Abstract
Incurable chronic viral infections are a major cause of morbidity and mortality worldwide. One potential approach to cure persistent viral infections is via the use of targeted endonucleases. Nevertheless, a potential concern for endonuclease-based antiviral therapies is the emergence of treatment resistance. Here we detect for the first time an endonuclease-resistant infectious virus that is found with high frequency after antiviral endonuclease therapy. While testing the activity of HIV pol-specific zinc finger nucleases (ZFNs) alone or in combination with three prime repair exonuclease 2 (Trex2), we identified a treatment-resistant and infectious mutant virus that was derived from a ZFN-mediated disruption of reverse transcriptase (RT). Although gene disruption of HIV protease, RT and integrase could inhibit viral replication, a chance single amino acid insertion within the thumb domain of RT produced a virus that could actively replicate. The endonuclease-resistant virus could replicate in primary CD4+ T cells, but remained susceptible to treatment with antiretroviral RT inhibitors. When secondary ZFN-derived mutations were introduced into the mutant virus’s RT or integrase domains, replication could be abolished. Our observations suggest that caution should be exercised during endonuclease-based antiviral therapies; however, combination endonuclease therapies may prevent the emergence of resistance.
Keywords: zinc finger nuclease, endonuclease, resistance, reverse transcriptase
1. INTRODUCTION
Traditional antiviral therapy is unable to cure many persistent infections. Therefore, alternative treatments are being developed to eliminate replication-competent viral genomes within cells. In one such approach, highly specific endonucleases have been used to directly cleave and mutate viral genomes, thus preventing virus replication and persistence (For review see [1–4]). Several classes of endonucleases, including zinc finger nucleases (ZFNs), meganucleases, TAL effector nucleases (TALENs) and CRISPR/Cas9 proteins, can now be engineered to cleave DNA at a precise sequence [5, 6], and this has enabled specific disruption of viral genomes, without host cell damage.
Recently a number of persistent viruses have been targeted for antiviral endonuclease therapy using HEs, ZFNs, TALENs or CRISPR/Cas9. Encouraging results have been achieved against all viruses targeted, with antiviral activity demonstrated using in vitro and in vivo models of viral replication. Endonucleases have been used to target HBV [7–13], HCV [14], the herpes viruses EBV and HSV [15–19], HPV [20–27], polyomavirus JC [28], and the retroviruses HIV and HTLV [29–36]. This body of data suggests that when highly active virus-specific endonucleases are efficiently delivered to infected cells, their antiviral activity is robust. Nevertheless, an important concern for any endonuclease-based antiviral therapy is the emergence of treatment-resistant infectious virus [2, 37], and so far this has not been described.
Here we report for the first time the appearance of infectious treatment-resistant virus in cells receiving antiviral endonuclease therapy. During experiments evaluating the antiviral activity of HIV pol-specific ZFNs, an RT mutant virus was identified that remained infectious despite ZFN target site disruption. This treatment-resistant virus, with an insertion of 3 nucleotides within a ZFN target site in RT, provided direct evidence for a previously-suggested mechanism of viral escape [2, 37]. Importantly, treatment-resistant virus was able to replicate in primary human CD4+ T cells, but could be rendered replication incompetent by the introduction of secondary mutations at other ZFN target sites.
2. MATERIAL AND METHODS
2.1 HIV-specific ZFNs
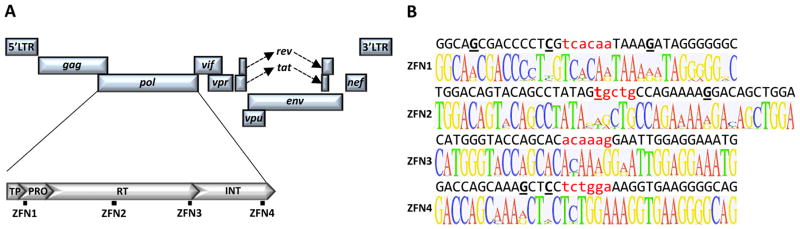
Zinc finger nucleases targeting sequences in HIV protease (ZFN1), reverse transcriptase (ZFN2 & ZFN3) and integrase (ZFN4) were generated by Sigma Life Science (Fig 1A). Heterogeneity analysis was performed using 3445 HIV polymerase sequences from the 2013 Los Alamos National Laboratory (LANL) HIV database (http://www.hiv.lanl.gov/). Logo plots were generated using GeneiousPro by Biomatters (www.genious.com).
Fig. 1.
HIV-specific zinc finger nucleases. (A) Location of ZFN binding sites within the HIV-1 genome. (B) Logo plot heterogeneity analysis of ZFN target sites (above) across 3445 HIV pol sequences within the 2013 Los Alamos National Laboratory database. HIV target site sequences are from NL4-3. Spacer nucleotides between the left and right zinc finger pairs are shown in lower case red and nucleotides that differ from the consensus sequence are underlined. TP – Transframe peptide; PRO – Protease; RT – Reverse transcriptase; INT – Integrase.
2.2 Cell lines and drugs
SupT1 cells (ATCC# CRL-1942) were grown in RPMI 1640 (Invitrogen) with 10% FBS. HEK293 [38], 293T (ATCC# CRL-3216) and TZM-bl cells [39] were grown in DMEM (Invitrogen) with 10% FBS. Tenofovir and AZT were provided through the NIH AIDS Reagent Program, NIH. Nevirapine and Etravirine were obtained from Santa Cruz Biotechnology and Selleckchem respectively.
2.3 AAV vectors and in vitro transductions
AAV plasmid cloning, vector production, and purification are described in the supplemental materials. Optimization of SupT1 cell AAV delivery was performed using iodixanol-purified virus stocks. All other AAV experiments were performed using viral lysates.
2.4 Plasmids
To analyze ZFN cleavage efficiency, target sites were amplified using primers ZFN1-F/ZFN1-R, ZFN2-F/ZFN2-R, ZFN3-F/ZFN3-R or ZFN4-F/ZFN4-R (Supplemental materials) and then cloned using the Zero Blunt TOPO PCR cloning kit (Invitrogen). The plasmid pNL4-3 was used to generate ZFN target-site mutants and was obtained through the NIH AIDS Reagent Program, NIH from Dr. Malcolm Martin and contains a full length NL4-3 strain HIV molecular clone [40]. The plasmid pDHIV3 contains the NL4-3 strain with a small deletion in env [41].
2.5 Production of HIV
Replication-defective and replication-competent NL4-3 derived virus was produced as described in the supplemental materials.
