Abstract
Oligodendroglioma represents a distinctive neoplasm in adults but similar neoplasms occur rarely in children. We studied 20 cases of pediatric oligodendroglioma by SNP array (median age 9 years, range 1–19; 15 grade II and 5 grade III). Cytogenetic abnormalities were present in 8 (53%) grade II and all five anaplastic oligodendrogliomas. Most changes were in the form of deletion and copy neutral loss of heterozygosity (LOH). The most common abnormality was 1p deletion (n = 5). Whole arm 1p19q co‐deletion was present in three cases from adolescent patients and 9p loss in 3, including one low‐grade oligodendroglioma with CDKN2A homozygous deletion. Common losses were largely limited to the anaplastic subset (n = 5) and included 3q29 (n = 3), 11p (n = 3), 17q (n = 3), 4q (n = 2), 6p (n = 2), 13q (n = 2), 14q (n = 2), 17p (n = 2) and whole Ch 18 loss (n = 2). Gains were non‐recurrent except for whole Ch 7 (n = 2) and gain on 12q (n = 2) including the MDM2 locus. Possible germ line LOH (or uniparental disomy) was present in seven cases (35%), with one focal abnormality (22q13.1‐13.2) in two. BRAF‐KIAA1549 fusions and BRAF p.V600E mutations were absent (n = 13 and 8). In summary, cytogenetic alterations in pediatric oligodendrogliomas are characterized mostly by genomic losses, particularly in anaplastic tumors.
Keywords: BRAF, cytogenetics, FISH, oligodendroglioma, pediatric glioma, SNP array
Introduction
Oligodendrogliomas represent an important clinicopathologic and molecular subset of infiltrating gliomas that have been well studied in adults. These tumors are characterized at the molecular level by the presence of whole‐arm 1p19q co‐deletion, a cytogenetic abnormality mediated by a t(1;19) translocation 11, 16. Whole‐exome sequencing studies have identified mutations in putative suppressor genes located in these regions, particularly FUBP1 and CIC 4, 43 in some, but not all, cases. Like other diffuse gliomas, these tumors also have frequent mutations in IDH1 or IDH2 42. Oligodendrogliomas with 1p19q co‐deletion and IDH1 or IDH2 mutation demonstrate the best prognosis in the diffuse glioma category, and are particularly responsive to chemotherapy, as highlighted by long‐term follow‐up in clinical trials 6, 41.
Tumors with all the histologic features of oligodendroglioma also occur in children. These tumors are usually low grade, and often recur but have a low rate of histologic progression to anaplasia 35. They usually lack the molecular changes typical of adult oligodendroglioma, that is, 1p19q co‐deletion and IDH1 or 2 mutations, particularly when developing in patients less than 15 years old 22, 31, 35. The differential diagnosis of oligodendroglioma in the pediatric population encompasses a variety of close mimics, including dysembryoplastic neuroepithelial tumor, neurocytoma, pilocytic astrocytoma and more recently, disseminated oligodendroglioma‐like neoplasm (DOLN) 1, 30, 34, 36, 38. However, cytogenetic abnormalities other than 1p19q have not been adequately examined in a systematic fashion in these tumors. In the current study, we assessed single nucleotide polymorphism microarray data from pediatric oligodendrogliomas to characterize cytogenetic changes in these tumors.
Materials and Methods
Patients
We studied 28 cases of pediatric oligodendroglioma examined at Johns Hopkins Hospital with sufficient formalin‐fixed paraffin‐embedded tissue for molecular analysis. The clinicopathologic features of 26 of these cases have been previously published 35. Two additional cases were included and reviewed by two of the authors (PCB and FJR) to confirm the diagnosis of pediatric oligodendroglioma. Patient ages ranged from <1 to 19 years of age at diagnosis, with nine females and 19 males. There were 20 WHO grade II and 8 WHO grade III (anaplastic) oligodendrogliomas. The study was performed under Institutional Review (IRB) Board approval at respective sites.
Single nucleotide polymorphism (SNP) array analysis
Areas containing tumor were identified and isolated using Pinpoint reagents (ZymoResearch, Orange, CA, USA). Extraction of DNA from recovered tissue was done with QIAmp DNA MiniKit (Qiagen, Valencia, CA, USA) and quantified by optical density. Extracted DNA was treated with the Infinium HD FFPE DNA Restore Kit (Illumina, San Diego, CA, USA). SNP analysis was performed on the CytoSNP‐12 BeadChip platform (Illumina). The platform provides information on 298,563 SNPs. The data were visualized in KaryoStudio software (Illumina), with all determinations of LOH performed manually. Single nucleotide polymorphism (SNP) analysis produced interpretable data in 20 cases. We evaluated B‐allele frequency (BAF) and log‐R ratio (LRR) for chromosomes 1–22 (q arm only for acrocentric chromosomes 13, 14, 15, 21 and 22), as well as the X chromosome. Some alterations were interpreted as likely representing germ line change. Briefly, in areas of increased spread of B‐allele frequency (BAF) caused by LOH in the tumor, normal log‐R ratio (LRR) would suggest copy‐neutral change. However, the presence of genetically normal cells such as lymphocytes and endothelia in the sample results in a “thickening” of BAF plots at the edges, whereas in some instances of widespread BAF with normal LRR such thickening was not seen. This suggests that all cells in the sample show the same LOH, and hence that it is likely present in the germ line. We have interpreted these regions as representing possible germ line LOH (or uniparental disomy).
