Abstract
Background
Peripheral epigenetic marks hold promise for understanding psychiatric illness and may represent fingerprints of gene–environment interactions. We conducted an initial examination of CpG methylation variation in children with or without attention-deficit/hyperactivity disorder (ADHD).
Methods
Children age 7–12 were recruited, screened, evaluated and assigned to ADHD or non-ADHD groups by defined research criteria. Two independent age-matched samples were examined, a discovery set (n = 92, all boys, half control, half ADHD) and a confirmation set (n = 20, half ADHD, all boys). 5-methylcytosine levels were quantified in salivary DNA using the Illumina 450 K HumanMethylation array. Genes for which multiple probes were nominally significant and had a beta difference of at least 2% were evaluated for biological relevance and prioritized for confirmation and sequence validation. Gene pathways were explored and described.
Results
Two genes met the criteria for confirmation testing, VIPR2 and MYT1L; both had multiple probes meeting cutoffs and strong biological relevance. Probes on VIPR2 passed FDR correction in the confirmation set and were confirmed through bisulfite sequencing. Enrichment analysis suggested involvement of gene sets or pathways related to inflammatory processes and modulation of monoamine and cholinergic neurotransmission.
Conclusions
Although it is unknown to what extent CpG methylation seen in peripheral tissue reflect transcriptomic changes in the brain, these initial results indicate that peripheral DNA methylation markers in ADHD may be promising and suggest targeted hypotheses for future study in larger samples.
Keywords: ADHD, methylation, epigenetic
Introduction
ADHD, like most psychiatric disorders, lacks either a full account of its pathophysiology and mechanisms of development, or biological measures that reliably predict either liability or illness. Although ADHD has substantial heritability, the search for biological targets is increasingly informed by interest in whether the disorder is shaped by interactions between genotype and environmental factors (Nigg, Nikolas, & Burt, 2010; Purcell, 2002). The mechanism by which environmental factors or genotype by environment interactions might influence ADHD remains unknown.
One possibility is via dynamic epigenetic modifications to gene expression. Epigenetic modification, a normal part of tissue development, can occur not only spontaneously but also in response to environmental effects (implying genotype by environment interactions). As a result, an integrated genetic/epigenetic approach to understanding complex disease is emerging (Rakyan, Down, Balding, & Beck, 2011), although not without important cautions (Heijmans & Mill, 2012). The best understood epigenetic modification is DNA methylation, which influences gene expression via the disruption of transcription factor binding and the attraction of methyl-binding proteins that initiate chromatin compaction and gene silencing.
Epigenetic variation cannot be assessed in brain tissue from living people, so studies of living individuals must be undertaken using peripheral tissue. Although the degree of cross-tissue concordance in DNA methylation remains in question, these studies still hold potential for novel discovery of liability markers or identify targets for future investigation in neural systems (Aberg et al., 2013; Horvath, 2013). Recent studies of peripheral tissue DNA methylation have identified epigenetic variation associated with autism (Berko et al., 2014; Wong et al., 2013) and psychosis (Aberg et al., 2014; Dempster et al., 2011), with promising results. In ADHD, a small number of prior studies have examined DNA methylation in candidate genes (van Mil et al., 2014; Xu et al., 2015) with encouraging results. Methylome-wide epigenetic exploration in ADHD has yet to be conducted. The current report therefore describes the first methylome-wide study in childhood ADHD, using DNA derived from saliva samples in a pilot study design.
Methods
Participants and case identification
Families were recruited by soliciting community volunteers with public advertisements and mass mailings using commercial mailing lists. The local Institutional Review Board approved the studies. Parents provided written informed consent for themselves and their children and children provided written informed assent. All families underwent a multigate screening process to establish eligibility and diagnostic group assignment that included a parent–clinician clinical structured diagnostic parent interview (Schedule for Affective Disorders and Schizophrenia for School-Age Children—Epidemiologic Version [KSAD-S-E]; Puig-Antich & Ryan, 1996), parent and teacher standardized rating forms that assessed symptoms and impairment, clinician observations, and child completion of a short from of the Wechsler Intelligence Scales for Children-Fourth Edition (WISC-IV; Wechsler, 2003). All of this information was then presented to a clinical diagnostic team in order to implement a best estimate diagnostic procedure. Their agreement rate was acceptable for ADHD (k > .80; all subtypes were allowed) and for all disorders with base rate >5% in the study (k > .75). Disagreements were resolved by discussion.
Exclusion criteria
Exclusion criteria include an estimated Full Scale IQ < 75; diagnosis of current major depressive episode, lifetime mania or psychosis, pervasive developmental disorder (including autism), learning disability or major medical/neurological disorders or injuries.
Medication
Current prescription of any psychoactive medication, including any stimulant or nonstimulant preparation, with a half-life greater than 7 hr was exclusionary, due to an accompanying study of cognitive performance requiring medication washout. DNA samples obtained for the current analysis were obtained without medication washout in medicated cases (29 subjects, all on various stimulant preparations: Daytrana Patch, Adderall, Dexedrine Tablets, Focalin Tablets, Metadate ER Capsules, Methylin ER Tablets, Methylin Tablets, Adderal XR, Ritalin LA, Concerta, Vyvanse). Medication status was handled in covariance analysis as described in the data analysis section.
Generation of subsamples
For the present report, two subsamples were selected from the larger cohort for purposes of hypothesis generation (‘discovery’) and testing (‘confirmation’). The discovery sample comprised 92 boys, half with ADHD, (age range 7–11.8 and a mean of 8.7 years), of whom 85 were retained in analysis after DNA quality control. Of the 85, 55% had been treated currently or in the past with stimulant medication. These individuals were allowed in to maximize representativeness, because medication status is positively correlated with ADHD severity in our sample as in others. Medication history is addressed statistically as explained below and by exclusion in the confirmation sample. A small confirmation sample was selected that comprised 20 never-medicated boys, 10 with ADHD and 10 without ADHD, with an age range of 7–11 and a mean of 9.0 years (Table 1, below).
Table 1.
