducible manner resulting in impaired respiration. We subsequently genetically reconstituted either the oxidative TCA cycle or the mitochondrial membrane potential without restoring ATP production to examine cell proliferation, epigenetics, and HIF-1 activation. This allowed us to detect the primary mechanisms by which respiration-linked mitochondrial processes control biological outcomes.
RESULTS
Inducible decrease of mitochondrial DNA diminishes respiration, mitochondrial membrane potential and cell proliferation
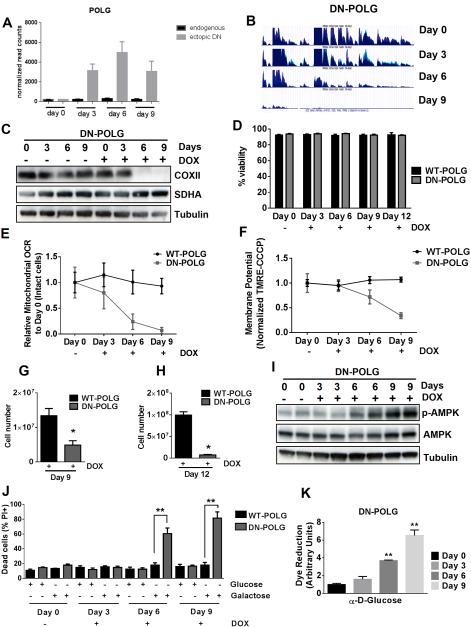
Nuclear DNA encodes the majority of mitochondrial proteins. However, there are 13 proteins encoded by the mtDNA that are critical subunits of the electron transport chain (ETC). DNA polymerase-γ (POLG) localized to the mitochondrial matrix is necessary for the replication of mtDNA. Therefore, inhibition of POLG promotes the loss of mtDNA without affecting nuclear DNA. Previous attempts to examine how mitochondrial respiration controls biological outcomes have utilized ethidium bromide, an inhibitor of POLG, to deplete mtDNA resulting in generation of ρ° cells over a few weeks (King and Attardi, 1989). Limitations of this strategy are that it allows for selection due to metabolic adaptation as well as potential off-target effects of ethidium bromide including intercalation into nuclear DNA. Thus, we stably expressed a doxycycline inducible dominant negative form of POLG in HEK293 cells (DN-POLG cells) to genetically remove mtDNA from cells (Wanrooij et al., 2007). Doxycycline caused an increase in the ectopic expression of DN-POLG, without affecting endogenous POLG expression (Fig. 1A). RNA sequencing demonstrated the loss of mitochondrial transcripts in DN-POLG cells following doxycycline-induced expression of the DN-POLG transgene within 6 days (Fig. 1B). In contrast, expression of wild-type POLG in HEK293 cells (WT-POLG cells) continued to express mitochondrial transcripts on day 9 in the presence of doxycycline (Fig. S1A). We confirmed the loss of mtDNA by doxycycline treatment in DN-POLG cells by real time PCR (Fig. S1B-C). The expression of the mtDNA-encoded protein COXII, but not the expression of the nuclear-encoded SDHA, was absent following the loss of mtDNA at day 6 and 9 in doxycycline-treated DN-POLG cells but not WT-POLG cells (Fig. 1C and Fig. S1D). Importantly, treatment of the DN-POLG cells with doxycycline for 12 days did not affect cell viability (Fig. 1D). However, loss of mtDNA resulted in a decrease in mitochondrial oxygen consumption (Fig. 1E) and mitochondrial membrane potential (Fig. 1F).
Figure 1. Inducible expression of DN-POLG in HEK293 cells diminishes mitochondrial respiration, mitochondrial membrane potential, and cell proliferation.
(A) Expression of the ectopic DN-POLG increases in cells treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=3).
(B) RNA-seq analysis of mitochondrial transcripts in three independent samples of DN-POLG cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days.
(C) Representative western blots of the expression of COXII (mtDNA encoded) and SDHA (nuclear encoded) in DN-POLG cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days from 3 independent experiments.
(D) WT and DN-POLG cells were left untreated or treated with doxycycline (10 ng/ml) for 3, 6, 9 for 12 days and viability was measured. Mean ± SEM (n=4).
(E-F) Relative mitochondrial oxygen consumption rate (OCR) (E) and mitochondrial membrane potential (F) of intact WT and DN-POLG cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=4).
(G-H) WT and DN-POLG cells were treated with doxycycline (10 ng/ml) and cell number was assessed at days 9 (G) and 12 (H). Mean ± SEM (n=3).
(I) Western blot analysis of p-AMPK, AMPK and Tubulin proteins in DN-POLG cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days.
(J) WT and DN-POLG untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days were grown in media containing 10 mM glucose or 10 mM galactose for 48h and assessed for cell death. Mean ± SEM (n=3).
(K) Glucose oxidation by DN-POLG cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=3).
* indicates significance p < 0.05. ** indicates significance p <0.01 throughout figure 1. See also Figure S1.
A functional consequence of mtDNA depletion was a significant decrease in proliferation of DN-POLG cells compared to WT-POLG cells at days 9 and 12 of doxycycline treatment (Fig. 1G-H). The presence of doxycycline for 3 and 6 days had a minimal effect on the proliferation rate of DN-POLG cells (Fig. S1E-F). Additionally, the proliferation defect of DN-POLG cells in the presence of doxycycline was confirmed using a CFSE assay (Fig. S1G). It is important to note that this defect in proliferation occurred even though cells had ample glucose, pyruvate and uridine, three key metabolic substrates required for ρ° cells to proliferate (King and Attardi, 1989). Since mitochondrial ATP generation ceases with loss of mtDNA, phosphorylation of AMPK in DN-POLG cells treated with doxycycline began to increase by day 6 (Fig. 1I and S1H). Moreover, DN-POLG cells treated with doxycycline for 6 or 9 days underwent cell death when glucose was replaced by galactose, which forces cells to rely primarily on mitochondrial ATP for survival (Fig. 1J). Consistent with this observation, glucose utilization was significantly higher in DN-POLG cells treated with doxycycline for 6 and 9 days (Fig. 1K).