BRAF:KIAA1549 FISH
In situ hybridization studies were performed on formalin‐fixed paraffin‐embedded sections using a dual color fusion strategy as previously reported 36, with probe RP11‐355D18 (fluorescein isothiocyanate labeled) targeting KIAA1549 and probe RP4‐726N20 (rhodamine labeled) targeting BRAF.
Immunohistochemistry for BRAF p.V600E
Formalin‐fixed paraffin embedded sections were stained with a mouse monoclonal antibody specific for the human BRAF p.V600E mutant protein (Clone VE1, 1:100 dilution). Detailed methods have been previously published 15.
Results
Somatic chromosomal alterations
Somatic cytogenetic abnormalities were present in 13 (of 20) cases (Figure 1 and Table 1). The predominant alterations were deletion, copy neutral loss of heterozygosity (LOH) or LOH indeterminate, particularly 1p deletion (n = 5). Other recurrent losses (involving two or more tumors) included 1q (n = 3), 3q (n = 3), 4q (n = 2), distal 6p (n = 2), 9p (n = 3), distal 11p (n = 3), 13q (n = 2), 17p (n = 3), 17q (n = 2), whole 18 (n = 2) and 19q (n = 4). Recurrent gains were rare and involved the whole Ch 7 (n = 2) and 12q (n = 2, whole chromosome in one and central/proximal region in one, the latter including the MDM2 oncogene but probably not CDK4)(Figure 2A). One low‐grade oligodendroglioma had a unique karyotype characterized by a combination of whole chromosome gains (triploidy) of 6, 7, 8, 11 and 12, with no other abnormalities. This case also lacked BRAF alterations (BRAF : KIAA1549 and BRAF p.V600E). Some of the alterations appeared to involve a subpopulation rather than the whole tumor. Homozygous deletion, frequent in diffuse gliomas, was identified in one low‐grade oligodendroglioma involving 9p.21.3 (Figure 2B). Genes in this region include CDKN2A, CDKN2B, CDKN2BAS, C9orf53 and MTAP.
Figure 1.
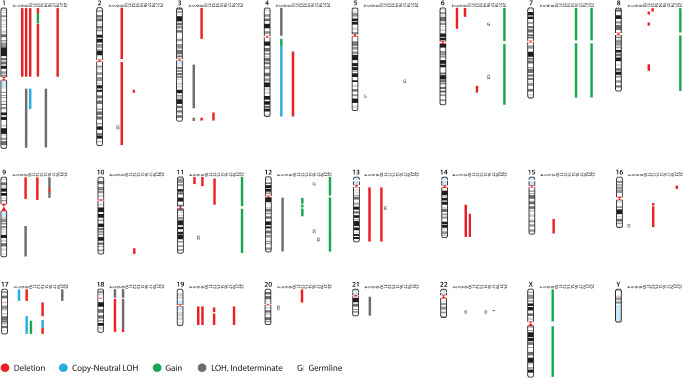
Molecular genetic alterations detected in pediatric oligodendrogliomas by SNP array profiling. 14 (of 20) cases showed loss of heterozygosity, which could be attributed in many cases to deletion, gain or copy neutral change. However, in some cases, definitive determination could not be made. Possible germ line LOH (or uniparental disomy) were present in seven cases.
Table 1.
Clinicopathologic and genetic features of pedicatric oligodendrogliomas
| Age/sex | Location | Grade | Prior therapy | Somatic alterations by SNP array | Possible germ line alterations by SNP array | Genes | BRAF‐KIAA1549 FISH | BRAF V600E | IDH1 (R132H) IHC | Molecular other |
|---|---|---|---|---|---|---|---|---|---|---|
| 2/M | Frontal lobe | II | 0 | Normal karyotype | NA | Negative | Negative | |||
| 1/M | Frontal lobe | II | 0 | Normal karyotype | NA | Negative | Negative (IHC) | Negative | MGMT not methylated | |
| 10/F | Frontal lobe | II | 0 | Normal karyotype | NA | Negative | Negative (IHC) | |||
| 13/M | Frontal lobe | II | 0 | No somatic alterations | 5q33.3, 16q24.1qtel(CNN), 20qcenq11.23(CNN) | NA | Negative | |||
| 19/F | Temporal lobe | II | 0 | del 1p (whole arm), del 19q (whole arm) | CN‐LOH 2q33.3q35, CN‐LOH 11q22.3q22.3(CNN), | Chr 1p (FUBP1), 19q (CIC) | Positive | |||
| 1/M | Parietal lobe | II | 0 | Normal karyotype | NA | |||||
| 1/M | Frontal lobe | II | 0 | Normal karyotype | NA | |||||
| 8/F | Temporal lobe | II | 0 | del8p23.1p23.1, del8p21.2p12, del8q13.2q13.3, del8q22.1q23.1 | 13q | Negative | Negative (IHC) | Negative | ||
| 15/M | Temporal lobe | II | 0 | LOH 1q (whole arm) | 22q13.1q13.2 | 22q (germ line XRCC6, RANGAP1, SGSM3,MKL1, ST13,DNAJB7, TEF, TOB2, NHP2L1, SREBF2, CENPM, TNFRSF13C, CYP2D6, TCF20, CYP2D7P1, RBX1, MIR4766, MIR1281, and MCHR1. ) | Negative | |||