Descriptive and diagnostic statistics for ADHD and control groups
| Sample A (N = 85) | Sample B (N = 20) | |||
|---|---|---|---|---|
| Variable | ADHDc (n = 43) | Control (n = 42) | ADHD (n = 10) | Control (n = 10) |
| Age (months; mean, SD) | 116.2 (17.1) | 116.7 (17.1) | 98.2 (7.4) | 100.1 (9.1) |
| Gender (% male) | 100% | 100% | 100% | 100% |
| Race (% White) | 97.60% | 92.90% | 90% | 70% |
| Family income ($K; mean, SD) | 67.9 (31.0) | 72.2 (30.5) | 68.5 (25.6) | 68.5 (30.0) |
| WISC-IVa FSIQb (mean, SD) | 113.5 (11.9) | 117.0 (12.8) | 110.2 (13.9) | 117.6 (9.6) |
| Conner’s 3rd edition (T-score) | ||||
| Parent-rated cognitive (Inattention) | 73.4 (11.2) | 45.6 (5.8)* | 74.6 (12.7) | 46.1 (5.9)* |
| Parent-rated hyperactive/Imp. | 77.8 (10.3) | 46.1 (7.4)* | 78.1 (10.5) | 46.7 (8.0)* |
| Teacher-rated cognitive (Inattention) | 65.7 (10.0) | 45.5 (4.9)* | 63.4 (8.7) | 44.9 (4.7)* |
| Teacher-rated hyperactive/Imp. | 73.5 (13.7) | 45.0 (4.9)* | 75.4 (8.9) | 44.1 (4.7)* |
| Comorbid disorders (K-SADS-Ed) | ||||
| Lifetime any mood disorder | 11.60% | 0.00% | 10.00% | 0.00% |
| Any anxiety disorder | 9.30% | 7.00% | 10.00% | 10.00% |
| Conduct disorder | 0.00% | 0.00% | 0.00% | 0.00% |
| Oppositional defiant disorder | 23.30% | 2.3%* | 30.00% | 0.0%* |
WISC-IV: Wechsler Intelligence Scales for Children.
Full-Scale Intelligence Quotient (estimated).
Attention-Deficit/Hyperactivity Disorder Rating Scale.
Kiddie Schedule of Affective Disorders and Schizophrenia.
Group difference within sample at p < .05.
Laboratory methods
DNA was extracted from salivary samples collected in Oragene® cups and extracted using the DNA Genotek PrepIt kit (DNA Genotek Inc., Kanata, ON, Canada) as per manufacturer’s protocols. Specimen concentration was determined using Pico-Green (Ahn, Costa, & Emanuel, 1996). Genome-wide DNA methylation was assessed with the Illumina Infinium HumanMethylation450 BeadChip (Bibikova et al., 2011) following sodium bisulfite treatment of genomic DNA. The first sample data set (n = 92) was assayed by Illumina, Inc. (San Diego, CA), and the second (N = 20) by Expression Analysis, Inc. (Durham, NC). Each data set was processed and analyzed independently.
Data preprocessing and quality control
Specimen and data processing
DNA was processed at the respective facilities using in-house QC protocols for assay performance. Raw data were imported into Genome Studio (Illumina, Inc.) to investigate the sample hybridization quality and to extract the signal intensities for each probe.
Data preprocessing
Raw methylation values were examined for acceptable quality of probes and samples using custom scripts within the R statistical programming language (http://www.r-project.org) using packages from the Bioconductor software project (http://www.bioconductor.org (Gentleman et al., 2004). Poorly performing probes and samples were removed from the dataset using the R package wateRmelon (Pidsley et al., 2013), including removal of probes with detection p-values >.05 for >1% across all samples; nonspecific probes and those containing polymorphic SNPs (Chen et al., 2013). Specimen samples were removed from the analysis if >1% of the total number of probes for that sample had detection p-values >.01. Six specimens failed in the first data set and one overlooked sibling was removed, leaving N = 85 for analysis (44 ADHD and 41 controls). The final probe number available after QC was 384,464 in the discovery sample and 392,598 in the confirmation sample.
Within each data set, samples passing QC were normalized together across all batches, using wateRmelon (Pidsley et al., 2013). Raw intensity values for each chip were quantile-normalized separately for the methylated and unmethylated signals. The average DNA methylation level (beta value) for each probe was calculated as the ratio of the normalized signal for the methylated signal to the sum of the normalized values of the methylated and unmethylated signals. Therefore, beta values range from 0 (unmethylated) to 1 (methylated).
Data analysis
Confounders and covariates
To correct for potential confounders, principal components analysis (PCA) was conducted on the normalized methylation values for each probe. The first principal component loading (99.1% of the variation for the discovery data set and 97% for the confirmation data set) was added to the analytic model. As it took up so much variation, it is likely that this principal component largely accounted for variation due to technical variation, spatial variation on the chip, saliva cell type variation between groups, ancestry variation between groups, as well as age and medication status. Supporting that impression, medication status, age, and race were unrelated to the first principle component, suggesting trivial associations with methylation values. Further supporting this point, secondary analyses were conducted with single covariates with no change in the primary conclusions. Based on the large amount of variance accounted for by the first principal component, additional corrections (such as statistical corrections for cell type or ancestry (Barfield et al., 2014; Houseman, Molitor, & Marsit, 2014), were not conducted to avoid power loss and collinearity as recommended by Barfield (Barfield et al., 2014).
Analysis
Differentially methylated probes (DMPs) were identified using custom R scripts. Each CpG site was tested individually in a linear regression model(Barfield, Kilaru, Smith, & Conneely, 2012) in which normalized beta values represent the dependent variable and the ADHD status the independent predictor, adjusted for the first principal component.