2.6 NL4-3 inhibition assay
HEK293 cells in 6 well plates were transfected with 1μg of pNL4-3, 0.75μg of each ZFN subunit-expressing plasmid and 0.5μg of a Trex2-expressing plasmid. At 72 hours post transfection supernatants were harvested and p24 levels measured by ELISA (Advanced Bioscience Laboratories, Rockville, MD). TZM-bl cells were then incubated with 2ng of p24 for 48 hours to determine infectious virus levels.
2.7 T7 endonuclease I assay and PCR amplicon sequencing
The T7 endonuclease I cleavage assay and amplicon-sequencing protocols have been described [15]. Primers to generate target site-specific PCR products are described in the supplemental materials.
2.8 Primary CD4+ T cell culture
CD4+ T cells were isolated by negative selection from PBMC obtained from healthy HIV-negative donors using the CD4+ isolation kit Human (Miltenyi Biotec). Isolated CD4+ cells were then activated using CD3/CD28 beads (Life Technologies) for 3 days according to the manufacturers protocol. Activated T cells were plated at 105 cells/well in 48-well plates and infected with 2–50ng of NL4-3 or mutant viruses by spinoculation in 500μl of RPMI containing 10% FBS at 2900 rpm for 2 hours. Infected cells were washed twice with 1X PBS, plated in 48-well plates with 30U/ml of IL-2, and incubated for an additional 72 hours before analysis of intracellular p24 levels.
2.9 Intracellular p24 staining in primary CD4+ T cells
At 72 hours post infection cells were fixed and permeabilized using the BD Cytofix/Cytoperm Fixation/Permeabilization Kit (BD Biosciences). Cells were then stained for HIV-1 p24 for 30 minutes at room temperature with 5μl of KC57-FITC monoclonal antibody (Beckman Coulter) immediately prior to flow cytometry analysis.
2.10 Illumina sequencing
Next generation sequencing of wild type or mutant ZFN target sites was performed using a MiSeq sequencer (Illumina) as described in the supplemental materials.
2.11 Statistical analysis
Analysis was performed using an unpaired t-test with two-tailed p-values.
3. RESULTS
3.1 HIV-specific ZFNs
HIV-specific ZFNs were generated against target sites within the NL4-3 HIV pol sequence (Fig 1A). Targets were within the protease (ZFN1), RT (ZFN2 & ZFN3) and integrase (ZFN4) coding sequences (Supplemental Fig 1A–C). To predict ZFN activity against different HIV isolates, 3445 pol sequences from the LANL HIV genome database were aligned and analyzed for target-site conservation. Target sites were highly conserved, with 3 (ZFN1), 1 (ZFN2), 0 (ZFN3) and 2 (ZFN4) differences from the consensus sequence ZFN DNA-binding domains respectively (Fig 1B).
3.2 Optimization of ZFN delivery
As a means to deliver ZFNs, AAV vectors were investigated to determine the optimal serotype, promoter and MOI for gene delivery to the SupT1 cell line (Supplemental data, Supplemental Figure 2). scAAV1 vectors utilizing the EF1α short (EFS) promoter were selected for further studies as they could deliver ZFNs to SupT1 cells by co-transduction of vectors expressing individual subunits at levels of >99%.
3.3 Gene disruption of HIV pol target sites
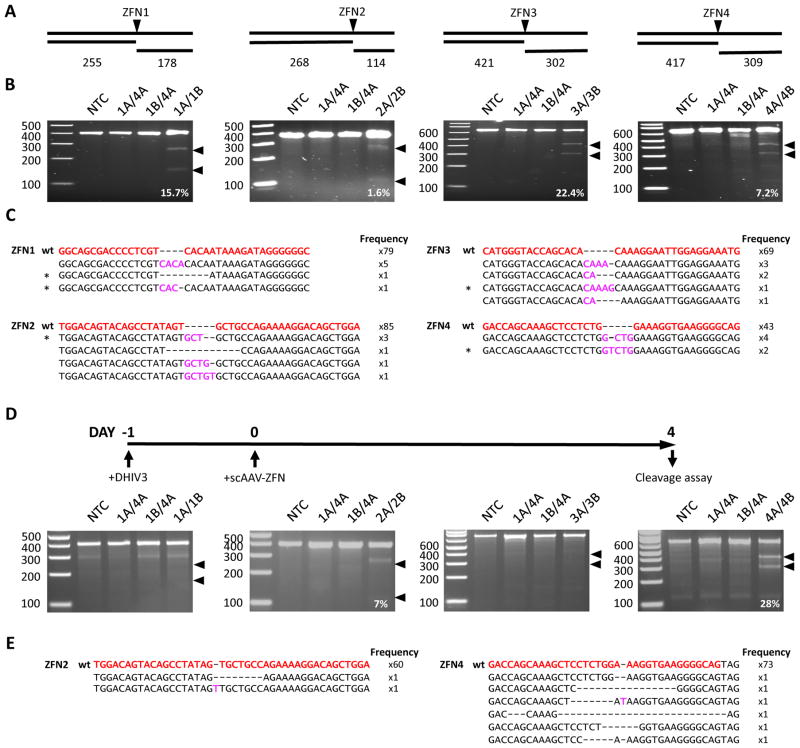
HIV-specific gene disruption was first analyzed in HEK293 cells transfected with ZFN expression plasmids and a plasmid containing a replication-incompetent NL4-3 derived provirus (pDHIV3). After 48 hours target site disruption was analyzed by T7 endonuclease I assay and clonal sequencing (Fig 2A–C). Cells treated with each ZFN pair showed evidence of target site disruption by T7 endonuclease I assay, at levels of 15.7%, 1.6%, 22.4% and 7.2% respectively for pairs 1–4. By clonal sequencing gene disruption levels were 8.1% (7 of 86 clones sequenced), 6.6% (6 of 91), 9.2% (7 of 76) and 12.2% (6 of 49). We then performed Illumina sequencing on target site PCR amplicons to more accurately assess the mutation landscape following treatment with each ZFN pair (Table 1, Supplemental data). We found that 9.8%, 9.1%, 10.6% and 7.5% of filtered reads contained mutations within the ZFN target site after treatment with ZFN pairs 1–4 respectively. Of the unique mutations detected 29.8% (59/198), 27.2% (53/195), 32.0% (33/103), and 31.1% (28/90) produced an in-frame pol ORF for ZFN pairs 1–4 respectively. For each ZFN pair insertions were more prevalent than deletions, and interestingly the most common insertions for each ZFN pair contained duplications of sequences found within the spacer region present between the ZFN binding domains of each ZFN pair (Supplemental Table 1).
Fig. 2.