| 9/F | Brain | II | 0 | Gain whole Chr7, LOH 9ptelp21.3, del9p21.3p21.3 (Homozygous), LOH 9p21.3p13.3 | 6p22.1p22.1, 6q22.2q22.31; 12p13.2p13.1, 12q23.1q23.3 | Ch 7 (EGFR, MET, HGF, BRAF), 9p (CDKN2A, CDKN2B, CDKN2BAS, C9orf53, MTAP) | ||||
| 1/M | brainstem | II | 0 | chr22q13.1 | 5q23.3q31.2; 12q24.11q24.13 | DDX17, CSNK1E, DNC1, CBY1, SUN2, CBX6, CBX7, PDGFB | Negative | |||
| 2/F | Parietal lobe | II | 0 | Normal karyotype | NA | Negative | Negative (IHC) | Negative | ||
| 16/M | Frontal lobe | II | 0 | del 1p (whole arm), del 19q (whole arm) | FUBP1, CIC | Negative | ||||
| 4/M | Medial temporal | II | 0 | ND | NA | Negative | ||||
| 14/M | Frontal lobe | II | 0 | del 16p13.11p12.3, LOH 17p (whole arm) | Chr 17p (TP53) | |||||
| 12/M | Temporal lobe | II | 0 | ND | NA | Negative | Negative (IHC) | Negative | Negative for 1p19q LOH (STR) | |
| 2/M | Frontal lobe | II | 0 | ND | NA | Negative | Negative | Negative for 1p19q LOH (STR) | ||
| 16/M | Parietal lobe | II | 0 | ND | NA | Negative | Negative (IHC) | Positive | ||
| 11/M | Posterior fossa | II | 0 | Gain whole Chr6, Chr7, Chr8, Chr11, Chr12 | Ch 7 (EGFR, MET, HGF, BRAF), FGFR1 | Negative | Negative (PCR) | Negative for 1p19q LOH (STR) | ||
| 19/M | Frontal lobe | II | 0 | ND | NA | Negative | 1p19q loss (STR) | |||
| 16/M | Frontal lobe | III | 0 | LOH 3q12.1q26.31, LOH 3q29qtel, LOH 4p (whole arm), dup4q12q13.1, CN‐LOH 4q13.1qtel, del6ptelp21.3, del6p21.3p21.31 (small population subset), del11ptelp15.3, LOH 12q (whole arm), del13q (whole arm), CN‐LOH 17ptelp11.2, LOH 18p (whole arm), del18q (near whole arm), LOH 21q (whole arm) | Chr 11p (CDKN1C, NUP98, SLC22A18), 13q (RB1), 17p (TP53), Chr 18 (DCC) | Negative (PCR) | Negative | MGMT methylated | ||
| 11/F | Frontal lobe | III | 0 | del 1p (whole arm), LOH 1q (whole arm), del 2 (whole Chr deletion), del 3ptelp21.1, del 3q29qtel, del6ptelp22.3, del9ptelp13.2, LOH 9q21.33qtel, del 11ptelp15.1, del 14q21.3qtel, del 15q24.3qtel, del 17ptelp11.2, CN‐LOH 17q22qtel, heterogeneous whole Ch18 loss, del 19q (whole arm), gain X (whole Chr) | 22q13.1q13.2 | Chr 1p (FUBP1), 11p (CDKN1C, NUP98, SLC22A18), 19q (CIC), 17p(TP53), Chr 18 (DCC), 22q(germ line XRCC6, RANGAP1, SGSM3,MKL1, ST13,DNAJB7, TEF, TOB2, NHP2L1, SREBF2, CENPM, TNFRSF13C, CYP2D6, TCF20, CYP2D7P1, RBX1, MIR4766, MIR1281, and MCHR1) | ||||
| 17/F | Frontal lobe | III | Chemotherapy and radiation 10 years prior for leukemia with CNS involvement | del 1p (whole arm), CN‐LOH 1qcentq25.2, del 4q21.1qtel, del 13q (whole arm), del14q23.3qtel, gain 17q23.2qtel | Chr 1p (FUBP1), Chr 13 (RB1) | |||||
| 3/M | cerebellum | III | RT+Chemo | del 1ptelp36.23, gain 1p36.23p36.13, gain 1p36.12p36.11, del 1p36.11cent, del 2q22.3p23.1, del 3q26.32qtel, del 6q23.3q25.1, del 8ptelp23.2, del 9ptelp13.1, del 10q26.3qtel, del 11p15.4p11.2, gain 12q12q13.11, gain 12q13.13q13.2, gain 12q13.2q13.3, gain 12q14.1q15, del 16q12.1qtel, del 19q12q13.11, del 19q13.13qtel, del 20p (whole arm) | 12q (CDK4, MDM2) | Negative | ||||
| 10/F | Parietal lobe | III | 0 | del17qcentq21.33, CN‐LOH 17q22q24.2(CNN), del17q24.3qtel | NF1 | Negative | Negative | |||
| 18/F | Frontal lobe | III | 0 | ND | NA | Negative | Positive | 1p19q loss (STR) | ||
| 7/M | Frontal lobe | III | 0 | ND | NA | Negative | Negative | Negative for 1p19q LOH (STR) | ||
| 11/M | Occipital lobe | III | 0 | ND | NA | Negative | Negative | Negative for 1p19q LOH (STR) |
Figure 2.