Selection of DMPs for validation
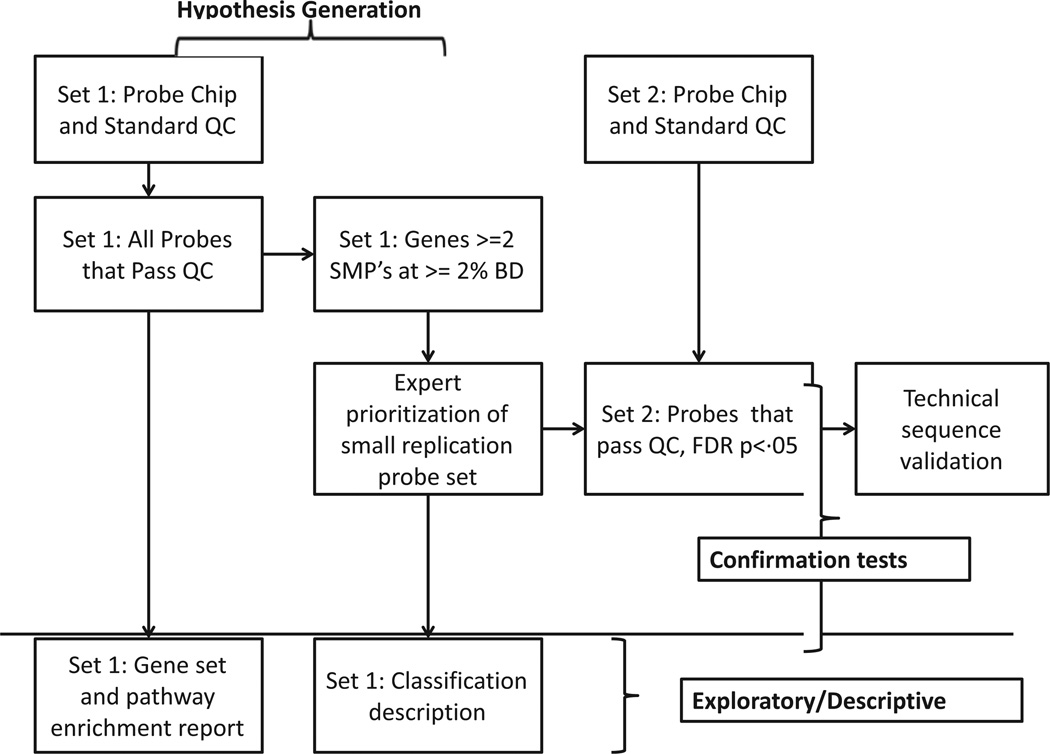
Figure 1 schematizes the probe prioritization and testing strategy adopted for testing in our small sample to balance Type I and Type II error. We prioritized loci with multiple qualifying probes in the discovery set at nominal significance and beta difference between groups of at least 2% in the discovery set. We then focused on differentially methylated probes (DMPs) with identifiable biological relevance to ADHD through examination of the information libraries available through the Wiezman Institute of Science (http://www.genecards.org/) and NIH (www.ncbi.nih.-gov/IEB/Research/Acembly). Potentially relevant genes were then scored on their relevance and prioritized based on prior literature on psychiatric disorders, animal models of knockouts related to ADHD symptoms, and expression in brain regions of particular interest in ADHD. Those selected were then limited to 10 probes to preserve power on the replication, based on qualitative examination of their theoretical relevance. They in turn were examined for nominal and FDR-corrected significance in the confirmation data set. Surviving probes underwent technical validation by sequencing.
Figure 1.
Work flow for probe prioritization. In the discovery set genes were prioritized if more than one probe was nominally significant at a moderate effect size (beta difference ≥.02). These were further prioritized based on brain expression and animal models. A small probe set was selected for confirmation testing in a second cohort of children. Surviving probes underwent technical sequence validation
Probe sequence validation
Targeted bisulfite sequencing was used to validate the DMPs. Primer design and validation were performed by Zymo Research Corporation (Irvine, CA) on bisulfite-converted control DNA. Primers were designed to flank each targeted CpG site in 100–300 nucleotide regions. Following primer validation, study specimen DNA was bisulfite converted using the EZ DNA Methylation-LightningTM Kit (Zymo Research). Samples were amplified and barcoded using the Fluidigm Access ArrayTM System and sequenced (MiSeq, Illumina, Inc., San Diego, CA) using the paired-end sequencing protocol according to the manufacturer’s guidelines.
Sequence reads were identified using standard Illumina base-calling software. Low quality nucleotides and adapter sequences were trimmed off during analysis QC. Sequence reads were aligned back to the reference genome using default Bismark parameters (Krueger & Andrews, 2011). The methylation level of each sampled cytosine was estimated as the number of reads reporting a C, divided by the total number of reads reporting a C or T.
Quality control was performed on all specimens before and following bisulfite conversion. Two specimens had low sequencing depth and were discarded (see Table S1, available online, for details). Differential methylation was assessed per CpG site including samples with at least 10 reads. Multiple testing correction was done by the False Discovery Rate (FDR) procedure.
Pathway (enrichment) analysis
As noted in Figure 1, to explore biological significance, we conducted pathway or gene set enrichment analysis using the gene sets defined in (a) the Gene Ontology Biological Process Category (www.http://geneontology.org/, http://www.broadinstitute.org/gsea/msigdb/index.jsp, 6/82014: c5 GO gene sets) and (b) the ConsensusPathdb (‘pathways’) (Kamburov, Stelzl, Lehrach, & Herwig, 2013), which includes protein–protein interactions, protein-DNA, and biochemical pathways such as signaling and metabolic pathways. Several methods exist for quantifying the association between functional groups of genomic variables and a clinical phenotype (Hung, Yang, Hu, Weng, & DeLisi, 2012; Khatri, Sirota, & Butte, 2012; Mooney, Nigg, McWeeney, & Wilmot, 2014). The methods used here were as follows.
Probes on the array were mapped to genes using the Illumina annotations. Because of the differences in number of probes per gene and the structure among them, we utilized a procedure that is robust to correlation structure among probes to generate gene level p-values for each gene (Wu et al., 2010) resulting in 19,984 single gene p-values For enrichment analysis, we used a commonly used framework (Goeman & Buhlmann, 2007) wherein a ‘competitive’ gene set test compares genes in the test set relative to all other genes (Gatti, Barry, Nobel, Rusyn, & Wright, 2010).