Inducible loss of mtDNA decreases acetylation of specific histone H3 marks
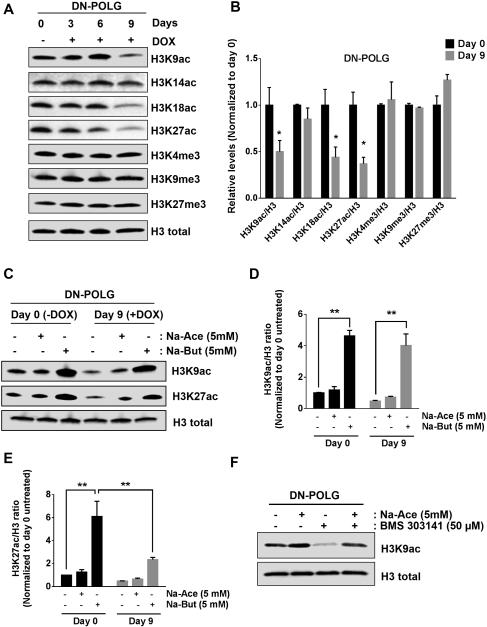
Next, we investigated how inducible loss of mtDNA, modifies histone acetylation and methylation. We performed high-resolution mass spectrometry analysis of histones H2B, H3 and H4 in combination with stable isotope labeling with amino acids in cell culture (SILAC). We quantitatively tracked the changes in post-translational modifications in DN-POLG cells in the presence or absence of doxycycline after 6 days. Loss of mtDNA invoked a decrease in multiple acetylation marks of histone H3, histone H2B and H4 within 6 days of doxycycline treatment in DN-POLG cells (S2A-C). These changes were more noticeable when the cells were treated with doxycycline for 9 days (Fig. S2D). We did not detect significant differences in methylation or other post-translational modifications (data not shown). The decrease in specific histone H3 acetylation marks (H3K9, H3K18 and H3K27) in doxycycline treated DN-POLG cells was verified by western blot (Fig. 2A, B). The methylation of histone H3 marks was not affected by the loss of mtDNA (Fig. 2A, B). Interestingly, DN-POLG cells treated with doxycycline for 9 days and exposed to acetate did not exhibit a rescue of the acetylation of H3K9 and H3K27 (Fig. 2C-E). However, acetate was able to rescue histone H3K9 acetylation when cells were treated with an ATP citrate lyase inhibitor (Fig. 2F). Butyrate, an inhibitor of histone deacetylases (HDACs), promoted a significant increase in H3K9 and H3K27 acetylation (Fig. 2C-E). DN-POLG cells treated with doxycycline for 9 days and exposed to butyrate had a significantly smaller increase in the acetylation of H3K27 compared to untreated cells (Fig. 2E). The activity of HATs and HDACs was not affected in WT-POLG cells (S2E, G) compared with DN-POLG cells treated with doxycycline for 9 days, which had lower levels of HATs and HDACs activity (Fig. S2F, H).
Figure 2. Inducible expression of DN-POLG in HEK293 cells diminishes specific histone H3 acetylation marks.
(A-B) Western blot analysis and quantification of histone 3 acetylation and methylation marks in DN-POLG cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=3).
(C) Acetylation state of H3K9 and H3K27 after treating DN-POLG cells at days 0 and 8 of doxycycline (10 ng/ml) treatment with 5 mM sodium acetate (Na-Ace) or 5 mM sodium butyrate (Na-But) for 24h.
(D-E) Quantification of the western blots assessing the acetylation of H3K9 (D) and H2K27
(E) of DN-POLG cells at days 0 and 8 of doxycycline (10 ng/ml) treatment in the presence or absence of 5 mM sodium acetate or sodium butyrate for 24h. Mean ± SEM (n=3).
(F) Acetylation state of H3K9 after treating DN-POLG cells with 5 mM sodium acetate (Na-Ace), 50 μM BMS 303141 (ACL inhibitor) and the combination of both treatments for 48h.
* indicates significance p < 0.05. ** indicates significance p <0.01 throughout Figure 2. See also Figure S2.
TCA cycle restoration is sufficient to reestablish the acetylation of specific histone H3 marks but not cell proliferation in mtDNA-depleted cells
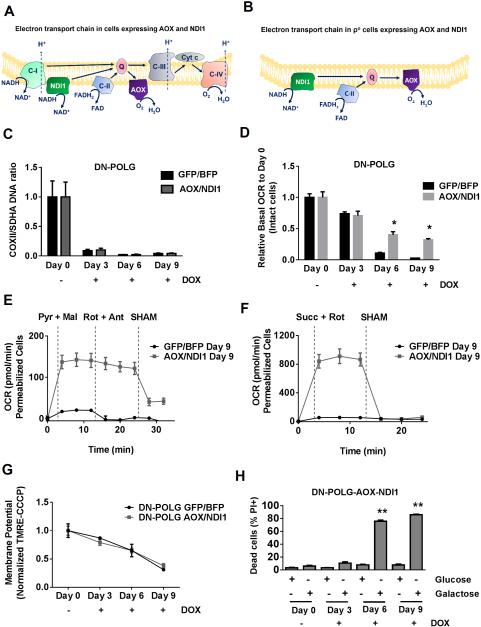
An intact ETC generates mitochondrial membrane potential through proton pumping and also allows for the oxidative TCA cycle to function by regenerating NAD+ and FAD. To specifically restore the oxidative TCA cycle in mtDNA-depleted cells, we expressed two non-mammalian proteins, S. cerevisiae NDI1 (alternative NADH dehydrogenase) and C. intestinalis AOX (alternative oxidase), which can conduct electron flux but not proton pumping (Perales-Clemente et al., 2008). NDI1 can oxidize NADH and thus restore the electron transport activity of mitochondrial complex I. AOX has the ability to transport electrons from ubiquinol to oxygen, allowing mitochondrial ETC complex III and IV activities to be bypassed. Thus, in wild-type cells, NDI1 and AOX compete for electrons with the endogenous ETC complexes (Fig. 3A). In mtDNA-depleted cells, endogenous complex I, III, and IV are not functional. Hence, the presence of NDI1 and AOX in these ρ° cells allows electrons to flow through the ETC, replenishing NAD+ and FAD to restore TCA cycle activity (Fig. 3B and Fig S3A-B). The expression of NDI1 and AOX proteins in mtDNA-depleted cells does not rescue proton pumping, therefore the mitochondrial membrane potential is still impaired (Fig. 3B). The treatment of both cell lines with doxycycline promoted the loss of mtDNA (Fig. 3C). AOX and NDI1 expression increased cellular oxygen consumption in intact DN-POLG cells treated with doxycycline (Fig. 3D). AOX and NDI1 expression in permeabilized DN-POLG cells treated for 9 days with doxycycline was sufficient to increase mitochondrial oxygen consumption when complex I substrates pyruvate and malate or complex II substrate succinate were added in the presence of ADP (Fig. 3E-F and S3D-E). Oxygen consumption rate of doxycycline-treated AOX and NDI1 expressing DN-POLG cells at day 3, 6 and 9 was decreased in the presence of salicylhydroxamic acid (SHAM), an inhibitor of AOX (Fig. 3E-F and S3D-E). Consistent with the inability of NDI1 and AOX to pump protons, we observed a loss of mitochondrial membrane potential similar to that seen in control cells in doxycycline-treated DN-POLG cells expressing AOX and NDI1 (Fig. 3G). The diminished mitochondrial membrane potential in DN-POLG cells expressing AOX and NDI1 impairs the ability to produce mitochondrial ATP resulting in their death when glucose was substituted for galactose (Fig. 3H and S3F). Glucose utilization was also increased in NDI1 and AOX expressing DN-POLG cells at day 6 of doxycycline treatment (Fig. S3G-J). These results indicate that ρ° cells containing AOX and NDI1 are completely dependent on glycolysis for survival.