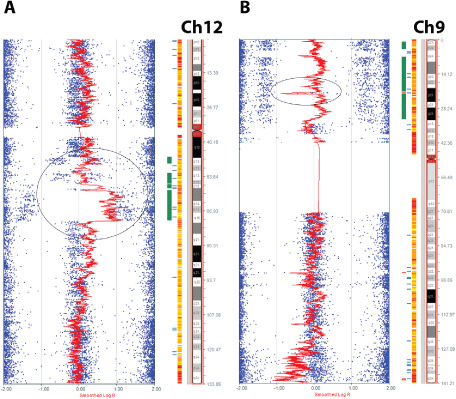
Recurrent genetic alterations in pediatric oligodendrogliomas. Common gains were very rare with two cases showing chromosome 12 gain, including the whole chromosome or a central portion of 12q (circle) in the region containing the known oncogene MDM2 (A). Recurrent alterations in these tumors were predominantly in the form of losses, such as 9p deletion, which in one case was homozygous and included the CDKN2A gene region (circle), with most of the remaining 9p arm showing LOH (B).
1p19q co‐deletion
The classic whole‐arm co‐deletions of 1p and 19q were present in three cases, patients aged 11, 16 and 19 years (Figure 3A,B). Two of these cases were classified as low grade, and the only other defects were possible germ line alterations on 2q and 11q in one of the two. The third case was diagnosed as anaplastic and demonstrated numerous other abnormalities similar to those previously reported as common in anaplastic ODG, including hemizygosity of 9p and LOH on 14q and 15q 28, as well as gain of X. Two other cases with noteworthy alterations included one low‐grade oligodendroglioma with whole‐arm deletion of 1p but intact 19q, and one anaplastic oligodendroglioma with complex cytogenetic changes on 1p characterized by deletion of most of the arm, and gain of the portion from 1p36.23 to 1p36.11 along with complex changes in 19q (Figure 3C,D).
Figure 3.
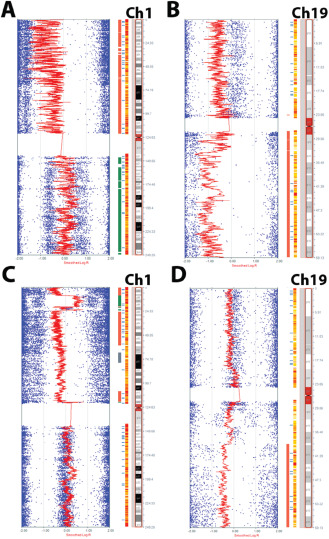
1p19q co‐deletion occurs in a small subset of pediatric oligodendrogliomas. Classic whole arm co‐deletion of 1p19q occurred in three cases (A, B). One unique case had a noteworthy complex abnormality of 1p characterized by a large area of loss and a more focal area of gain near the telomere end (C), as well as LOH in areas of 19q (D).
Increased number of cytogenetic alterations by grade
Next, we stratified somatic cytogenetic alterations by grade. These were present in 8 of 15 (53%) grade II oligodendrogliomas and all five anaplastic oligodendrogliomas studied. Common losses, other than 1p19q co‐deletion, were absent in low‐grade pediatric oligodendrogliomas. Gain in chromosome 7 was present in two cases as the only recurrent abnormality in this group.
Among the anaplastic cases (n = 5), the number of somatic alterations was higher (mean 11.6, range 3–19), compared with grade II oligodendrogliomas (mean 1.4, range 0–5), a difference that was statistically significant (P = 0.002, Wilcoxon rank‐sum; Figure 4). 1p19q co‐deletion was present in two cases, and isolated 1p deletion in one. Alterations in anaplastic tumors are outlined in Table 1 by case, but encompassed all common losses other than 1p and 19q, including 3q29 (n = 3), 4q (n = 2), 6p (n = 2), 11p(n = 3), 9p (n = 2), 13q (n = 2), 14q (n = 2), 17p(n = 2), 17q (n = 3) and chromosome 18 loss (n = 2).
Figure 4.
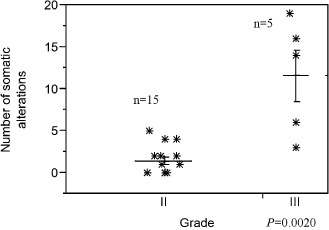
Anaplasia in pediatric oligodendrogliomas is associated with an increased number of cytogenetic abnormalities. The mean number of cytogenetic abnormalities was higher in anaplastic pediatric oligodendrogliomas compared with low‐grade (II) tumors (Wilcoxon rank‐sum test).
Possible germ line cytogenetic alterations
Possible germ line alterations were present in seven cases (35%) (Table 1). One focal possible germ line LOH (or uniparental disomy) on 22q13.1‐13.2 overlapped in two tumors (Figure 5), one anaplastic oligodendroglioma with whole arm 1p19q co‐deletion in an 11‐year‐old girl and the second in a low‐grade oligodendroglioma lacking 1p19q alterations in a 15‐year‐old boy. This region of interest included several genes: XRCC6, RANGAP1, SGSM3, MKL1, ST13, DNAJB7, TEF, TOB2, NHP2L1, SREBF2, CENPM, TNFRSF13C, CYP2D6, TCF20, CYP2D7P1, RBX1, MIR4766, MIR1281 and MCHR1.