However, intergene correlations among genes in different pathways (gene sets) inflate the p-value and the apparent false discovery rate (Gatti et al., 2010). To address this, we utilized a method that adjusts for the correlation among the genes to allow for differences in the correlation structure of each gene set (Wu & Smyth, 2012). Normalized methylation values for all genes in the data set were used to generate the correlation among all genes. A variation inflation factor was then calculated and used to adjust for these correlations. Each defined gene set was tested for enrichment of differentially methylated genes (hypermethylated and hypomethylated combined) relative to genes not in the gene set using the function ‘camera’ in the limma R package (Ritchie et al., 2015). After analysis, results were separated into gene sets containing >50% genes that were hypomethylated in ADHD and gene sets containing >50% genes that were hypermethylated in ADHD for ease of viewing and interpretation. We interpreted gene sets with nominal p ≤ .01 for purposes of developing hypotheses for future studies.
Results
Demographic and clinical descriptions of the discovery and confirmation cohorts are provided in Table 1. The sample is typical of many ADHD studies in clinical severity, comorbidity, and demographic profile.
Prioritization
We identified 95 genes with >1 annotated DMP (n = 245 DM probes) (at a nominal p-value of p < .05 and a beta difference >2%) (Table S2). Among the 95 genes were several known to be expressed in brain and to have prior associations with behavior, neurodevelopment, and/or psychiatric illness and thus to be particularly biologically relevant to ADHD, including: neuron navigator 1 (NAV1), ninjurin2 (NINJ2), solute carrier family 12, member 9 (SLC12A9), vasoactive intestinal peptide receptor 2 (VPR2), myelin transcription factor 1-like (MYT1L), oxytocin receptor (OXTR), histone deacetylase 4 (HDAC4), and major histocompatibility complex genes (HLA-A, HLA-B, HLA-C). With the goal of limiting the replication test to enhance power for multiple testing correction, we selected a small subset of these based on literature review and those having the most elements relevant to our study, including (1) relevant animal models, (b) prior association to psychiatric illness, (c) association with neural development, (d) known expression in well studied brain regions identified in ADHD. The top-rated two genes chosen were VIPR2 and MYT1L. VIPR2 (7q36.3) was prioritized because it met the following criteria: (a) expression in the caudate, a key ADHD-relevant brain area, (b) animal knockout model in which underexpression is associated with hyperactivity (Sheward et al., 2010), and (c) associated with psychiatric disorder (Levinson et al., 2011). MYT1L (2p25.3) was selected for (a) its potential involvement in myelin formation and nervous system formation, of interest to our group due to our work on white matter formation in ADHD (Nagel et al., 2011), and (b) because of its prior association with both major depressive disorder (Wang et al., 2010) and schizophrenia (Pidsley et al., 2014).
Confirmation set
Six of the probes from these two genes were present in the second, confirmation cohort (Table 2). The MYT1L probes failed to meet our confirmation standard of FDR p < .05, but we did confirm the association in the same direction for probe cg13444538 in VIPR2 (discovery beta difference = −.059, p = .0345; confirmation beta difference = −.0964, p = .0064, FDR = .0386). Two other VIPR2 probes trended close to confirmation: cg08479516 (discovery beta difference = −.0321, p = .0256; confirmation beta difference = −.0472, p = .0169, FDR = .058) and cg05554000 (discovery beta difference = −.0537, p = .0199; confirmation beta difference = −.0692, p = .0608, FDR = .093). All probes indicated hypomethylation in VIPR2 for ADHD samples. Differences in probe cg13444538 methylation value distributions for each cohort set are shown in Figure S1.
Table 2.
Cross-sample test of discovery probes
| Discovery set | Confirmation set | |||||
|---|---|---|---|---|---|---|
| Probe ID | Gene Symbol | p-Value | Beta diff.a | p-Value | Beta diff.a | FDR |
| cg05180887 | MYT1L | .0407 | −.0448 | .0840 | −.0922 | .0903 |
| cg06201514 | MYT1L | .0197 | −.0764 | .0903 | −.1374 | .0903 |
| cg10075506 | MYT1L | .0375 | −.0807 | .0822 | −.1484 | .0903 |
| cg13444538b | VIPR2 | .0345 | −.0588 | .0064 | −.0964 | .0386 |
| cg08479516c | VIPR2 | .0256 | −.0321 | .0169 | −.0472 | .0508 |
| cg05554000 | VIPR2 | .0199 | −.0537 | .0608 | −.0692 | .0903 |
Beta difference where a negative beta difference indicates hypomethylation in ADHD samples.
Probe cg13444538 was significantly associated with ADHD in both data sets.
Probe cg08479516 trended toward significance in the confirmation set.
Technical validation by sequencing
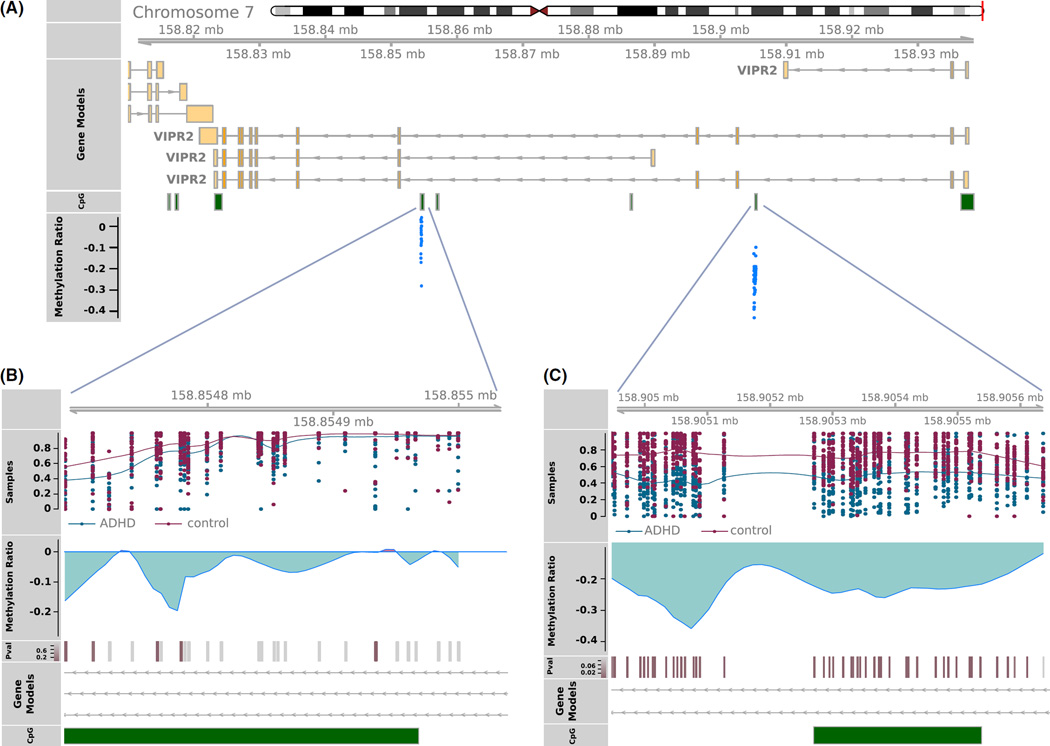
Amplicons were generated to cover 250 base pairs 5′ and 3′ of the three VIPR2 replicated probes. Targeted bisulfite sequencing was used to profile DNA methylation across 73 CpG sites, for which 43 sites were significantly hypomethylated in ADHD subjects after correction for multiple testing (FDR p = .000007 to p = .041, Table S3). Two intronic regions of VIPR2 were covered by sequencing, (Figure 2).