ZFN-mediated disruption of HIV target sites. (A) PCR amplicon and T7 endonuclease cleavage product sizes indicative of HIV-specific ZFN cleavage and mutagenesis. ZFN-mediated gene disruption was analyzed in HEK293 cells transfected with the plasmid pDHIV3 using the T7 endonuclease cleavage assay (B) and clonal amplicon sequencing (C). HEK293 cells were co-transfected with plasmids expressing ZFN pairs 1–4, a mismatch ZFN homodimer (1A/4A), or a mismatch heterodimer (1A/4B), and 48 hours later total DNA was isolated for analysis of target site disruption. ZFN-mediated gene disruption within the integrated provirus was also analyzed in SupT1 cells infected with DHIV3 using the T7 endonuclease cleavage assay (D) and clonal amplicon sequencing (E). SupT1 cells were infected with DHIV3 (MOI=2 transducing units/cell) on day 0 and then co-transduced on day 1 with scAAV1 vectors (MOI = 50,000 genomes/vector/cell) expressing ZFN pairs 1–4, a mismatch ZFN homodimer (1A/4A), or a mismatch heterodimer (1A/4B) and target site analysis was performed using cells harvested at day 4. Arrows indicate expected cleavage product locations (B, D). NTC – No treatment control. Wild type ZFN target sequences are shown in red, deletions are indicated by dashes, and insertions are shown in pink (C, E). Black asterisks indicate mutant sequences that were later introduced into the pNL4-3 molecular clone. wt – wild type.
Table 1.
Illumina sequencing of ZFN target sites in HEK293 cells. ZFN target sites from the experiment shown in figure 1 were amplified by PCR and then subjected to Illumina sequencing. Target site sequence variations are shown for each sample as the percentage of filtered reads containing an insertion, deletion or either variant.
| Mutations in target site (% of filtered reads) | ||||||
|---|---|---|---|---|---|---|
| Total | Insertion | Deletion | In frame | Frame shift | ||
| ZFN1A/1B | control | 0.3 | 0.1 | 0.2 | 0.0 | 0.3 |
| treatment | 9.8 | 8.9 | 0.9 | 0.9 | 8.9 | |
|
| ||||||
| ZFN2A/2B | control | 0.1 | 0.0 | 0.1 | 0.0 | 0.1 |
| treatment | 9.1 | 7.9 | 1.2 | 3.3 | 5.8 | |
|
| ||||||
| ZFN3A/3B | control | 0.1 | 0.0 | 0.0 | 0.0 | 0.1 |
| treatment | 10.6 | 9.9 | 0.7 | 1.3 | 9.3 | |
|
| ||||||
| ZFN4A/4B | control | 0.1 | 0.1 | 0.0 | 0.0 | 0.1 |
| treatment | 7.5 | 7.0 | 0.5 | 1.6 | 5.9 | |
We next tested ZFN disruption of integrated HIV using a T-cell model of heterogeneous integration. SupT1 cells were infected with replication-defective DHIV3 virus at a MOI of 2 to ensure low provirus copy numbers (Fig 2D) and the following day co-transduced with scAAV1 vectors expressing ZFN pairs 1–4, control homodimer (ZFN1A/4A) or heterodimer (ZFN1B/4A) mismatch ZFN pairs, or control GFP- and mCherry-expressing vectors. ZFN target sites were analyzed for levels of mutation by T7 endonuclease assay and clonal sequencing (Fig 2D, E). Flow cytometry for GFP and mCherry at day 3-post scAAV treatment showed that over 90% of cells were co-transduced (data not shown). At day 4-post treatment, cells receiving ZFN2 and ZFN4 showed mutation frequencies of 7% and 28% respectively by T7 endonuclease assay, or 3.22% (2 of 62) and 7.5% (6 of 79) by amplicon sequencing. No evidence of mutations was seen at the target sites from cells treated with ZFN1 and ZFN3 using either assay. Provirus gene disruption in ZFN2 and ZFN4 treated SupT1 cells could be increased up to 2-fold upon co-expression of the 3′ to 5′ exonuclease Trex2 (Supplemental data, Supplemental Figure 3). We also performed Illumina sequencing of target site PCR amplicons from ZFN-treated SupT1 cells (Table 2, Supplemental data). By Illumina sequencing we were able to detect target site mutations in SupT1 cells treated with all 4 ZFN pairs. We saw mutations in 2.9%, 7.5%, 1.7% and 12.3% of Illumina reads for ZFN 1–4 respectively. Of the unique mutations detected 36.0% (9/25), 27.7% (18/65), 15.8% (6/38), and 29.3% (54/184) produced an in-frame pol ORF for ZFN pairs 1–4 respectively. In contrast to HEK293 cells, mutations in ZFN treated SupT1 cells were predominantly deletions and not insertions (Supplemental Table 2).
Table 2.
Illumina sequencing of ZFN target sites in SupT1 cells. ZFN2 and ZFN4 treated SupT1 cells from the experiment described in Figure 2D, E were analyzed for target site mutations. ZFN target sites were amplified by PCR and then used for Illumina sequencing. Target site sequence variations are shown for each sample as the percentage of filtered reads containing an insertion, deletion or either variant.