Figure 5.
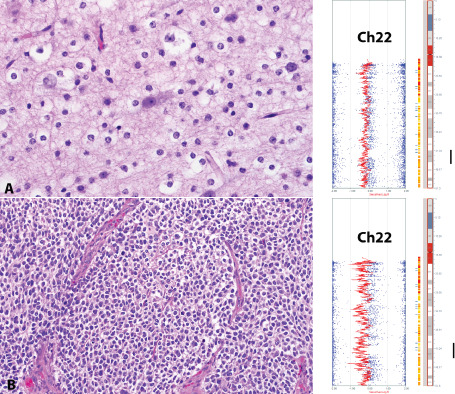
Possible germ line alterations in pediatric oligodendrogliomas. Possible germ line alterations (or uniparental disomy) were present in seven cases. However, in two unrelated cases (A and B), the germ line alteration involved a focal, overlapping region in 22q. This raises the possibility of a pediatric oligodendroglioma‐predisposing gene (s) in this region.
Pediatric oligodendrogliomas usually lack BRAF alterations
Fluorescence in situ hybridization studies were negative for BRAF‐KIAA1549 fusion in all cases tested (n = 13). BRAF p.V600E mutation tested by immunohistochemistry (n = 6) or sequencing (n = 2) was also absent.
Discussion
Pediatric oligodendroglioma is emerging as a neoplasm with distinct biology from its adult counterpart. Numerous studies have demonstrated a lack of 1p19q co‐deletion in most pediatric oligodendrogliomas 22, 31, 35, and more recently, a lack of IDH1 (R132H) mutations 35, 39. In the largest clinicopathologic series to date focusing on pediatric oligodendrogliomas and critically excluding morphologic mimics as well as tumors with ambiguous histology, 1p19q co‐deletion was present in 10 (25%), and isolated 1p loss in one (2%), while the majority lacked these alterations 35.
Array‐based methods such as SNP arrays and array CGH are powerful techniques, with excellent performance in formalin‐fixed paraffin‐embedded tissues 12, 26, that are increasingly finding clinical applications in the study of diffuse gliomas. We searched for global cytogenetic alterations in pediatric oligodendroglioma, which emerged as molecularly distinct from adult oligodendroglioma. Our current study of pediatric tumors using SNP array profiling in formalin‐fixed paraffin‐embedded tissue showed molecular cytogenetic alterations in 8 of 15 (53%) grade II and all 5 (100%) grade III tumors. In contrast, a prior study of 35 adult oligodendroglial tumors by Kitange et al using array comparative genomic hybridization in frozen tissue demonstrated genetic alterations in 18 of 20 (90%) grade II and 9 of 9 (100%) grade III oligodendrogliomas 18. Recurrent alterations present in this and other CGH studies included losses involving 4q, 9p, 11p and 13q 5, 21, 23, 37, 40.
Of interest, almost half of low‐grade oligodendrogliomas in our group lacked gross somatic cytogenetic alterations (47%), a much higher frequency compared with adult oligodendrogliomas. This suggests that smaller genetic alterations are the genetic drivers in a significant subset of these tumors, as has been reported in other low‐grade pediatric gliomas. For example, in a recent whole‐genome sequencing study, Zhang et al found FGFR1 tyrosine kinase duplications in three, combined 1p19q co‐deletion, CIC and IDH1 mutation in one, and a MYB‐MAML fusion in one of their five pediatric oligodendrogliomas 44. Conversely, all anaplastic tumors in our cohort had identifiable cytogenetic alterations, consistent with a higher degree of chromosome instability in these tumors.
In our cohort, 9p loss was present in three cases (two anaplastic and one low grade). The low‐grade oligodendroglioma showed a homozygous CDKN2A deletion. In a recent study, CDKN2A deletions were frequent in a subset of pediatric low‐grade gliomas that progressed to high grade over time and this appeared to be an early event 25. Unfortunately, this patient was lost to follow‐up and it is therefore unclear if malignant progression developed over time. RB1 pathway alterations, including RB1 and CDKN2A, have been associated with worse outcome in adult low‐grade diffuse gliomas 33, and appear to be frequent in those tumors lacking more common diffuse glioma alterations (e.g. 1p19q co‐deletion, IDH1/2 and TP53 mutations) 19.
Another recent study reported a high frequency of 1q gain and 6q loss in diffuse high‐grade astrocytomas of infancy 9, alterations that were in general absent in our group. Loss of 10q is a frequent event in adult diffuse gliomas, may be associated with worse prognosis in oligodendrogliomas 14, and occurs in a subset of pediatric gliomas 27. This alteration was absent in our cases, except for a small distal 10q26.3qtel deletion in one case, not involving DMBT1, PTEN or MGMT, relevant genes in adult gliomas. These findings suggest that pediatric oligodendrogliomas have a distinct genetic cytogenetic profile from other adult and pediatric gliomas.