Figure 2.
VIPR2 Targeted bisulfate sequence validation. (Panel A) VIPR2 gene models indicating the genomic locations for the probes targeted for validation. All probes were located in two CpG islands shown in green. (Panel B and C) Individual sample methylation ratios for ADHD (red) and control (blue) specimens. p-Values for differential methylation between ADHD and controls is shown per CpG tested
Gene pathways and enrichment analysis
The functional relationship among the differentially methylated genes was assessed for enrichment of a particular functional category using the GO Biological Process gene and pathways as defined in the ConsensusPathDB. For the GO Biological Process category gene sets, we found six signatures associated with ADHD using an unadjusted p ≤ .01 (Table 3). The hypomethylated set revealed sets involved in anti-oxidant activity. The top two hypermethylated sets involved nicotinic receptor activity. ConsensusPathDB pathways were tested for enrichment in the same manner. Of 3669 Canonical pathways tested, 34 showed nominal (p < .01) differences in methylated genes (Table S4). Hypomethylated pathways included homocysteine, cysteine, glutathione, and fatty acid oxidation pathways, all related to inflammatory mechanisms and thus potentially related to the hypomethylated process categories. Hypermethylated pathways included glutamine degradation and G-protein signaling pathways, and thus potentially related to the hypermethylated process categories.
Table 3.
GO Biological Process (BP). (A) Gene sets more hypomethylated in ADHD than other gene sets in the GO BP and (B) GO Genes sets more hypermethylated in ADHD than other GO Gene sets
| N Genesa | Correlationb | p-valuec | GO gene set |
|---|---|---|---|
| (A) | |||
| 151 | .0055 | .000 | OXIDOREDUCTASE_ACTIVITY_ACTING_ON_PEROXIDE_AS_ACCEPTOR |
| 239 | .0013 | .001 | NEGATIVE_REGULATION_OF_ANGIOGENESIS |
| 247 | .0047 | .003 | ANTIOXIDANT_ACTIVITY |
| 205 | .0037 | .009 | CASPASE_REGULATOR_ACTIVITY |
| (B) | |||
| 154 | .0001 | .009 | NICOTINIC_ACETYLCHOLINE_GATED_RECEPTOR_CHANNEL_COMPLEX |
| 154 | .0001 | .009 | NICOTINIC_ACETYLCHOLINE_ACTIVATED_CATION_SELECTIVE_CHANNEL_ACTIVITY |
Number of genes in the gene set.
Correlation among the genes in the gene set.
p-values are uncorrected; none of these effects would be significant at FDR p < .05.
Covariate assessment
To confirm our assumptions about the PCA approach, post hoc checks on the surviving final probes were conducted while directly covarying age, history of simulant medication use, and race. These results did not change results or conclusions meaningfully and so were not presented. Sequence validation results were also retested post hoc with these same covariates (PCA is inapplicable to the targeted gene approach), with no meaningful change in the results presented earlier.
Discussion
The examination of peripheral DNA methylation patterns in psychiatric disease is of interest as a potential liability or disease biomarker and as a clue to etiology. Here, we present the first methylome-wide exploration of DNA methylation in peripheral tissue, examining salivary DNA from children with ADHD relative to non-ADHD controls.
Probes in VIPR2 showed lower CpG methylation in the saliva of children with ADHD, a finding that was supported by our additional tests. VIPR2 encodes the Vasoactive intestinal peptide receptor 2 and is expressed throughout the central nervous system and the periphery—a useful feature of a marker to be studied in this way. SNPs in VIPR2 have been associated previously with mood disorder (Soria et al., 2010) while duplications in VIPR2 have been seen in schizophrenia (Levinson et al., 2011; Vacic et al., 2011). Low-copy repeats within VIPR2 have been identified as having a role in the 7q36.3 genomic rearrangements in a patient with mild mental retardation and were also found as both simple and complex structural polymorphisms associated with the VIPR2-LCR region (Beri, Bonaglia, & Giorda, 2013). Mutations in the VIPR2 homolog in the mouse cause hypoactivity, as well as disruptions in circadian rhythm (Eppig, Blake, Bult, Kadin, & Richardson, 2012), thus decreased methylation might be speculated to increase hyperactivity. Circadian hormones are of ongoing interest in relation to ADHD (Chaste et al., 2011) as well.