Figure 3. NDI and AOX expression in inducible mtDNA depleted cells restores oxygen consumption but not mitochondrial membrane potential.
(A-B) Schematic representation of the electron transport chain in cells expressing the non-mammalian proteins NDI1 and AOX in wild-type (A) and ρ° cells (B). NDI1 can rescue electron transport capacity but not the proton pumping ability of complex I. Thus is can oxidize NADH to NAD+. AOX can accept electrons from ubiquinol and transfer them to oxygen thus bypassing complexes III and IV.
(C) mtDNA content in DN-POLG expressing AOX and NDI1 and control cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=3).
(D) Relative mitochondrial oxygen consumption rate (OCR) of intact DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=3).
(E-F) Complex I (E) or Complex II (F) driven oxygen consumption rate of saponin permeabilized Control-DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 9 days. Rotenone (1 μM) and antimycin A (1 μM) were used to inhibit complex I and III respectively. SHAM (2 mM) was used to inhibit AOX activity. Mean ± SEM (n=3).
(G) Mitochondrial membrane potential assessed by TMRE (50nM) staining and corrected by CCCP (50 μM) of DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=3).
(H) DN-POLG-AOX/NDI1 untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days were grown in media containing 10 mM glucose or 10 mM galactose for 48h and subsequently viability was assessed. Mean ± SEM (n=3).
* Indicates significance p < 0.05. ** Indicates significance p <0.01 throughout figure 3. See also Figure S3.
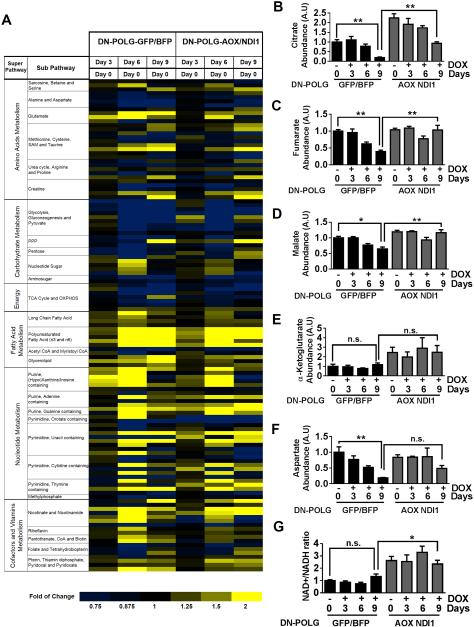
To demonstrate that expression of NDI1 and AOX is sufficient to restore TCA cycle activity, we performed metabolomics of NDI1 and AOX expressing DN-POLG cells and control cells treated with or without doxycycline for 3, 6 and 9 days. Compared to control cells, expression of NDI1 and AOX in DN-POLG cells promoted changes in the abundance of metabolites involved in key metabolic pathways (Fig. 4A). Importantly, at day 9 of doxycycline treatment, the abundance of TCA cycle metabolites such as citrate was higher in NDI1 and AOX expressing DN-POLG cells compared to control cells (Fig. 4B-E). An essential function of respiration is to maintain the NAD+/NADH ratio for aspartate synthesis (Birsoy et al., 2015; Sullivan et al., 2015). NDI1 and AOX increased the abundance of aspartate and the NAD+/NADH ratio in DN-POLG cells treated with doxycycline compared to control cells (Fig. 4F-G). A complete summary of the changed metabolites in doxycycline-treated DN-POLG cells expressing NDI1 and AOX compared to control cells is displayed in Figure S4 and Table S1. Taken together, these results indicate that non-mammalian proteins NDI1 and AOX can restore oxidative TCA cycle function without restoring mitochondrial membrane potential.
Figure 4. NDI1 and AOX expression in inducible mtDNA depleted cells restores the levels of TCA cycle metabolites.
(A) The heat map displays the fold of change in metabolites involved in important metabolic pathways in DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells treated with doxycycline (10 ng/ml) for 3, 6 and 9 days relative to untreated cells. A yellow-blue color scale depicts the abundance of the metabolites (Yellow: high, Blue: low). Data represent the average of 4 independent experiments.
(B-G) Intracellular citrate (B), fumarate (C), malate (D), α-Ketoglutarate (E), aspartate (F) levels and NAD+/NADH ratio (G) in DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days. Mean ± SEM (n=4).
* indicates significance p < .05. ** indicates significance p <0.01 throughout figure 4. See also Figure S4.
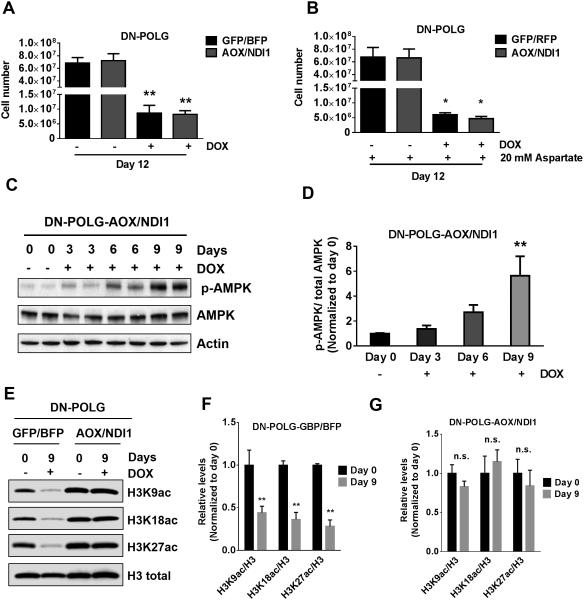
The expression of NDI1 and AOX did not rescue cell proliferation in DN-POLG cells treated with doxycycline even with pyruvate, uridine and aspartate supplementation (Fig. 5A-B). Since NDI1 and AOX expression does not restore mitochondrial ATP production, doxycycline-treated DN-POLG-AOX/NDI1 cells continued to exhibit an increase in phosphorylation of AMPK (Fig. 5C-D). However, NDI1 and AOX expression did restore acetylation of H3K9, H3K18 and H3K27 in DN-POLG cells treated with doxycycline (Fig. 5E-G). Interestingly, AOX and ND1 did not restore HATs or HDACs activity indicating that the restoration of TCA cycle metabolites by AOX and NDI1 is the mechanism by which histone H3 acetylation marks are reestablished (Fig. S5).