Chromosomal gains were infrequent in the current cohort, although small gains involving putative oncogenes may be potentially missed given the increased noise involved in karyotyping FFPE tissue. Other studies of low‐grade adult oligodendrogliomas have also found a predominance of deletions, with common gains in the study by Rossi et al involving mostly chromosome 7 37. Whole chromosome 7 gains were present in two cases in our study, a relatively frequent alteration in a variety of pediatric gliomas, including pilocytic astrocytomas 3. Chromosome 12 gains were present in two cases, which has also been previously reported in adult gliomas, gains including the known oncogene MDM2 32. Additionally, 8q gains, previously associated with a subset of diffuse astrocytomas and oligodendroglial tumors with worse outcome 10, 20, were absent.
In addition, we identified possible germ line LOH (or uniparental disomy) in approximately one‐third of this group, which appears to be higher than in adult cases using the same platform 28, and raises the possibility that these contain pediatric oligodendroglioma‐predisposition loci. In adult patients, specific SNPs in 8q24.21 have been associated with an increased risk of IDH1 or 2 mutant glioma development, including oligodendroglioma, in genotyping studies 17. Interestingly, in our cohort, two patients had a focal overlapping area of possible germ line alteration at 22q13.1‐13.2, which included a number of interesting genes, involved in cell biology, including DNA repair (XRCC6), nuclear transport and other nuclear proteins (RANGAP1, NHP2L1, CENPM), cell signaling (SGSM3, MKL1, TOB2, TNFRSF13C, RBX1, MCHR1), transcription (TEF, SREBF2, TCF20), molecular chaperones associating with heat shock proteins (ST13, DNAJB7), cytochrome P450 (CYP2D6, CYP2D7P1) and microRNAs (MiR‐4766, MiR‐1281). ST13 is also a candidate tumor suppressor in colorectal carcinoma 2. LOH in these and nearby 22q regions are not uncommon in glial tumors 13. CYP2D6 abnormalities/polymorphisms been associated with the development of glial tumors, including oligodendrogliomas 8, 18. However, these preliminary observations using only neoplastic tissue should be validated in future studies using matched non‐neoplastic tissues.
The main differential diagnosis with pediatric oligodendroglioma is dysembryoplastic neuroepithelial tumor (DNT), a tumor that may be difficult if not impossible to distinguish from, particularly in small biopsies. Recurrent genetic alterations have not been described in these tumors, other than BRAF p.V600E in a subset 7. Interestingly, in the whole‐genome sequencing study of Zhang et al, the single DNT studied had an FGFR1 tyrosine kinase duplication, as did 3 of 5 pediatric oligodendrogliomas, suggesting that these two lesions may be difficult to distinguish even at the molecular level 44. However, no alterations were present by array CGH in a recent study of six DNT and grade II pediatric gliomas 29.
A primarily low‐grade, pediatric neoplasm with oligodendroglioma‐like cytology but extensive and disproportionate superficial parenchymal and leptomeningeal involvement has been studied in several series and case reports. These disseminated oligodendroglioma‐like leptomeningeal neoplasms (DOLN) have been recently found to be characterized by a high frequency of combined 1p deletion and BRAF : KIAA1549 fusion 36, a molecular signature that separates them both from adult and pediatric oligodendrogliomas. Our current data reinforce this molecular distinction in the pediatric setting, as isolated 1p loss was relatively rare and BRAF : KIAA1549 fusion was absent in all cases tested (n = 13). It must be noted, however, that rare case reports have also described BRAF : KIAA1549 fusions in pediatric oligodendroglioma 24.
In summary, we report global molecular cytogenetic alterations in pediatric oligodendrogliomas present in approximately half of low‐grade tumors but in all anaplastic oligodendrogliomas studied. A subset of these tumors is characterized by predominantly genomic losses. Deletion of 1p, usually combined with 19q deletion, is relatively rare in the group overall [5 of 20 (25%)], but represented the most common identifiable abnormality, with the caveat that these alterations are relatively rare compared with their adult counterparts and other pediatric low‐grade gliomas. BRAF alterations do not appear to play a prominent role in pediatric oligodendroglioma, and lack of BRAF fusions or BRAF p.V600E separate these tumors from several other pediatric low‐grade gliomas.
Acknowledgments
This work was supported in part by the Childhood Brain Tumor Foundation and the Pilocytic/Pilomyxoid Research Fund including Lauren's First and Goal.