The pathway enrichment analysis here were exploratory only; they suggested that future studies might hypothesize differential methylation gene sets that (a) regulate inflammation and cell damage via oxidation and anti-oxidation, (b) lie in the biological pathways related to the methionine cycle and glutathione metabolism and (c) in relation to cross-modulation of Ach, glutamatergic, and/or G-protein modulated neurotransmitter systems. Recent work on the effects of maternal and child nutrition on neural development (Bhatia et al., 2011; Chen & Su, 2013; Janssen et al., 2015) suggests that anti-inflammatory long chain fatty acid intake is protective against inflammation risk. A recent meta-analysis reports that children with ADHD have reduced blood levels of these important nutrients and appear to benefit from supplementation (Gow & Hibbeln, 2014; Hawkey & Nigg, 2014; Widenhorn-Muller, Schwanda, Scholz, Spitzer, & Bode, 2014). Second, G-protein coupled receptors are of interest in relation to brain neurotransmitter function and have recently been proposed to have implications for dopaminergic and glutamatergic neurotransmission (Garcia-Olivares et al., 2013; Lesch, Merker, Reif, & Novak, 2013; Xie et al., 2012). The cross-talk of cholinergic and glutamatergic systems is already widely studied (Grupe et al., 2013; Gu et al., 2014; Nelson, Bussert, Kreitzer, & Seal, 2014; Pancani et al., 2014; Pidoplichko, Prager, Aroniadou-Anderjaska, & Braga, 2013), but likewise underexamined in ADHD.
Thus, a third more focused hypothesis generated by those explorations involves epigenetic modulation of cholinergic or nicotinic receptor functioning. This focus may be particularly intriguing in light of the recent interest in those receptors’ role in prefrontal cortical function, impulsivity, and attention (Lee, Fuemmeler, McClernon, Ashley-Koch, & Kollins, 2013; Ohmura, Tsutsui-Kimura, & Yoshioka, 2012; Sterley, Howells, & Russell, 2014; Wallace & Bertrand, 2013), recent genetic findings related to ADHD and cholinergic receptor genes (Williams et al., 2012), and interest in the role of nicotinic receptor intervention as an effective and novel treatment target for ADHD in animal and human trials (Bain et al., 2012; Potter, Dunbar, Mazzulla, Hosford, & Newhouse, 2014).
ADHD status in the children from whom saliva was analyzed in this study was very well characterized. Although half of the children in the initial sample were treated with stimulant medication, methylation differences were not dependent on medication status.
The results should be viewed with caution for several reasons in addition to the small samples, which may have resulted in failure to confirm MYT1L. The decision to prioritize probes for replication and validation based on the significance of multiple probes per gene could result in overlooking important signals in the data from single probes. Relatedly, we opted to use a larger sample for discovery than replication to maximize our power to identify putative methylation affects, knowing that we would be testing only a very small set of probes for replication. However, a larger confirmation study might result in more reliable estimates (Duncan & Keller, 2011). Second, although we excluded probes known to be located in SNPs, a full accounting of epigenetic effects requires formal genetic control, by either analysis of genotyped data or twin studies, which we are currently undertaking. Likewise, we did not examine mRNA expression in relation to sites identified; that is, a study we are undertaking in future. While it is likely that the PCA accounted for cell type variation (Barfield et al., 2014), cell types were not directly examined and should be directly evaluated to fully understand the meaning of peripheral tissue findings in future work. Likewise, ancestry was not examined genetically although is unlikely to account for effects based again on the large amount of variation accounted for in the first principal component here (Barfield et al., 2014).
Other analytic methods such as differential methylation region analysis, could also be considered in future larger studies. Sex differences do not account for results because only boys were included, but results cannot be generalized to girls. Finally, we cannot draw conclusions from these data about patterns of DNA methylation, nor changes in gene function, within the brain (Davies et al., 2012; Horvath, 2013).
In conclusion, the present results provide initial evidence that ADHD is associated with differential methylation in genes relevant to children’s activity levels. The findings provide hypotheses to guide further study and help open the door for new work in this area. We conclude that VIPR2 is an interesting novel target for ADHD research and that further study of peripheral tissue methylation in ADHD is promising.
Key points.
Study of DNA methylation is a growing strategy for understanding roots of psychopathology; DNA methylation can occur in response to early experience and suggest one mechanism for gene by environment interplay in development.
Prior studies have looked at candidate genes but this is the first methylome-wide study of DNA methylation in child psychopathology.
Results identified and validated an association of ADHD diagnosis with reduced DNA methylation in the VIPR2 gene, which is associated with circadian rhythm and with activity level in an animal model and has also been previously identified in a study of DNA methylation in schizophrenia.
Gene pathways related to neurotransmitters and neural development were identified for future research.
This is a pilot study; results are strictly preliminary due to a very small sample. However, they illustrate that this approach to understanding ADHD may be fruitful and suggest promising hypotheses for follow up work.
Acknowledgements
This study was supported by the Abracadabra Foundation of Portland Oregon.
Footnotes
The authors have declared that they have no competing or potential conflicts of interest.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table S1. Quality control of targeted bisulfite sequencing.
Table S2. List of probes for which there was more than one probe per gene that was nominally significant.
Table S3. Sequencing validation results for VIPR2.
Table S4. Pathways enrichment analysis.
Figure S1. Distributions of VIPR2 cg 13444538 normalized methylation values of ADHD and non-ADHD boys.