Figure 5. Restoration of the oxidative TCA cycle activity in inducible mtDNA depleted cells is sufficient to rescue specific histone 3 acetylation marks but not cell proliferation.
(A-B) DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells were grown in the presence or absence of doxycycline (10 ng/ml) for 12 days and cell number was assessed. Cells were supplemented with pyruvate and uridine in the presence (B) or absence (A) of aspartate (20 mM). Mean ± SEM (n=3).
(C) Western blot analysis of p-AMPK, AMPK and Tubulin in DN-POLG-AOX/NDI1 untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 days.
(D) Quantification of the western blots assessing the phosphorylation state of AMPK in DN-POLG-AOX/NDI1. Data is represented relative to total levels of AMPK and the mean value in untreated cells. Mean ± SEM (n=3).
(E-G) Western blot analysis and quantification of the acetylation of H3K9, H3K18 and H3K27 and the total expression of H3 in DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 9 days. Data was normalized to the mean value in untreated cells. Mean ± SEM (n=3).
* indicates significance p < .05. ** indicates significance p <0.01 throughout figure 5. See also Figure S5.
Restoration of the mitochondrial membrane potential increases cell proliferation but not specific histone H3 acetylation marks in mtDNA-depleted cells
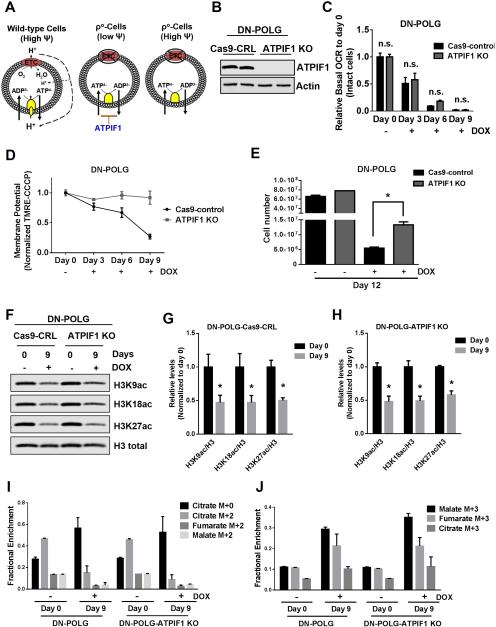
An intact ETC generates the mitochondrial membrane potential required for the synthesis of ATP (Fig. 6A). In the absence of a functional ETC, cells can maintain mitochondrial membrane potential by reversing the F0F1-ATPase, which utilizes glycolytic ATP to pump protons into the intermembrane space. However, mitochondrial DNA encodes subunits of the F0 component, thus ρ° cells cannot utilize this mechanism to generate the mitochondrial membrane potential. Instead, ρ° cells attempt to maintain the mitochondrial membrane potential by using an incomplete F0F1-ATPase where the F1 component can utilize glycolytic ATP and the electrogenic exchange of ATP4- for ADP3- by the adenine nucleotide translocator (Appleby et al., 1999; Buchet and Godinot, 1998). However, the presence of ATPIF1, an endogenous physiological inhibitor of the F0F1-ATPase, reduces the activity of the F1 component resulting in a diminished mitochondrial membrane potential in ρ° cells (Chen et al., 2014) (Fig. 6A). Thus, we utilized CRISPR technology to knockout ATPIF1 in our doxycycline-inducible DN-POLG cells to generate ρ° cells that sustain a mitochondrial membrane potential (Fig. 6B). Doxycycline treatment still induced the loss of mtDNA and decreased the mitochondrial oxygen consumption in Cas9-control and ATPIF1 knockout DN-POLG cells (Fig. 6C and S6A). DN-POLG ATPIF1 knockout cells compared with DN-POLG Cas9-control cells treated with doxycycline were able to maintain their mitochondrial membrane potential (Fig. 6D). To demonstrate that the maintenance of the membrane potential is directly due to the loss of ATPIF1, we reconstituted DN-POLG-ATPIF1 knockout cells with ATPIF1 cDNA (Fig. S6B) and observed that ATPIF1 expression was sufficient to decrease mitochondrial membrane potential following doxycycline treatment (Fig. S6C). The loss of ATPIF1 significantly increased cell proliferation of DN-POLG cells treated with doxycycline (Fig. 6E). Importantly, reconstitution of ATPIF1 cDNA in DN-POLG ATPIF1 knockout cells decreased cell proliferation upon doxycycline treatment (Fig. S6D). We pharmacologically validated the importance of ATPIF1 in controlling cell proliferation by administering a combination of inhibitors against complex I (piericidin), III (antimycin) and V (oligomycin) to Cas9-control and ATPIF1 knockout DN-POLG cells. These inhibitors decreased mitochondrial membrane potential and cell proliferation in Cas9-control but not in ATPIF1 knockout cells (Figure S6E-F). However, loss of ATPIF1 was not sufficient to re-establish the acetylation of specific histone H3 marks in DN-POLG cells treated with doxycycline (Fig. 6F-H).
Figure 6. Restoration of the mitochondrial membrane potential in inducible mtDNA depleted cells partially rescues cell proliferation but not specific histone 3 acetylation marks.
(A) A functional ETC generates a mitochondrial membrane potential that is used to produce ATP. In ρ° cells, an impaired ETC is not able to generate a mitochondrial membrane potential. ρ° cells are able to sustain mitochondrial membrane potential through the electrogenic exchange of ATP4- for ADP3- by the adenine nucleotide carrier. This mechanism relies on the F1-ATPase activity of an incomplete F0F1-ATPase that is loosely associated with the membrane. However, the ATPIF1 protein, an inhibitor of the F1-ATPase prevents the maintenance of the mitochondrial membrane potential. The loss of ATPIF1 increases the F1-ATPase activity to sustain the mitochondrial membrane potential.
(B) Western blot analysis of ATPIF1 protein levels in DN-POLG-Cas9-control cells and DN-POLG-ATPIF1 KO cells.