| Mutations in target site (% of filtered reads) | ||||||
|---|---|---|---|---|---|---|
| Total | Insertion | Deletion | In frame | Frame shift | ||
| ZFN1A/1B | control | 0.4 | 0.1 | 0.3 | 0.0 | 0.4 |
| treatment | 2.9 | 0.1 | 2.7 | 0.8 | 2.0 | |
|
| ||||||
| ZFN2A/2B | control | 0.3 | 0.2 | 0.1 | 0.0 | 0.3 |
| treatment | 7.5 | 1.3 | 6.2 | 4.0 | 3.5 | |
|
| ||||||
| ZFN3A/3B | control | 0.1 | 0.0 | 0.1 | 0.0 | 0.1 |
| treatment | 1.7 | 0.0 | 1.7 | 0.5 | 1.2 | |
|
| ||||||
| ZFN4A/4B | control | 0.3 | 0.1 | 0.2 | 0.0 | 0.3 |
| treatment | 12.3 | 2.9 | 9.5 | 2.7 | 9.6 | |
3.4 ZFN-mediated disruption of pol inhibits HIV replication
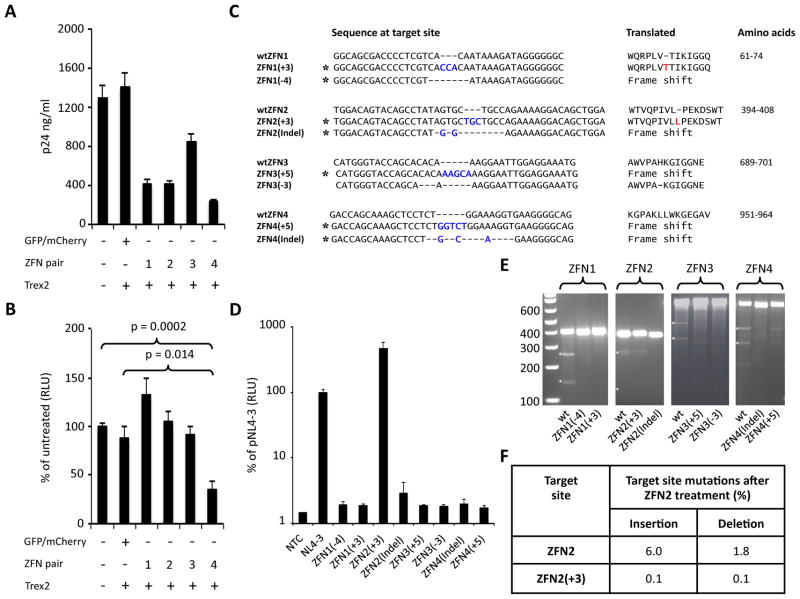
Antiviral activity of ZFNs was first analyzed in HEK293 cells transfected with pNL4-3 in combination with ZFN- and Trex2-expressing plasmids. At 48 hours post transfection supernatants from all HIV-specific ZFN treated cells contained fewer virus particles as indicated by levels of p24 when compared to control cells transfected with pNL4-3 alone or in combination with mCherry/GFP and Trex2 (Fig 3A), indicating that ZFN treatment could disrupt viral replication. When TZM-bl cells were incubated with equalized levels of detected viral particles (2ng p24/well) no loss of infectivity was seen after treatment with ZFNs 1–3. In contrast, ZFN4 treatment significantly reduced the infectivity of detected viral particles by 64% (Fig 3B, p=0.0002 vs untreated; p=0.0139 vs mCherry/GFP treated), suggesting that ZFN4 treatment rendered some virus particles replication incompetent.
Fig. 3.
Mutations in pol inhibit HIV replication. (A) p24 levels in supernatants from 293T cells transfected with pNL4-3 and plasmids expressing HIV-specific ZFN pairs in combination with Trex2 at 48 hours post transfection. (B) Levels of infectious HIV were analyzed by incubating TZM-bl cells with ZFN treated cell supernatants from A (2ng of p24/well). Cells were analyzed for luciferase activity 48 hours after supernatant treatment. (C) Nucleotide mutations introduced at each ZFN target site in pNL4-3. Asterisks indicate mutants identified in ZFN treated cells. Introduced nucleotides are shown in blue and introduced amino acids in red along with the location in the pol polypeptide. All sequences and coordinates are from the pNL4-3 sequence (Genbank Accession: AF324493). (D) Wild type and mutant pNL4-3 plasmids were transfected into HEK293 cells and 48 hours later supernatants were harvested for p24 quantification. TZM-bl cells were then inoculated with supernatant containing 2ng of p24 and 48 hours later luciferase activity was measured. (E) T7 endonuclease cleavage analysis of plasmid-derived mutant ZFN target sites in HEK293 cells 48 hours after co-transfection with plasmids expressing corresponding ZFN pairs. (F) Summary of mutations detected by Illumina sequencing at wtZFN2 or ZFN2(+3) target sites in HEK293 48 hours after treatment with ZFN2A/B. NTC – No treatment control; wt – wild type.
Viruses with non-functional protease, RT, or integrase should not undergo productive infection. To determine whether the individual mutations we observed would disrupt HIV replication, pol mutations that were identified in ZFN-treated HEK293 cells or SupT1 cells (Fig 2C, Supplemental Fig 3D) were introduced into pNL4-3. Mutations were chosen that either altered the pol reading frame, deleted a single codon, or introduced a single extra codon (Fig 3C). We selected predominantly small mutations as we reasoned that viruses with small mutations would be the most likely to retain infectivity. Wild-type and mutant NL4-3 plasmids were then transfected into HEK293 cells and production of infectious virus analyzed using TZM-bl cells. NL4-3 mutants representing each of the 4 ZFN target sites (7 out of 8) had viral infectivity reduced to background levels (Fig 3D). Unexpectedly, mutant NL4-3-ZFN2(+3), which contains a TGC duplication within the spacer region of the ZFN2 target site that introduces an additional leucine residue adjacent to Leu246 of RT (Fig 3C), retained infectivity comparable to wild-type NL4-3. Illumina sequencing data showed that in the HEK293 cells from which mutant ZFN2(+3) was initially identified, the TGC mutation could be found in 2.4% (498/20406) of all reads, 26.9% of reads containing mutations in the target site (498/1854), and 74.0% (498/673) of reads with in-frame mutations within the ZFN2 target site (Supplemental Table 4). To determine whether the frequency of this TGC mutation was reproducible, we repeated the analysis of pDHIV3 mutation in ZFN2 treated HEK293 cells in 2 additional experimental replicates. Illumina sequencing was performed using PCR amplicons from HEK293 cells harvested 48 hour after ZFN2A/2B treatment. In all 3 experimental replicates the pattern of mutations present in ZFN2 target site was comparable (Supplemental Table 3). Furthermore, the TGC insertion was either the most or second most prevalent mutation detected in each replicate (Supplemental data), and its frequency in all reads, target site mutations, and in-frame target site mutations was highly comparable (Supplemental Table 4). Illumina sequencing data was then used to determine the frequency of the TGC insertion in SupT1 cells infected with DHIV3 and treated with ZFN2 (Supplemental Table 4). In SupT1 cells the TGC insertion was the most frequently found insertion and was seen in 0.4% (62/14238) of all reads covering the target region, 5.8% (62/1067) of reads containing target site mutations, and 10.8% (63/585) of reads containing in-frame target site mutations respectively.
ZFNs consist of a pair of binding arrays, targeting the + and − DNA strands, with each array linked to one half of the FokI nuclease cleavage domain. Dimerization is required for DNA cleavage [42] and mutagenesis typically occurs between the array binding sites, which are often not themselves disrupted. Because the array-binding sequences in many of our mutants were not changed, it was possible that the mutant sequences might still be cleaved by our ZFNs. We therefore determined whether each mutant target site could be re-cleaved by its cognate ZFN pair in HEK293 cells, to determine whether the mutants were resistant to additional rounds of ZFN treatment. After 48 hours no target-site cleavage was detected by T7 endonuclease I assay in cells transfected with 6 of the 8 mutants. Cleavage of the infectious mutant ZFN2(+3) and the non-infectious mutant ZFN4(+5) was observed, but at considerably lower levels than in cells containing wild-type ZFN target sites (Fig 3E). To quantitate the resistance of the ZFN2(+3) target site relative to the wild-type ZFN2 site, we again performed Illumina sequencing analysis (Fig 3F). Mutations were found in the wild-type ZFN target site in 7.8% of reads (1565/20190), compared to only 0.2% (52/21638) of ZFN2(+3) target site reads. Thus, the virus containing the NL4-3-ZFN2(+3) mutant would be almost completely resistant to cleavage by ZFN2 compared to the wild-type virus. Similarly, mutant target sites ZFN1(+3) and ZFN3(+5) respectively were not cleaved despite also having intact ZFN DNA-binding sequences and similar target-site insertions. These data support prior observations that ZFN cleavage at mutant target sites can occur, but is unpredictable and can be context dependent [43, 44].