References
- Aberg KA, McClay JL, Nerella S, Clark S, Kumar G, Chen W, Van den Oord EJ. Methylome-wide association study of schizophrenia: Identifying blood biomarker signatures of environmental insults. JAMA Psychiatry. 2014;71:255–264. doi: 10.1001/jamapsychiatry.2013.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberg KA, Xie LY, McClay JL, Nerella S, Vunck S, Snider S, Van den Oord EJ. Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics. 2013;5:367–377. doi: 10.2217/epi.13.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SJ, Costa J, Emanuel JR. PicoGreen quantitation of DNA: Effective evaluation of samples pre- or post-PCR. Nucleic Acids Research. 1996;24:2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain EE, Apostol G, Sangal RB, Robieson WZ, McNeill DL, Abi-Saab WM, Saltarelli MD. A randomized pilot study of the efficacy and safety of ABT-089, a novel alpha4beta2 neuronal nicotinic receptor agonist, in adults with attention-deficit/hyperactivity disorder. Journal of Clinical Psychiatry. 2012;73:783–789. doi: 10.4088/JCP.10m06719. [DOI] [PubMed] [Google Scholar]
- Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, Conneely KN. Accounting for population stratification in DNA methylation studies. Genetic Epidemiology. 2014;38:231–241. doi: 10.1002/gepi.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: An R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28:1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beri S, Bonaglia MC, Giorda R. Low-copy repeats at the human VIPR2 gene predispose to recurrent and nonrecurrent rearrangements. European Journal of Human Genetics. 2013;21:757–761. doi: 10.1038/ejhg.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berko ER, Suzuki M, Beren F, Lemetre C, Alaimo CM, Calder RB, Greally JM. Mosaic epigenetic dysregulation of ectodermal cells in autism spectrum disorder. PLoS Genetics. 2014;10:e1004402. doi: 10.1371/journal.pgen.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia HS, Agrawal R, Sharma S, Huo YX, Ying Z, Gomez-Pinilla F. Omega-3 fatty acid deficiency during brain maturation reduces neuronal and behavioral plasticity in adulthood. PLoS ONE. 2011;6:e28451. doi: 10.1371/journal.pone.0028451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- Chaste P, Clement N, Botros HG, Guillaume JL, Konyukh M, Pagan C, Bourgeron T. Genetic variations of the melatonin pathway in patients with attention-deficit and hyperactivity disorders. Journal of Pineal Research. 2011;51:394–399. doi: 10.1111/j.1600-079X.2011.00902.x. [DOI] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8:203–209. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HF, Su HM. Exposure to a maternal n-3 fatty acid-deficient diet during brain development provokes excessive hypothalamic-pituitary-adrenal axis responses to stress and behavioral indices of depression and anxiety in male rat offspring later in life. Journal of Nutritional Biochemistry. 2013;24:70–80. doi: 10.1016/j.jnutbio.2012.02.006. [DOI] [PubMed] [Google Scholar]
- Davies MN, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Mill J. Functional annotation of the human brain methylome identifies tissue-specific epigenetic variation across brain and blood. Genome Biology. 2012;13:R43. doi: 10.1186/gb-2012-13-6-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempster EL, Pidsley R, Schalkwyk LC, Owens S, Georgiades A, Kane F, Mill J. Disease-associated epigenetic changes in monozygotic twins discordant for schizophrenia and bipolar disorder. Human Molecular Genetics. 2011;20:4786–4796. doi: 10.1093/hmg/ddr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. American Journal of Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The Mouse Genome Database (MGD): Comprehensive resource for genetics and genomics of the laboratory mouse. Nucleic Acids Research. 2012;40(Database issue):D881–D886. doi: 10.1093/nar/gkr974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Olivares J, Torres-Salazar D, Owens WA, Baust T, Siderovski DP, Amara SG, Torres GE. Inhibition of dopamine transporter activity by G protein betagamma subunits. PLoS ONE. 2013;8:e59788. doi: 10.1371/journal.pone.0059788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti DM, Barry WT, Nobel AB, Rusyn I, Wright FA. Heading down the wrong pathway: On the influence of correlation within gene sets. BMC Genomics. 2010;11:574. doi: 10.1186/1471-2164-11-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Zhang J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biology. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, Buhlmann P. Analyzing gene expression data in terms of gene sets: Methodological issues. Bioinformatics. 2007;23:980–987. doi: 10.1093/bioinformatics/btm051. [DOI] [PubMed] [Google Scholar]
- Gow RV, Hibbeln JR. Omega-3 fatty acid and nutrient deficits in adverse neurodevelopment and childhood behaviors. Child and Adolescent Psychiatric Clinics of North America. 2014;23:555–590. doi: 10.1016/j.chc.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe M, Paolone G, Jensen AA, Sandager-Nielsen K, Sarter M, Grunnet M. Selective potentiation of (alpha4)3(beta2)2 nicotinic acetylcholine receptors augments amplitudes of prefrontal acetylcholine- and nicotine-evoked glutamatergic transients in rats. Biochemical Pharmacology. 2013;86:1487–1496. doi: 10.1016/j.bcp.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Cheng J, Zhong P, Qin L, Liu W, Yan Z. Abeta selectively impairs mGluR7 modulation of NMDA signaling in basal forebrain cholinergic neurons: Implication in Alzheimer’s disease. Journal of Neuroscience. 2014;34:13614–13628. doi: 10.1523/JNEUROSCI.1204-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkey E, Nigg JT. Omega-3 fatty acid and ADHD: Blood level analysis and meta-analytic extension of supplementation trials. Clinical Psychology Review. 2014;34:496–505. doi: 10.1016/j.cpr.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Mill J. Commentary: The seven plagues of epigenetic epidemiology. International Journal of Epidemiology. 2012;41:74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biology. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–1439. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung JH, Yang TH, Hu Z, Weng Z, DeLisi C. Gene set enrichment analysis: Performance evaluation and usage guidelines. Briefings in Bioinformatics. 2012;13:281–291. doi: 10.1093/bib/bbr049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen CI, Zerbi V, Mutsaers MP, de Jong BS, Wiesmann M, Arnoldussen IA, Kiliaan AJ. Impact of dietary n-3 polyunsaturated fatty acids on cognition, motor skills and hippocampal neurogenesis in developing C57BL/6J mice. Journal of Nutritional Biochemistry. 2015;26:24–35. doi: 10.1016/j.jnutbio.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Research. 2013;41(Database issue):D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri P, Sirota M, Butte AJ. Ten years of pathway analysis: Current approaches and outstanding challenges. PLoS Computational Biology. 2012;8:e1002375. doi: 10.1371/journal.pcbi.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CT, Fuemmeler BF, McClernon FJ, Ashley-Koch A, Kollins SH. Nicotinic receptor gene variants interact with attention deficient hyperactive disorder symptoms to predict smoking trajectories from early adolescence to adulthood. Addictive Behaviors. 