(C-D) Relative mitochondrial oxygen consumption rate (OCR) (C) and mitochondrial membrane potential (D) of intact DN-POLG-Cas9-control cells and DN-POLG-ATPIF1 KO cells untreated or treated with doxycycline (10 ng/ml) for 3, 6 and 9 day. Mean ± SEM (n=3).
(E) DN-POLG-Cas9-control cells and DN-POLG-ATPIF1 KO cells were grown in the presence or absence of doxycycline (10 ng/ml) for 12 days and assessed for cell number. Mean ± SEM (n=3).
(F-H) Western blot analysis and quantification of the acetylation of H3K9, H3K18 and H3K27 and the total expression of H3 in DN-POLG-Cas9-nt-control and DN-POLG-ATPIF1 KO cells untreated or treated with doxycycline (10 ng/ml) for 9 days. Data was normalized to the mean value in untreated cells. Mean ± SEM (n=3).
(I-J) DN-POLG and DN-POLG-ATPIF1 KO cells untreated or treated with doxycycline (10 ng/ml) for 9 days were labeled for six hours with [U-13C]glucose and subsequently metabolite pools were examined. m+0 pools represent unlabeled fractions. m+2 and m+3 pools results from glucose flow through pyruvate dehydrogenase and pyruvate carboxylase, respectively. Mean ± SEM (n=3).
* Indicates significance p < 0.05 throughout figure 6. See also Figure S6.
An intriguing aspect of our findings was the lack of a complete rescue of cell proliferation upon restoration of the mitochondrial membrane potential. The DN-POLG ATPIF1 knockout cells and DN-POLG Cas9-control cells both lose the ability to oxidize glucose through the TCA cycle when treated with doxycycline as assessed by reduced m+2 isotopologues and by enhanced fractional abundances of M+3 isotopologues (Fig. 6I-J and S6G-I). These labeling patterns suggest that pyruvate carboxylation (PC) is involved in producing TCA cycle metabolites required for cell proliferation after prolonged DN-POLG expression. Enhanced PC-dependent labeling has been previously observed in metabolically adapted proliferating cells with defective complex III (Mullen et al., 2014). Thus, it was possible that ρ° cells required oxidative TCA cycle in addition to reestablishment of mitochondrial membrane potential to completely restore cell proliferation. NDI1 and AOX proteins expression in DN-POLG ATPIF1 knockout cells reestablished these cells’ ability to consume oxygen in the presence of doxycycline thereby restoring oxidative TCA cycle (Fig S6J). Surprisingly, the ability of these cells to conduct oxidative TCA cycle did not confer a proliferative advantage, as their proliferation rate was comparable to control cells when treated with doxycycline (Fig. S6K). It is important to note that ATPIF1 knockout DN-POLG cells are unable to produce ATP from the mitochondria and therefore are dependent on glycolysis for survival as they undergo cell death in galactose media (Fig. S6L-M). Consequently, these cells displayed AMPK phosphorylation upon doxycycline treatment (Fig. S6N-O). AMPK under certain conditions negatively regulate cell proliferation and might prevent the complete rescue of proliferation in ρ° cells that exhibit a mitochondrial membrane potential.
Mitochondrial membrane potential dependent ROS production is necessary for cell proliferation and HIF-1 activation
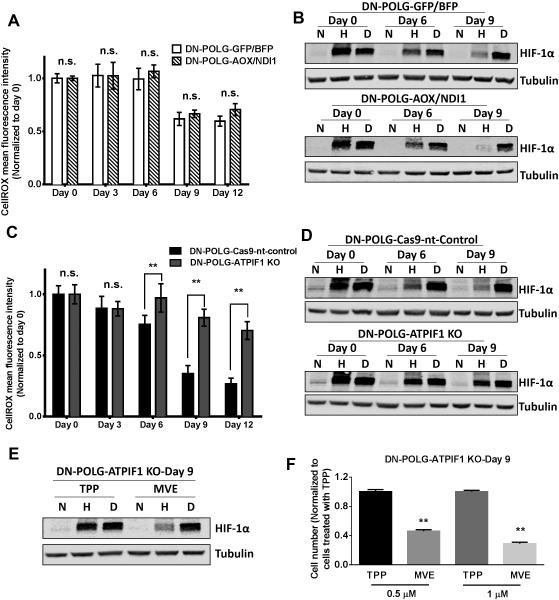
An important consequence of respiration is the generation of ROS, which has been implicated in activation of the transcription factor HIF-1 (Chandel et al., 1998). However, the factors that control mitochondrial ROS in intact cells are not fully understood. HIF-1 is a heterodimer containing an oxygen sensitive HIF-1α subunit and an oxygen insensitive HIF-1β subunit (Semenza, 2012). Under normoxia, the HIF-1α subunit is hydroxylated at specific proline residues by prolyl hydroxylases (PHDs), thereby targeting it for proteasomal-dependent degradation (Kaelin and Ratcliffe, 2008). Hypoxia diminishes hydroxylation of the HIF-1α subunit, resulting in its stabilization and dimerization with the HIF-1β subunit to activate metabolic genes that confer adaptation to hypoxia. DN-POLG control cells and DN-POLG cells expressing NDI1 and AOX displayed a decrease in ROS levels as well as diminished hypoxic stabilization of HIF-1α protein upon doxycycline treatment (Fig. 7A-B and S7A). Importantly, the PHD inhibitor DMOG was able to stabilize HIF-1α protein under normoxic conditions in both cell types upon doxycycline treatment indicating that loss of ETC function does not directly cause loss of HIF-1α protein expression in ρ° cells. Doxycycline-treated DN-POLG ATPIF1 knockout cells were capable of maintaining ROS levels indicating that the mitochondrial membrane potential was required for ROS release into cytosol (Fig. 7C). Additionally, in contrast to control cells, DN-POLG ATPIF1 knockout cells were able to stabilize HIF-1α protein under hypoxia upon doxycycline treatment (Fig. 7D). To test whether mitochondrial membrane potential dependent ROS production is necessary for cell proliferation and HIF-1α protein stabilization during hypoxia, we administered the mitochondrial targeted antioxidant Mito-Vitamin E (MVE) or the control compound TPP. MVE is a vitamin E conjugated to the cation TPP, which allows accumulation of compounds into mitochondria based on mitochondrial membrane potential. DN-POLG ATPIF1 knockout cells untreated or treated with doxycycline for 9 days showed diminished stabilization of HIF-1α protein and cell proliferation in the presence of MVE (Fig. 7E-F and S7B-C).