References
- 1. Agamanolis DP, Katsetos CD, Klonk CJ, Bartkowski HM, Ganapathy S, Staugaitis SM et al (2012) An unusual form of superficially disseminated glioma in children: report of 3 cases. J Child Neurol 27:727–733. [DOI] [PubMed] [Google Scholar]
- 2. Bai R, Shi Z, Zhang JW, Li D, Zhu YL, Zheng S (2012) ST13, a proliferation regulator, inhibits growth and migration of colorectal cancer cell lines. J Zhejiang Univ Sci B 13:884–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bar EE, Lin A, Tihan T, Burger PC, Eberhart CG (2008) Frequent gains at chromosome 7q34 involving BRAF in pilocytic astrocytoma. J Neuropathol Exp Neurol 67:878–887. [DOI] [PubMed] [Google Scholar]
- 4. Bettegowda C, Agrawal N, Jiao Y, Sausen M, Wood LD, Hruban RH et al (2011) Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science 333:1453–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blesa D, Mollejo M, Ruano Y, de Lope AR, Fiano C, Ribalta T et al (2009) Novel genomic alterations and mechanisms associated with tumor progression in oligodendroglioma and mixed oligoastrocytoma. J Neuropathol Exp Neurol 68:274–285. [DOI] [PubMed] [Google Scholar]
- 6. Cairncross G, Wang M, Shaw E, Jenkins R, Brachman D, Buckner J et al (2013) Phase III trial of chemoradiotherapy for anaplastic oligodendroglioma: long‐term results of RTOG 9402. J Clin Oncol 31:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chappe C, Padovani L, Scavarda D, Forest F, Nanni‐Metellus I, Loundou A et al (2013) Dysembryoplastic neuroepithelial tumors share with pleomorphic xanthoastrocytomas and gangliogliomas BRAF(V600E) mutation and expression. Brain Pathol 23:574–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Elexpuru‐Camiruaga J, Buxton N, Kandula V, Dias PS, Campbell D, McIntosh J et al (1995) Susceptibility to astrocytoma and meningioma: influence of allelism at glutathione S‐transferase (GSTT1 and GSTM1) and cytochrome P‐450 (CYP2D6) loci. Cancer Res 55:4237–4239. [PubMed] [Google Scholar]
- 9. Gielen GH, Gessi M, Buttarelli FR, Baldi C, Hammes J, Zur Muehlen A et al (2015) Genetic analysis of diffuse high‐grade astrocytomas in infancy defines a novel molecular entity. Brain Pathol 25:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gorovets D, Kannan K, Shen R, Kastenhuber ER, Islamdoust N, Campos C et al (2012) IDH mutation and neuroglial developmental features define clinically distinct subclasses of lower grade diffuse astrocytic glioma. Clin Cancer Res 18:2490–2501. [DOI] [PubMed] [Google Scholar]
- 11. Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, Murphy KM (2006) Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol 65:988–994. [DOI] [PubMed] [Google Scholar]
- 12. Harada S, Henderson LB, Eshleman JR, Gocke CD, Burger P, Griffin CA, Batista DA (2011) Genomic changes in gliomas detected using single nucleotide polymorphism array in formalin‐fixed, paraffin‐embedded tissue: superior results compared with microsatellite analysis. J Mol Diagn 13:541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hartmann C, Numann A, Mueller W, Holtkamp N, Simon M, von Deimling A (2004) Fine mapping of chromosome 22q tumor suppressor gene candidate regions in astrocytoma. Int J Cancer 108:839–844. [DOI] [PubMed] [Google Scholar]
- 14. Horbinski C, Nikiforova MN, Hobbs J, Bortoluzzi S, Cieply K, Dacic S, Hamilton RL (2012) The importance of 10q status in an outcomes‐based comparison between 1p/19q fluorescence in situ hybridization and polymerase chain reaction‐based microsatellite loss of heterozygosity analysis of oligodendrogliomas. J Neuropathol Exp Neurol 71:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ida CM, Vrana JA, Rodriguez FJ, Jentoft ME, Caron AA, Jenkins SM, Giannini C (2013) Immunohistochemistry is highly sensitive and specific for detection of BRAF V600E mutation in pleomorphic xanthoastrocytoma. Acta Neuropathol Commun 1:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66:9852–9861. [DOI] [PubMed] [Google Scholar]
- 17. Jenkins RB, Xiao Y, Sicotte H, Decker PA, Kollmeyer TM, Hansen HM et al (2012) A low‐frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet 44:1122–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kelsey KT, Wrensch M, Zuo ZF, Miike R, Wiencke JK (1997) A population‐based case‐control study of the CYP2D6 and GSTT1 polymorphisms and malignant brain tumors. Pharmacogenetics 7:463–468. [DOI] [PubMed] [Google Scholar]
- 19. Kim YH, Lachuer J, Mittelbronn M, Paulus W, Brokinkel B, Keyvani K et al (2011) Alterations in the RB1 pathway in low‐grade diffuse gliomas lacking common genetic alterations. Brain Pathol 21:645–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kitange G, Misra A, Law M, Passe S, Kollmeyer TM, Maurer M et al (2005) Chromosomal imbalances detected by array comparative genomic hybridization in human oligodendrogliomas and mixed oligoastrocytomas. Genes Chromosomes Cancer 42:68–77. [DOI] [PubMed] [Google Scholar]
- 21. Koschny R, Koschny T, Froster UG, Krupp W, Zuber MA (2002) Comparative genomic hybridization in glioma: a meta‐analysis of 509 cases. Cancer Genet Cytogenet 135:147–159. [DOI] [PubMed] [Google Scholar]
- 22. Kreiger PA, Okada Y, Simon S, Rorke LB, Louis DN, Golden JA (2005) Losses of chromosomes 1p and 19q are rare in pediatric oligodendrogliomas. Acta Neuropathol 109:387–392. [DOI] [PubMed] [Google Scholar]