3.5 Characterization of infectious and endonuclease-resistant HIV
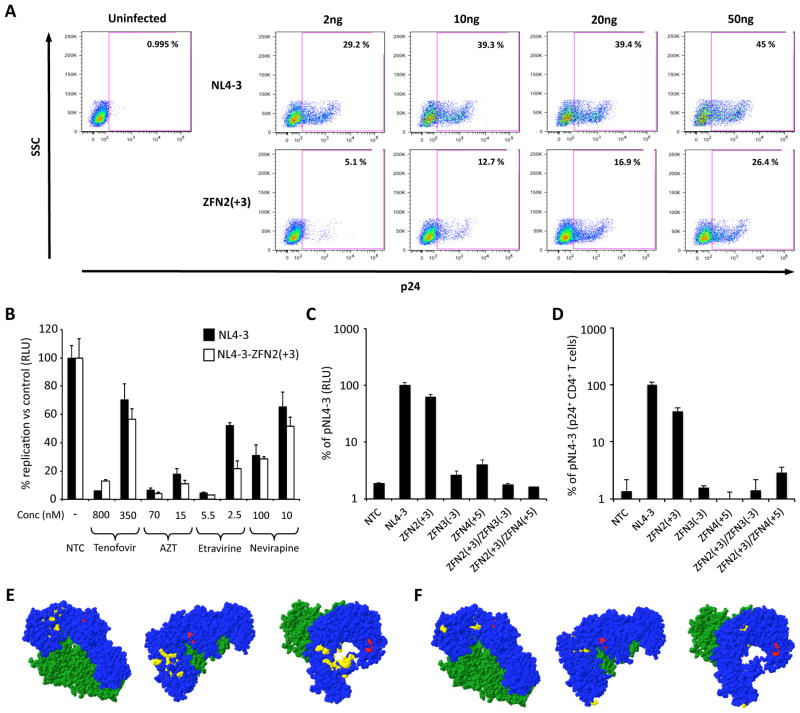
Studies in HEK293 cells or TZM-bl cells may not reflect replication in lymphoid cells. Therefore, we evaluated the NL4-3-ZFN2(+3) mutant virus in primary human T cells. Activated human CD4+ T cells were incubated with increasing amounts of NL4-3 or NL4-3-ZFN2(+3) virus and intracellular p24 levels analyzed as a surrogate for viral replication. CD4+ T cells incubated with both NL4-3 and NL4-3-ZFN2(+3) expressed intracellular p24 in a dose-dependent manner, although higher levels of p24 were seen in NL4-3 infected cells at each p24 dose (Fig 4A). Maximal levels of p24 resulted after incubation with 50ng of p24-containing supernatant, with 45% and 26.4% of CD4+ T cells positive for p24 after infection with NL4-3 and NL4-3-ZFN2(+3) respectively. This suggests that NL4-3-ZFN2(+3) can replicate in primary human CD4+ T cells, but with somewhat reduced efficiency compared to the parental NL4-3 virus.
Fig. 4.
Characterization of endonuclease resistant HIV. (A) Primary activated CD4+ T cells were incubated with the indicated amounts of NL4-3 or NL4-3-ZFN2(+3) viruses and then intracellular p24 expression was detected 72 hours later by flow cytometry. (B) TZM-bl cells were incubated with NRTi (Tenofovir; AZT) or NNRTi (Etravirine; Nevirapine) drugs for 2 hours prior to infection. Infections were then performed with normalized levels of infectious virus [2.5×105 luciferase producing units/well of NL4-3 or NL4-3-ZFN2(+3)] in the presence or absence of drugs, and levels of infection were analyzed 48 hours later by quantifying luciferase activity. (C) Wild type, single mutant or double mutant pNL4-3 plasmids were transfected into 293T cells and 48 hours later p24 levels in supernatants were quantified by ELISA. TZM-bl cells were then inoculated with supernatant containing 2ng of p24 and 48 hours later luciferase activity was measured. (D) Primary CD4+ T cells isolated from PBMC were activated by incubation with CD3/CD28 beads for 3 days. Activated CD4+ T cells were incubated with 50ng of wild type or NL4-3 mutant viruses and then intracellular p24 expression was detected by flow cytometry. NTC – No treatment control. The experiments shown in 4A and 4D were performed using cells from different donors. Structural location of the ZFN2(+3) RT mutation and amino acids known to convey NRTi or NNRTi resistance (E, F). HIV-1 Reverse Transcriptase without DNA (pdb structure 3KJV) is shown through 3 different orientations with NRTi (E) or NNRTi (F) resistance residues shown in yellow. Blue – p66 subunit amino acids 1-556; Green – p51 subunit amino acids 4-431; Red – amino acids L246 and P247. NRTi resistance residues M41, A62, K65, D67, T69, K70, L74, V75, F77, Y115, F116 Q151, M184, L210, T215 and K219 are shown (E). NNRTi resistance residues L100, K101, K103, V106, V108, E138, Y181, Y188, G190 and M230 are shown (F).
In mutant NL4-3-ZFN2(+3) a leucine-leucine (LL) motif is created in the p66 thumb domain of reverse transcriptase, adjacent to the polymerase domain active site (Fig 4E, F, Supplemental Fig 1B). We therefore determined whether susceptibility to clinically used nucleotide or non-nucleotide reverse transcriptase inhibitors (NRTi or NNRTi) would be altered. TZM-bl cells were treated with NRTi (Tenofovir or AZT) or NNRTi (Etravirine or Nevirapine) antivirals, and then exposed to equalized infectious levels of NL4-3 or NL4-3-ZFN2(+3) mutant virus. Levels of viral infection were determined after 48 hours. NL4-3 and the NL4-3-ZFN2(+3) mutant virus were inhibited at similar levels by Tenofovir, AZT, Etravirine or Nevirapine at all tested drug concentrations (Fig 4B), suggesting that the LL motif does not influence NRTi or NNRTi antiviral susceptibility. This may not be surprising since the LL motif does not overlap with known NRTi or NNRTi resistance mutations within reverse transcriptase (Fig 4E, F).