Figure 7. Mitochondrial membrane potential dependent ROS is essential for hypoxic stabilization of HIF-1α protein and cell proliferation.
(A) ROS levels were measured in DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 3, 6, 9 and 12 days. Mean ± SEM (n=3).
(B) Representative western blot from 3 independent experiments of DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells untreated or treated with doxycycline (10 ng/ml) for 6 and 9 days and placed in normoxia (21% O2), hypoxia (1.5% O2) or treated with 1 mM DMOG (21% O2) for 4 hours. Hypoxic induction of HIF-1α was analyzed by western blot.
(C) ROS levels were measured in DN-POLG-Cas9-nt-control and DN-POLG-ATPIF1 KO cells untreated or treated with doxycycline (10 ng/ml) for 3, 6, 9 and 12 days. Mean ± SEM (n=3).
(D) Representative western blot from 3 independent experiments of DN-POLG-Cas9-nt-control and DN-POLG-ATPIF1 KO cells untreated or treated with doxycycline (10 ng/ml) for 6 and 9 days and placed in normoxia (21% O2), hypoxia (1.5% O2) or treated with 1 mM DMOG (21% O2) for 4 hours. Hypoxic stabilization of HIF-1α was analyzed by western blot.
(E) Representative western blot from 3 independent experiments of DN-POLG-ATPIF1 KO cells treated with doxycycline (10 ng/ml) for 9 days pretreated with TPP or MVE for 2 hours and subsequently placed in normoxia (21% O2), hypoxia (1.5% O2) or treated with 1 mM DMOG (21% O2) for 4 hours. Hypoxic stabilization of HIF-1α was analyzed by western blot.
(F) DN-POLG-ATPIF1 KO cells treated with doxycycline (10 ng/ml) for 9 days were exposed to 0.5 μM or 1 μM TPP or MVE for the last 3 days of doxycycline treatment and cell number was assessed. Mean ± SEM (n=3).
** Indicates significance p <0.01 throughout figure 7. See also Figure S7.
To further support the necessity of mitochondrial membrane potential for cell proliferation we utilized complex III deficient cytochrome b null 143B osteosarcoma (143B CytB) cells. Recent studies demonstrated that 143B CytB cells are dependent on pyruvate to maintain the NAD+/NADH ratio to generate aspartate, which is necessary for cell proliferation (Birsoy et al., 2015; Sullivan et al., 2015). Consistent with these papers we demonstrate that α-ketobutyrate, known to maintain the NAD+/NADH ratio, can substitute for exogenous pyruvate (Fig. S7E). Complex III deficiency of 143B CytB cells results in dependency on an intact complex V to maintain the mitochondrial membrane potential. The addition of oligomycin decreases the mitochondria membrane potential and ROS release into cytosol thus diminishing cell proliferation (S7D-F). MVE also diminished proliferation of cytochrome b null cells (Fig. S7G). Collectively our data indicates that both the maintenance of NAD+/NADH ratio and mitochondrial membrane potential for aspartate and ROS production, respectively are necessary but not sufficient for cell proliferation.
Discussion
Here, we report that the maintenance of a mitochondrial membrane potential is essential for cell proliferation. Cells depleted of mitochondrial DNA (ρ°-cells) use glycolysis as their only source of ATP and must rely on a mechanism other than the ETC-dependent proton pumping to maintain a mitochondrial membrane potential. It has been suggested that ρ°-cells can potentially maintain a mitochondrial membrane potential through the electrogenic exchange of ATP4- for ADP3- by the adenine nucleotide carrier as well as from the ATPase activity of an incomplete F0F1-ATPase loosely associated with the membrane (Buchet and Godinot, 1998). It is estimated that 13% of ATP produced by glycolysis is utilized to maintain this mitochondrial membrane potential (Appleby et al., 1999). However, the inhibitor ATPIF1 limits the activity of the F1-ATPase component (Formentini et al., 2012). A recent genetic screen in mammalian cells identified that loss of ATPIF1 maintains the mitochondrial membrane potential to protect against antimycin induced cell death (Chen et al., 2014). Thus, in the presence of ATPIF1, ρ°-cells have a diminished membrane potential. Indeed, we observed inducible ρ°-cells in the absence of ATPIF1 were able to maintain mitochondrial membrane potential and increase proliferation. By contrast, ectopic expression of NDI1 and AOX that was sufficient to generate metabolites by the oxidative TCA cycle could not support proliferation of inducible ρ°-cells without a mitochondrial membrane potential. One possible reason for the failure of NDI1 and AOX expression to sustain proliferation in inducible ρ°-cells might be an inability to import nuclear DNA encoded proteins into the mitochondrial matrix due to the low mitochondrial membrane potential. However, we observed functional nuclear encoded succinate dehydrogenase activity in the inducible ρ°-cells. Previous isolated mitochondria studies demonstrated that as the mitochondrial membrane potential increases, ROS production elevates (Murphy, 2009). Indeed, we observed that restoration of a mitochondrial membrane potential but not the expression of NDI1 and AOX proteins in ρ°-cells increases the levels of ROS which are necessary for cell proliferation. Therefore, we propose that mitochondrial membrane potential provides sufficient ROS levels to sustain cell proliferation.
The stabilization of hypoxia-inducible factor (HIF-1α) protein was an early biological outcome linked to mitochondrial ROS (Chandel et al., 1998). Our data demonstrates that inducible ρ°-cells without a robust mitochondrial membrane potential do not stabilize HIF-1α protein during hypoxia. However, restoration of mitochondrial membrane potential in ρ°-cells by abolishing ATPIF1 protein expression increased ROS levels which reestablished hypoxic stabilization of HIF-1α protein. These results were surprising since they indicate that an incomplete ETC can generate ROS provided there is a robust mitochondrial membrane potential. Interestingly, ρ°-cells in the absence of a robust mitochondrial membrane potential are unable to stabilize HIF-1α protein during hypoxia despite restoration of the oxidative TCA cycle by ectopic expression of NDI1 and AOX, which is sufficient to generate TCA cycle metabolites and oxygen consumption. Collectively our findings indicate that an essential factor in regulating hypoxia-induced HIF-1α protein stabilization is the mitochondrial membrane potential through the production of ROS.