- 23. Kros JM, van Run PR, Alers JC, Beverloo HB, van den Bent MJ, Avezaat CJ, van Dekken H (1999) Genetic aberrations in oligodendroglial tumours: an analysis using comparative genomic hybridization (CGH). J Pathol 188:282–288. [DOI] [PubMed] [Google Scholar]
- 24. Kumar A, Pathak P, Purkait S, Faruq M, Jha P, Mallick S et al (2015) Oncogenic KIAA1549‐BRAF fusion with activation of the MAPK/ERK pathway in pediatric oligodendrogliomas. Cancer Genet 208:91–95. [DOI] [PubMed] [Google Scholar]
- 25. Mistry M, Zhukova N, Merico D, Rakopoulos P, Krishnatry R, Shago M et al (2015) BRAF mutation and CDKN2A deletion define a clinically distinct subgroup of childhood secondary high‐grade glioma. J Clin Oncol 33:1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mohapatra G, Engler DA, Starbuck KD, Kim JC, Bernay DC, Scangas GA et al (2011) Genome‐wide comparison of paired fresh frozen and formalin‐fixed paraffin‐embedded gliomas by custom BAC and oligonucleotide array comparative genomic hybridization: facilitating analysis of archival gliomas. Acta Neuropathol 121:529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakamura M, Shimada K, Ishida E, Higuchi T, Nakase H, Sakaki T, Konishi N (2007) Molecular pathogenesis of pediatric astrocytic tumors. Neuro‐Oncol 9:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nauen DW, Guajardo A, Haley L, Powell K, Burger PC, Gocke CD (2015) Chromosomal defects track tumor subpopulations and change in progression in oligodendroglioma. Convergent Sci Phys Oncol (adv online pub). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Padovani L, Colin C, Fernandez C, Maues de Paula A, Mercurio S, Scavarda D et al (2012) Search for distinctive markers in DNT and cortical grade II glioma in children: same clinicopathological and molecular entities? Curr Top Med Chem 12:1683–1692. [DOI] [PubMed] [Google Scholar]
- 30. Perilongo G, Gardiman M, Bisaglia L, Rigobello L, Calderone M, Battistella A et al (2002) Spinal low‐grade neoplasms with extensive leptomeningeal dissemination in children. Childs Nerv Syst 18:505–512. [DOI] [PubMed] [Google Scholar]
- 31. Raghavan R, Balani J, Perry A, Margraf L, Vono MB, Cai DX et al (2003) Pediatric oligodendrogliomas: a study of molecular alterations on 1p and 19q using fluorescence in situ hybridization. J Neuropathol Exp Neurol 62:530–537. [DOI] [PubMed] [Google Scholar]
- 32. Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP (1994) Amplification of multiple genes from chromosomal region 12q13‐14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res 54:4299–4303. [PubMed] [Google Scholar]
- 33. Reis GF, Pekmezci M, Hansen HM, Rice T, Marshall RE, Molinaro AM et al (2015) CDKN2A loss is associated with shortened overall survival in lower‐grade (World Health Organization Grades II‐III) astrocytomas. J Neuropathol Exp Neurol 74:442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rodriguez FJ, Perry A, Rosenblum MK, Krawitz S, Cohen KJ, Lin D et al (2012) Disseminated oligodendroglial‐like leptomeningeal tumor of childhood: a distinctive clinicopathologic entity. Acta Neuropathol 124:627–641. [DOI] [PubMed] [Google Scholar]
- 35. Rodriguez FJ, Tihan T, Lin D, McDonald W, Nigro J, Feuerstein B et al (2014) Clinicopathologic features of pediatric oligodendrogliomas: a series of 50 patients. Am J Surg Pathol 38:1058–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodriguez FJ, Schniederjan MJ, Nicolaides T, Tihan T, Burger PC, Perry A (2015) High rate of concurrent BRAF‐KIAA1549 gene fusion and 1p deletion in disseminated oligodendroglioma‐like leptomeningeal neoplasms (DOLN). Acta Neuropathol 129:609–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rossi MR, Gaile D, Laduca J, Matsui S, Conroy J, McQuaid D et al (2005) Identification of consistent novel submegabase deletions in low‐grade oligodendrogliomas using array‐based comparative genomic hybridization. Genes Chromosomes Cancer 44:85–96. [DOI] [PubMed] [Google Scholar]
- 38. Schniederjan MJ, Alghamdi S, Castellano‐Sanchez A, Mazewski C, Brahma B, Brat DJ et al (2013) Diffuse leptomeningeal neuroepithelial tumor: 9 pediatric cases with chromosome 1p/19q deletion status and IDH1 (R132H) immunohistochemistry. Am J Surg Pathol 37:763–771. [DOI] [PubMed] [Google Scholar]
- 39. Suri V, Jha P, Agarwal S, Pathak P, Sharma MC, Sharma V et al (2011) Molecular profile of oligodendrogliomas in young patients. Neuro‐Oncol 13:1099–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Talagas M, Marcorelles P, Uguen A, Redon S, Quintin‐Roue I, Costa S et al (2012) Identification of a novel population in high‐grade oligodendroglial tumors not deleted on 1p/19q using array CGH. J Neurooncol 109:405–413. [DOI] [PubMed] [Google Scholar]
- 41. van den Bent MJ, Brandes AA, Taphoorn MJ, Kros JM, Kouwenhoven MC, Delattre JY et al (2013) Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long‐term follow‐up of EORTC brain tumor group study 26951. J Clin Oncol 31:344–350. [DOI] [PubMed] [Google Scholar]
- 42. Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yip S, Butterfield YS, Morozova O, Chittaranjan S, Blough MD, An J et al (2012) Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J Pathol 226:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang J, Wu G, Miller CP, Tatevossian RG, Dalton JD, Tang B et al (2013) Whole‐genome sequencing identifies genetic alterations in pediatric low‐grade gliomas. Nat Genet 45:602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]