To establish whether secondary mutations could prevent emergence of infectious but endonuclease-resistant HIV, additional mutations were introduced into the NL4-3-ZFN2(+3) mutant at the ZFN3 and ZFN4 target sites within RT and integrase respectively. Mutations ZFN3(−3) or ZFN4(+5), which were able to block NL4-3 infectivity (Fig 3C, D), were selected and the infectivity of double mutants NL4-3-ZFN2(+3)/ZFN3(−3) and NL4-3-ZFN2(+3)/ZFN4(+5) compared with wild-type NL4-3, NL4-3-ZFN2(+3), NL4-3-ZFN3(−3), and NL4-3-ZFN4(+5) in TZM-bl cells (Fig 4C). The infectivity of both double mutants was reduced to below the levels seen with single ZFN3(−3) and ZFN4(+5) mutants, whereas mutant NL4-3-ZFN2(+3) retained infectivity similar to wild type NL4-3. These observations suggest that a combination endonuclease therapy could be used to overcome the emergence of endonuclease-resistant HIV.
Finally, we evaluated the infectivity of wild type, single-, or double-mutant NL4-3 viruses in activated human CD4+ T cells to confirm that mutant virus replication is also blocked in primary cells. Activated CD4+ T cells were incubated with 50ng of wild type, single mutant, or double mutant NL4-3 viruses, and levels of intracellular p24 determined by flow cytometry 72 hours later. As seen previously in a different donor (Fig 4A), p24 expression was detected in CD4+ T cells infected with ZFN2(+3) mutant virus, albeit at levels lower than wild type NL4-3 (Fig 4D). In contrast, cells infected with NL4-3 mutants ZFN3(−3), ZFN4(+5), NL4-3-ZFN2(+3)/ZFN3(−3) or NL4-3-ZFN2(+3)/ZFN4(+5) showed background levels of intracellular p24 expression (Fig 4D). These observations support the conclusion that infectious endonuclease resistant virus could emerge in HIV-infected patients, but treatment with multiple endonucleases could overcome resistance.
4. DISCUSSION
As an approach to treat persistent viral infections, we are targeting essential genes in viral genomes for disruption using targeted endonucleases [2–4]. Like conventional antiviral treatments, endonuclease therapies might create resistance [2, 37]. In this study we developed ZFNs targeting HIV pol, and during analysis of their efficacy we identified an infectious treatment-resistant virus. This RT mutant virus was resistant to endonuclease cleavage, was susceptible to antiviral RTi drugs, and maintained infectivity in primary human CD4+ T cells. These observations have important implications for the use of endonucleases as antiviral therapeutics.
First, to our knowledge the emergence of resistant infectious virus has not been evaluated in previous antiviral therapies targeting EBV, JCV, HBV, HCV, HCMV, HIV, HSV and HTLV using endonucleases or recombinases [45, 46]. Drug resistance influences clinical treatments against viral infections [47–50], so the emergence of resistance to novel endonuclease therapies should also be considered. This is especially important for HIV, which has a high mutation rate that drives virus evolution and treatment resistance. During endonuclease therapy treatment resistance could occur indirectly, through SNPs introduced during HIV replication that alter target site binding or cleavage, or directly, through endonuclease-mediated mutations that prevent target site binding or cleavage. In either scenario, resistance would only occur if the mutated viral gene remained functional. Endonuclease-mediated mutations are often large and likely to functionally inactivate target genes, so the frequency of resistance generated this way may be low despite the relatively high number of different in-frame mutations detected. For SNP-driven resistance, the frequency may be higher, but will differ for each endonuclease as target sequence heterogeneity tolerance varies [51]. Our data confirms that endonuclease-mediated mutations can produce treatment-resistant viruses, so further analyses of endonuclease resistance are warranted.
It is possible that endonuclease therapies could produce different levels of endonuclease resistance, as determined by target site re-cleavage. We have identified infectious virus that is largely resistant to cleavage at the ZFN target site. Under continued selective pressure it is possible that a mutant virus with partial endonuclease resistance could evolve to produce a completely endonuclease-resistant virus, as happens with low-level drug-resistant viruses in HIV patients [52–54]. Further studies are needed to determine whether increasing endonuclease resistance evolves during treatment.
This study shows that small indels are not always inactivating, and may be either resistant or susceptible to endonuclease cleavage, despite being highly similar. We deliberately studied the infectivity of mutants with small in-frame mutations, as we predicted they would be the most likely to maintain replicative capacity, and for mutant ZFN2(+3) this proved to be true. We did not expect such variation in ZFN re-cleavage at similar mutant target sites. The ZFN1(+3) and ZFN3(+5) target sites could not be re-cleaved, while target sites ZFN2(+3) and ZFN4(+5) could be re-cleaved with reduced efficiency, which suggests that ZFNs can tolerate different levels of target site heterogeneity. This is important, as replication-competent mutant viruses that can be re-cleaved by an endonuclease during therapy remain targets for further disruption and are thus less likely to persist.
The NL4-3-ZFN2(+3) mutant virus was originally identified in HEK293 cells. However, we found the same mutation repeatedly in HEK293 cells and in lymphoid CD4+ SupT1 cells. We have also previously seen similar small insertions in the lymphoid Jurkat cell line and primary human fibroblasts after endonuclease treatment [15, 55]. Together these data strongly suggest that a resistance mutation like the ZFN2(+3) RT mutation could occur in HIV-susceptible primary human cells. Future experiments will determine the frequency with which the ZFN2(+3) mutation, or similar resistance mutations, are found in ZFN-treated primary human CD4+ T cells.
Our study identifies a unique region of RT that can support amino acid insertion without eliminating viral infectivity. The ZFN2(+3) RT mutant has an extra leucine residue inserted adjacent to Leu246, located between the palm and thumb domains, close to the polymerase domain (Fig 4E, F). To our knowledge there have been no reports of HIV with altered activity due to mutations at Leu246. A widespread analysis of RT mutations showed that Leu246 is predominantly invariant in treatment-naïve and RTi-treated HIV-1 infected patients [56]. In contrast, a comparison of retroelement and retrovirus RT sequences showed no conservation of Leu246 [57]. Interestingly, an LL motif is present at the same location within the prototype foamy virus RT [57], so it is likely that an LL motif at this site in HIV RT retains functionality. Further studies of HIV-1 RT containing modifications around residue Leu246 may be warranted.