The TCA cycle intermediates acetyl-CoA and α-ketoglutarate are needed for histone acetylation and demethylation, respectively (Kaelin and McKnight, 2013). Surprisingly, using our inducible mtDNA depletion strategy to impair respiration, we did not observe significant global changes in histone methylation. This result is likely due to the presence of multiple mechanisms to sustain α-ketoglutarate in the cytosol including alanine and aspartate transaminases. Indeed, we did not observe significant changes in α-ketoglutarate levels in cells depleted of mtDNA. In contrast, the inducible loss of mtDNA decreased site-specific histone acetylation marks. The inducible loss of mtDNA did not display the widespread loss of acetylation on a variety of histones as it was observed in cells lacking cytosolic ACL enzyme, which would restrict the production of cytosolic acetyl-CoA from citrate. These ρ°-cells are able to generate a small pool of cytosolic citrate thus likely preventing widespread loss of histone acetylation, but yet still invoking some decrease in specific histone acetylation marks. The restoration of TCA cycle metabolites including citrate in inducible ρ°-cells with the ectopic expression of NDI1 and AOX was sufficient to restore specific histone H3 acetylation marks. However, these H3 acetylation marks were not re-established following restoration of the mitochondrial membrane potential. Furthermore, the presence of cytosolic acetate and pyruvate, which can both generate nuclear acetyl-coA (Shi and Tu, 2015; Sutendra et al., 2014), did not restore histone H3 acetylation marks in inducible ρ°-cells suggesting that in addition to acetyl-coA limitation invoked by impaired respiration there are likely other metabolites from TCA cycle that are necessary for histone acetylation. Collectively, these findings indicate that respiration regulates specific histone acetylation by allowing the oxidative TCA cycle to generate metabolites.
In summary, loss of mitochondrial respiration significantly impairs cellular proliferation, oxygen sensing, and acetylation of specific histone H3 marks without diminishing cell viability. Our study deciphers the distinct functions of mitochondria responsible for regulating various biological outcomes by separating the oxidative TCA cycle and the mitochondrial membrane potential. Here, we show that the contribution of mitochondria to control diverse biological outcomes extends beyond their bioenergetic function to include the biosynthetic function of the TCA cycle and the signaling function of mitochondrial ROS. Moving forward, it will be important to utilize similar approaches to dissect mitochondrial functions in vivo and decipher which mitochondrial-dependent metabolic requirements are required for proliferation, oxygen sensing, and epigenetics.
Experimental Procedures
Cell Culture
WT-POLG and DN-POLG T-REx293 cells were previously described (Wanrooij et al., 2007). Cells were grown in ρ° media (Dulbecco’s Modified Eagle’s Medium containing 4.5 g/L glucose, 4 mM L-glutamine, 10% fetal bovine serum, 1 mM sodium pyruvate, 100 μg/ml uridine, 1% HEPES, 1% antibiotic-antimycotic (Gibco), 150 μg/ml hygromycin and 5 μg/ml blasticidin) at 37 °C with 5% CO2. AOX, NDI1 and control pWPI vectors containing GFP and BFP (Cannino et al., 2012) as well as ATPIF1 mutated cDNA, NDI1 and pCDH vectors, containing GFP and RFP, were transfected into 293T cells using lipofectamine 2000 (Invitrogen) along with pMD2.G and psPAX2 packaging vectors to produce Control-GFP, Control-BFP, Control-RFP, AOX-GFP, NDI1-BFP, ATPIF1-GFP or NDI1-RFP lentivirus. DN-POLG-ATPIF1 KO cells were generated using the LentiCRISPR plasmid, which expresses Cas9 along with gene-specific RNA guides (gRNAs). 143B CytB cells were grown as previously described (Weinberg et al., 2010).
Proliferation and Cell Viability analysis
2.0×104 cells were plated on 24mm dishes. Cells were expanded and counted 3, 6, 9 and 12 days after plating in the presence or absence of doxycycline using AccuCount Fluorescent Particles from Fisher. Cell viability percentage was determined by DAPI or PI staining and analyzed by flow cytometry. To monitor distinct generations of proliferating cells, carboxyfluorescein succinimidyl ester (CFSE) was used to stain cells for 3 days and dilution of the dye was detected by flow cytometry. All flow cytometric analyses were performed using a LSR II flow-cytometer (Becton Dickinson, Franklin Lakes, NJ USA).
SILAC
DN-POLG cells treated with doxycycline for 6 days were grown in ‘light media’ containing [U-12C614N2]-L-lysine and [U-12C614N4]-L-arginine. Untreated DN-POLG cells were grown in ‘heavy media’ containing the isotopes [U-13C615N2]-L-lysine and [U-13C615N4]-L-arginine. All medias were supplemented with 10% dialyzed FBS. Before doxycycline treatment, cells were grown in ‘heavy media’ for more than 6 generations to achieve more than 99% labeling efficiency. The core histone proteins were prepared from the cell mixture using an acid-extraction procedure described previously (Shechter et al., 2007), resolved in an SDS-PAGE gel and visualized by Coomassie Brilliant Blue staining. The bands of histone H2A, H2B, H3, H4 were excised and in-gel digested using a procedure described previously to quantify histone proteins and to map PTMs (Zhao et al., 2003). All the modified peptide hits were manually verified. The final PTM site ratios were normalized by their protein ratios.
Metabolic Assays
OCR and ECAR were measured utilizing the XF24 Seahorse Biosciences Extracellular Flux Analyzer as previously described (Sullivan et al., 2013). The membrane potential of mitochondria was analyzed with the potential-dependent fluorescent dye tetramethylrhodamine (TMRE, Molecular Probes). ROS was measure using CellROX Deep Red or CellROX Orange reagents (Life Technologies). Metabolomics of DN-POLG-GFP/BFP and DN-POLG-AOX/NDI1 cells were conducted at Metabolon (Durham, N.C.). Glucose and glutamine uptake was determined using Biolog Assays. Carbon labeling experiments were performed as previously described (Mullen et al., 2014).
Immunoblot analysis
Protein was extracted using cell lysis buffer (Cell Signaling) plus PMSF (600μM) and inhibitor of phosphatases (100x). Protein concentration was quantified using the BCA Protein Assay (Pierce). Protein samples were resolved on SDS polyacrylamide gels (Bio-Rad) and subsequently transferred to nitrocellulose membranes by semi-dry transfer using the Trans-Blot Turbo (Bio-Rad). The following antibodies were utilized: subunit 70 kDa (SDHA) antibody from MitoSciences (abcam), COXII antibody from Invitrogen, anti-AMPK and anti-p-AMPK from Cell Signaling, anti-ATPIF1 from Sigma, HIF-1α (BD Transduction Laboratories, Clone 54/HIF-1), β-actin and α-tubulin (Sigma). To assess changes in epigenetics marks, nuclear extracts were prepared using the nuclear complex Co-IP kit from Active Motif and the following antibodies were used: anti-Histone H3K27ac and anti-Histone H3K14ac from Active Motif, anti-Histone H3K18ac, anti-Histone H3K9ac, anti-Histone H3 (tri methyl K4) and anti-Histone H3 (tri methyl K9) from Abcam, anti-Histone H3 (tri methyl K27) and anti-Histone 3 from Millipore. Anti-rabbit 800CW and anti-mouse 680RD from Licor were used as secondary antibodies. Image Studio Lite version 3.1 (Licor) was used for analysis and quantification of protein levels.