2013;38:2683–2689. doi: 10.1016/j.addbeh.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch KP, Merker S, Reif A, Novak M. Dances with black widow spiders: Dysregulation of glutamate signalling enters centre stage in ADHD. European Neuropsychopharmacology. 2013;23:479–491. doi: 10.1016/j.euroneuro.2012.07.013. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, Gejman PV. Copy number variants in schizophrenia: Confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. American Journal of Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney MA, Nigg JT, McWeeney SK, Wilmot B. Functional and genomic context in pathway analysis of GWAS data. Trends in Genetics. 2014;30:390–400. doi: 10.1016/j.tig.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel BJ, Bathula D, Herting M, Schmitt C, Kroenke CD, Fair D, Nigg JT. Altered white matter microstructure in children with attention-deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2011;50:283–292. doi: 10.1016/j.jaac.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AB, Bussert TG, Kreitzer AC, Seal RP. Striatal cholinergic neurotransmission requires VGLUT3. Journal of Neuroscience. 2014;34:8772–8777. doi: 10.1523/JNEUROSCI.0901-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg J, Nikolas M, Burt SA. Measured gene-by-environment interaction in relation to attention-deficit/hyperactivity disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 2010;49:863–873. doi: 10.1016/j.jaac.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura Y, Tsutsui-Kimura I, Yoshioka M. Impulsive behavior and nicotinic acetylcholine receptors. Journal of Pharmacological Sciences. 2012;118:413–422. doi: 10.1254/jphs.11r06cr. [DOI] [PubMed] [Google Scholar]
- Pancani T, Bolarinwa C, Smith Y, Lindsley CW, Conn PJ, Xiang Z. M4 mAChR-mediated modulation of glutamatergic transmission at corticostriatal synapses. ACS Chemical Neuroscience. 2014;5:318–324. doi: 10.1021/cn500003z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoplichko VI, Prager EM, Aroniadou-Anderjaska V, Braga MF. alpha7-Containing nicotinic acetylcholine receptors on interneurons of the basolateral amygdala and their role in the regulation of the network excitability. Journal of Neurophysiology. 2013;110:2358–2369. doi: 10.1152/jn.01030.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Viana J, Hannon E, Spiers HH, Troakes C, Al-Saraj S, Mill J. Methylomic profiling of human brain tissue supports a neurodevelopmental origin for schizophrenia. Genome Biology. 2014;15:483. doi: 10.1186/s13059-014-0483-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidsley R, Wong CCY, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter AS, Dunbar G, Mazzulla E, Hosford D, Newhouse PA. AZD3480, a novel nicotinic receptor agonist, for the treatment of attention-deficit/hyperactivity disorder in adults. Biological Psychiatry. 2014;75:207–214. doi: 10.1016/j.biopsych.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Puig-Antich J, Ryan N. Kiddie schedule for affective disorders and schizophrenia. Pittsburgh, PA: Western Psychiatric Institute; 1996. [Google Scholar]
- Purcell S. Variance components models for gene-environment interaction in twin analysis. Twin Research. 2002;5:554–571. doi: 10.1375/136905202762342026. [DOI] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nature Reviews Genetics. 2011;12:529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheward WJ, Naylor E, Knowles-Barley S, Armstrong JD, Brooker GA, Seckl JR, Harmar AJ. Circadian control of mouse heart rate and blood pressure by the suprachiasmatic nuclei: Behavioral effects are more significant than direct outputs. PLoS ONE. 2010;5:e9783. doi: 10.1371/journal.pone.0009783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria V, Martinez-Amoros E, Escaramis G, Valero J, Perez-Egea R, Garcia C, Urretavizcaya M. Differential association of circadian genes with mood disorders: CRY1 and NPAS2 are associated with unipolar major depression and CLOCK and VIP with bipolar disorder. Neuropsychopharmacology. 2010;35:1279–1289. doi: 10.1038/npp.2009.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterley TL, Howells FM, Russell VA. Nicotine-stimulated release of [3H]norepinephrine is reduced in the hippocampus of an animal model of attention-deficit/hyperactivity disorder, the spontaneously hypertensive rat. Brain Research. 2014;1572:1–10. doi: 10.1016/j.brainres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Sebat J. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mil NH, Steegers-Theunissen RP, Bouwland-Both MI, Verbiest MM, Rijlaarsdam J, Hofman A, Tiemeier H. DNA methylation profiles at birth and child ADHD symptoms. Journal of Psychiatric Research. 2014;49:51–59. doi: 10.1016/j.jpsychires.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Wallace TL, Bertrand D. Importance of the nicotinic acetylcholine receptor system in the prefrontal cortex. Biochemical Pharmacology. 2013;85:1713–1720. doi: 10.1016/j.bcp.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng Z, Li T, Liu J, Li J, Li Y, Shi Y. Common SNPs in myelin transcription factor 1-like (MYT1L): Association with major depressive disorder in the Chinese Han population. PLoS ONE. 2010;5:e13662. doi: 10.1371/journal.pone.0013662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D. Wechsler intelligence scale for children: Administration and scoring manual. 4th edn. San Antonio, TX: Psychological Corporation; 2003. [Google Scholar]
- Widenhorn-Muller K, Schwanda S, Scholz E, Spitzer M, Bode H. Effect of supplementation with long-chain omega-3 polyunsaturated fatty acids on behavior and cognition in children with attention deficit/hyperactivity disorder (ADHD): A randomized placebo-controlled intervention trial. Prostaglandins Leukotrienes and Essential Fatty Acids. 2014;91:49–60. doi: 10.1016/j.plefa.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Williams NM, Franke B, Mick E, Anney RJ, Freitag CM, Gill M, Faraone SV. Genome-wide analysis of copy number variants in attention deficit hyperactivity disorder: The role of rare variants and duplications at 15q13.3. American Journal of Psychiatry. 2012;169:195–204. doi: 10.1176/appi.ajp.2011.11060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, Mill J. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Molecular Psychiatry. 2013;23:41. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. ROAST: Rotation gene set tests for complex microarray experiments. Bioinformatics. 2010;26:2176–2182. doi: 10.1093/bioinformatics/btq401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Smyth GK. Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Research. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Ge S, Collins VE, Haynes CL, Renner KJ, Meisel RL, Martemyanov KA. Gbeta5-RGS complexes are gatekeepers of hyperactivity involved in control of multiple neurotransmitter systems. Psychopharmacology (Berl) 2012;219:823–834. doi: 10.1007/s00213-011-2409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Chen XT, Luo M, Tang Y, Zhang G, Wu D, Wang HL. Multiple epigenetic factors predict the attention deficit/hyperactivity disorder among the Chinese Han children. Journal of Psychiatric Research. 2015;64:40–50. doi: 10.1016/j.jpsychires.2015.03.006. [DOI] [PubMed] [Google Scholar]