The NL4-3-ZFN2(+3) mutant retained sensitivity to RT inhibitors, which has implications for endonuclease therapy. Since antiviral endonuclease therapy is not dependent on active viremia, it would be advantageous to concurrently administer HAART. Therefore, it is important that endonuclease therapy does not promote the emergence of drug-resistant virus. Our study shows that an infectious ZFN2-derived mutant virus remains susceptible to NRTi and NNRTi drugs. Moving forward, it will be important to monitor whether antiviral drug sensitivity is affected when endonucleases target genes encoding HIV drug targets such as protease, RT or integrase.
Our data illustrate the utility of mathematical modeling of treatment regimens in the context of endonuclease resistance. Previously we predicted that HBV resistance to endonucleases could be avoided by treatment with 3 or more virus-specific endonucleases, using a complex mathematical model [37]. The present study directly shows that endonuclease resistance and multiple endonuclease target sites can impact therapeutic success during HIV treatment. Further modeling studies should include therapy dosing, endonuclease cleavage efficiency, and off-target cleavage experiments to optimize future virus-directed endonuclease therapies.
The experiments performed in SupT1 cells provide a proof of principle that AAV-mediated endonuclease therapies could be used to introduce disabling mutations into the integrated provirus in HIV-infected T cells. To this end recent data suggests that AAV vectors can be used to transduce primary T cells with extremely high efficiency [58], which could enable efficient endonuclease delivery to target cells during an anti-HIV therapy. Future studies will be required to determine whether AAV-endonuclease therapy can be used to effectively target HIV in primary T cells.
In conclusion, we have demonstrated that treatment-resistant infectious HIV can be detected after endonuclease therapy. To our knowledge this is the first such demonstration and has implications for antiviral endonuclease development. In the future efforts should be made to identify and characterize resistant virus on a study-by-study basis.
Supplementary Material
Highlights.
Zinc finger nucleases (ZFNs) that cleave HIV pol were developed as antiviral agents
ZFNs could inhibit production of HIV virions and infectious HIV in vitro
An infectious and ZFN-resistant HIV provirus was identified in treated cells
ZFN-resistance HIV remained susceptible to NRTi and NNRTi drugs
Secondary mutations rendered the ZFN-resistance virus replication incompetent
Acknowledgments
This work was funded by NIH supported Martin Delaney Collaboratory grant U19 AI 096111 and in part by a developmental grant from the University of Washington Center for AIDS Research (CFAR), an NIH funded program under award number P30 AI 027757 which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA, NIGMS, NIDDK). HLP was supported by a Research Scholarship from the Mary Gates Endowment for Students.
We thank Vincente Planelles for providing the pDHIV3 and pLET-LAI plasmids, and Michael Emerman for providing SupT1 and TZM-bl cells. We thank Florian Hladik for helpful discussions and providing Tenofovir, and John McNevin for providing AZT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Harshana S. De Silva Feelixge, Email: hdesilva@fredhutch.org.
Daniel Stone, Email: dstone2@fredhutch.org.
Harlan L. Pietz, Email: harlan.pietz@nih.gov.
Pavitra Roychoudhury, Email: proychou@fredhutch.org.
Alex L. Greninger, Email: agrening@uw.edu.
Joshua T. Schiffer, Email: jschiffe@fredhutch.org.
Martine Aubert, Email: maubert@fredhutch.org.
Keith R. Jerome, Email: kjerome@fredhutch.org.
References
- 1.Manjunath N, et al. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5(11):2748–66. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schiffer JT, et al. Targeted DNA mutagenesis for the cure of chronic viral infections. Journal of virology. 2012;86(17):8920–36. doi: 10.1128/JVI.00052-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Current opinion in HIV and AIDS. 2013;8(3):217–23. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber ND, et al. DNA cleavage enzymes for treatment of persistent viral infections: recent advances and the pathway forward. Virology. 2014;454–455:353–61. doi: 10.1016/j.virol.2013.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends in biotechnology. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafez M, Hausner G. Homing endonucleases: DNA scissors on a mission. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada. 2012;55(8):553–69. doi: 10.1139/g2012-049. [DOI] [PubMed] [Google Scholar]
- 7.Bloom K, et al. Inactivation of hepatitis B virus replication in cultured cells and in vivo with engineered transcription activator-like effector nucleases. Molecular therapy: the journal of the American Society of Gene Therapy. 2013;21(10):1889–97. doi: 10.1038/mt.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J, et al. An efficient antiviral strategy for targeting hepatitis B virus genome using transcription activator-like effector nucleases. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22(2):303–11. doi: 10.1038/mt.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy EM, et al. Suppression of hepatitis B virus DNA accumulation in chronically infected cells using a bacterial CRISPR/Cas RNA-guided DNA endonuclease. Virology. 2014;476C:196–205. doi: 10.1016/j.virol.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin SR, et al. The CRISPR/Cas9 System Facilitates Clearance of the Intrahepatic HBV Templates In Vivo. Molecular therapy. Nucleic acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeger C, Sohn JA. Targeting Hepatitis B Virus With CRISPR/Cas9. Molecular therapy. Nucleic acids. 2014;3:e216. doi: 10.1038/mtna.2014.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber ND, et al. AAV-mediated delivery of zinc finger nucleases targeting hepatitis B virus inhibits active replication. PloS one. 2014;9(5):e97579. doi: 10.1371/journal.pone.0097579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhen S, et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene therapy. 2015 doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 14.Price AA, et al. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(19):6164–9. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aubert M, et al. In vitro Inactivation of Latent HSV by Targeted Mutagenesis Using an HSV-specific Homing Endonuclease. Molecular therapy. Nucleic acids. 2014;3:e146. doi: 10.1038/mtna.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbadawy HM, et al. Gene transfer of integration defective anti-HSV-1 meganuclease to human corneas ex vivo. Gene therapy. 2014;21(3):272–81. doi: 10.1038/gt.2013.82. [DOI] [PubMed] [Google Scholar]
- 17.Grosse S, et al. Meganuclease-mediated Inhibition of HSV1 Infection in Cultured Cells. Molecular therapy: the journal of the American Society of Gene Therapy. 2011;19(4):694–702. doi: 10.1038/mt.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(36):13157–62. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuen KS, et al. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. The Journal of general virology. 2014 doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 20.Ding W, et al. Zinc Finger Nucleases Targeting the Human Papillomavirus E7 Oncogene Induce E7 Disruption and a Transformed Phenotype in HPV16/18-Positive Cervical Cancer Cells. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(24):6495–503. doi: 10.1158/1078-0432.CCR-14-0250. [DOI] [PubMed] [Google Scholar]