Histones Acetyltransferases (HATs) and Histone deacetylases (HDACs) activity assay
Nuclear fractionation was performed using the nuclear/cytosol fractionation kit from Biovision (Mountain View, CA) according to the manufacturer’s directions. HATs and HDACs activity was measured using the HATs activity fluorometric assay kit (Biovision) and HDAC activity fluorometric assay kit (Biovision) respectively, following the protocol of the manufacturer.
Statistical Analysis
Data are presented as the mean +/− SEM. Statistical significance was determined using 1-way ANOVA with a Bonferroni posttest correction, 2-way ANOVA when two variables were present, or the students T-test comparing control to experimental conditions for p<0.05 or p<0.01.
Supplementary Material
Acknowledgements
This work was supported by NIH (RO1 CA12306708, PO1AG049665 and RO1 HL122062) to NSC, a postdoctoral fellowship by Ramon Areces Foundation of Spain to IMR, NIH (T32 CA009560) to MMM, NIH (T32 GM008061) to LPD, NIH (T32 T32HL076139) to SEW and NIH (T32 HL076139) to MS. JNS is supported by Netherlands Organization for Scientific Research (865.10.004). RJD was supported by RO1 CA157996 and Robert A. Welch foundation grant I17733. We thank Robert H. Lurie Cancer Center Flow Cytometry facility for their invaluable assistance. We thank the members of the Chandel lab for thoughtful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures, one table, and Supplemental Experimental Procedures and can be found with this article at http://
Author Contributions
IMR, LPD, HK, MS, MMM, SW performed cell proliferation, western blot, HAT, ROS, metabolic assay, and oxygen consumption experiments. TW and JS performed RNA sequencing. HE and IMR performed SILAC experiments. CH and IMR performed the isotopic carbon labeling experiments. ED and JS provided critical reagents. IMR, RD, YZ, MS, JS and RW and NSC provided insightful discussions and designed the study. IMR and NSC wrote the manuscript.
References
- Appleby RD, Porteous WK, Hughes G, James AM, Shannon D, Wei YH, Murphy MP. Quantitation and origin of the mitochondrial membrane potential in human cells lacking mitochondrial DNA. European journal of biochemistry / FEBS. 1999;262:108–116. doi: 10.1046/j.1432-1327.1999.00350.x. [DOI] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, Sabatini DM. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroughs LK, DeBerardinis RJ. Metabolic pathways promoting cancer cell survival and growth. Nature cell biology. 2015;17:351–359. doi: 10.1038/ncb3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchet K, Godinot C. Functional F1-ATPase essential in maintaining growth and membrane potential of human mitochondrial DNA-depleted rho degrees cells. The Journal of biological chemistry. 1998;273:22983–22989. doi: 10.1074/jbc.273.36.22983. [DOI] [PubMed] [Google Scholar]
- Cannino G, El-Khoury R, Pirinen M, Hutz B, Rustin P, Jacobs HT, Dufour E. Glucose modulates respiratory complex I activity in response to acute mitochondrial dysfunction. The Journal of biological chemistry. 2012;287:38729–38740. doi: 10.1074/jbc.M112.386060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413–416. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel N, Maltepe E, Goldwasser E, Mathieu C, Simon M, Schumacker P. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell metabolism. 2015 doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Chen WW, Birsoy K, Mihaylova MM, Snitkin H, Stasinski I, Yucel B, Bayraktar EC, Carette JE, Clish CB, Brummelkamp TR, et al. Inhibition of ATPIF1 ameliorates severe mitochondrial respiratory chain dysfunction in mammalian cells. Cell reports. 2014;7:27–34. doi: 10.1016/j.celrep.2014.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formentini L, Sanchez-Arago M, Sanchez-Cenizo L, Cuezva JM. The mitochondrial ATPase inhibitory factor 1 triggers a ROS-mediated retrograde prosurvival and proliferative response. Molecular cell. 2012;45:731–742. doi: 10.1016/j.molcel.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Molecular cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, et al. Oxidation of Alpha-Ketoglutarate Is Required for Reductive Carboxylation in Cancer Cells with Mitochondrial Defects. Cell reports. 2014 doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. How mitochondria produce reactive oxygen species. The Biochemical journal. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales-Clemente E, Bayona-Bafaluy MP, Perez-Martos A, Barrientos A, Fernandez-Silva P, Enriquez JA. Restoration of electron transport without proton pumping in mammalian mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18735–18739. doi: 10.1073/pnas.0810518105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieber M, Chandel NS. ROS function in redox signaling and oxidative stress. Current biology : CB. 2014;24:R453–462. doi: 10.1016/j.cub.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nature protocols. 2007;2:1445–1457. doi: 10.1038/nprot.2007.202. [DOI] [PubMed] [Google Scholar]
- Shi L, Tu BP. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Current opinion in cell biology. 2015;33:125–131. doi: 10.1016/j.ceb.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell. 2015;162:552–563. doi: 10.1016/j.cell.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, Licht JD, Deberardinis RJ, Chandel NS. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Molecular cell. 2013;51:236–248. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutendra G, Kinnaird A, Dromparis P, Paulin R, Stenson TH, Haromy A, Hashimoto K, Zhang N, Flaim E, Michelakis ED. A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell. 2014;158:84–97. doi: 10.1016/j.cell.2014.04.046. [DOI] [PubMed] [Google Scholar]
- Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–1258. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanrooij S, Goffart S, Pohjoismaki JL, Yasukawa T, Spelbrink JN. Expression of catalytic mutants of the mtDNA helicase Twinkle and polymerase POLG causes distinct replication stalling phenotypes. Nucleic acids research. 2007;35:3238–3251. doi: 10.1093/nar/gkm215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Finkel T. Mitohormesis. Cell metabolism. 2014;19:757–766. doi: 10.1016/j.cmet.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Zhang W, White MA, Zhao Y. Capillary high-performance liquid chromatography/mass spectrometric analysis of proteins from affinity-purified plasma membrane. Analytical chemistry. 2003;75:3751–3757. doi: 10.1021/ac034184m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.