- 21.Hu Z, et al. TALEN-mediated targeting of HPV oncogenes ameliorates HPVrelated cervical malignancy. The Journal of clinical investigation. 2014 doi: 10.1172/JCI78206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu Z, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. BioMed research international. 2014;2014:612823. doi: 10.1155/2014/612823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kennedy EM, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. Journal of Virology. 2014;88(20):11965–72. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mino T, et al. Gene- and protein-delivered zinc finger-staphylococcal nuclease hybrid for inhibition of DNA replication of human papillomavirus. PloS one. 2013;8(2):e56633. doi: 10.1371/journal.pone.0056633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mino T, et al. Inhibition of DNA Replication of Human Papillomavirus by Using Zinc Finger-Single-Chain FokI Dimer Hybrid. Molecular biotechnology. 2014 doi: 10.1007/s12033-014-9751-3. [DOI] [PubMed] [Google Scholar]
- 26.Yu L, et al. Disruption of human papillomavirus 16 E6 gene by clustered regularly interspaced short palindromic repeat/Cas system in human cervical cancer cells. OncoTargets and therapy. 2015;8:37–44. doi: 10.2147/OTT.S64092. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Zhen S, et al. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochemical and biophysical research communications. 2014;450(4):1422–6. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Wollebo HS, et al. CRISPR/Cas9 System as an Agent for Eliminating Polyomavirus JC Infection. PloS one. 2015;10(9):e0136046. doi: 10.1371/journal.pone.0136046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebina H, et al. A High Excision Potential of TALENs for Integrated DNA of HIV-Based Lentiviral Vector. PLoS ONE. 2015;10(3):e0120047. doi: 10.1371/journal.pone.0120047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebina H, et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Scientific reports. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(31):11461–6. doi: 10.1073/pnas.1405186111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao HK, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nature communications. 2015;6:6413. doi: 10.1038/ncomms7413. [DOI] [PubMed] [Google Scholar]
- 33.Qu X, et al. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic acids research. 2013;41(16):7771–82. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu X, et al. Zinc finger nuclease: a new approach for excising HIV-1 proviral DNA from infected human T cells. Molecular biology reports. 2014;41(9):5819–27. doi: 10.1007/s11033-014-3456-3. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka A, et al. A novel therapeutic molecule against HTLV-1 infection targeting provirus. Leukemia. 2013;27(8):1621–7. doi: 10.1038/leu.2013.46. [DOI] [PubMed] [Google Scholar]
- 36.Zhu W, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12(1):22. doi: 10.1186/s12977-015-0150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiffer JT, et al. Predictors of hepatitis B cure using gene therapy to deliver DNA cleavage enzymes: a mathematical modeling approach. PLoS computational biology. 2013;9(7):e1003131. doi: 10.1371/journal.pcbi.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham FL, et al. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. The Journal of general virology. 1977;36(1):59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 39.Wei X, et al. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrobial agents and chemotherapy. 2002;46(6):1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adachi A, et al. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. Journal of Virology. 1986;59(2):284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Andersen JL, et al. HIV-1 Vpr-induced apoptosis is cell cycle dependent and requires Bax but not ANT. PLoS pathogens. 2006;2(12):e127. doi: 10.1371/journal.ppat.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wirt SE, Porteus MH. Development of nuclease-mediated site-specific genome modification. Current opinion in immunology. 2012;24(5):609–16. doi: 10.1016/j.coi.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Molecular Cell. 2008;31(2):294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sander JD, Maeder ML, Joung JK. Engineering designer nucleases with customized cleavage specificities. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit12. Chapter 12. 2011. p. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karpinski J, et al. Universal Tre (uTre) recombinase specifically targets the majority of HIV-1 isolates. Journal of the International AIDS Society. 2014;17(4 Suppl 3):19706. doi: 10.7448/IAS.17.4.19706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarkar I, et al. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316(5833):1912–5. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 47.Glebe D, Geipel A. Selected phenotypic assays used to evaluate antiviral resistance and viral fitness of hepatitis B virus and its variants. Intervirology. 2014;57(3–4):225–31. doi: 10.1159/000360950. [DOI] [PubMed] [Google Scholar]
- 48.Pawlotsky JM. New hepatitis C therapies: the toolbox, strategies, and challenges. Gastroenterology. 2014;146(5):1176–92. doi: 10.1053/j.gastro.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 49.Pham QD, et al. Global burden of transmitted HIV drug resistance and HIV-exposure categories: a systematic review and meta-analysis. AIDS. 2014;28(18):2751–62. doi: 10.1097/QAD.0000000000000494. [DOI] [PubMed] [Google Scholar]
- 50.Piret J, Boivin G. Antiviral drug resistance in herpesviruses other than cytomegalovirus. Reviews in medical virology. 2014;24(3):186–218. doi: 10.1002/rmv.1787. [DOI] [PubMed] [Google Scholar]
- 51.Pattanayak V, Guilinger JP, Liu DR. Determining the specificities of TALENs, Cas9, and other genome-editing enzymes. Methods in enzymology. 2014;546:47–78. doi: 10.1016/B978-0-12-801185-0.00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boucher CA, et al. Ordered appearance of zidovudine resistance mutations during treatment of 18 human immunodeficiency virus-positive subjects. The Journal of infectious diseases. 1992;165(1):105–10. doi: 10.1093/infdis/165.1.105. [DOI] [PubMed] [Google Scholar]
- 53.Harrigan PR, Bloor S, Larder BA. Relative replicative fitness of zidovudine-resistant human immunodeficiency virus type 1 isolates in vitro. Journal of Virology. 1998;72(5):3773–8. doi: 10.1128/jvi.72.5.3773-3778.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kellam P, et al. Zidovudine treatment results in the selection of human immunodeficiency virus type 1 variants whose genotypes confer increasing levels of drug resistance. The Journal of general virology. 1994;75(Pt 2):341–51. doi: 10.1099/0022-1317-75-2-341. [DOI] [PubMed] [Google Scholar]
- 55.Aubert M, et al. Successful targeting and disruption of an integrated reporter lentivirus using the engineered homing endonuclease Y2 I-AniI. PloS one. 2011;6(2):e16825. doi: 10.1371/journal.pone.0016825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ceccherini-Silberstein F, et al. High sequence conservation of human immunodeficiency virus type 1 reverse transcriptase under drug pressure despite the continuous appearance of mutations. Journal of Virology. 2005;79(16):10718–29. doi: 10.1128/JVI.79.16.10718-10729.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nowak E, et al. Ty3 reverse transcriptase complexed with an RNA-DNA hybrid shows structural and functional asymmetry. Nature structural & molecular biology. 2014;21(4):389–96. doi: 10.1038/nsmb.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sather BD, et al. Efficient modification of CCR5 in primary human hematopoietic cells using a megaTAL nuclease and AAV donor template. Science translational medicine. 2015;7(307):307ra156. doi: 10.1126/scitranslmed.aac5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.