SUMMARY
The regulation of 5’ end resection at DSBs and telomeres prevents genome instability. DSB resection is positively and negatively regulated by ATM signaling through CtIP/MRN and 53BP1-bound Rif1, respectively. Similarly, telomeres lacking TRF2 undergo ATM-controlled CtIP-dependent hyper-resection when the repression by 53BP1/Rif1 is alleviated. However, telomere resection in absence of 53BP1/Rif1 is more extensive upon complete removal of shelterin, indicating additional protection against resection by shelterin. Here we show that TPP1 and POT1a/b in shelterin block a resection pathway distinct from that repressed by TRF2. This second pathway is regulated by ATR signaling, involves Exo1 and BLM, and is inhibited by 53BP1/Rif1. Thus, mammalian cells have two distinct 5’ end resection pathways that are regulated by DNA damage signaling, in part through Rif1-mediated inhibition. The data show that telomeres are protected from hyper-resection through the repression of the ATM and ATR kinases by TRF2 and TPP1-bound POT1a/b, respectively.
Keywords: 53BP1, Rif1, telomere, resection
INTRODUCTION
The mechanism and regulation of DNA 5’ end resection is of interest given its contribution to high fidelity homology-directed repair (HDR) and the maintenance of a stable genome. DSB resection requires the ATM DNA damage response (DDR) kinase and its target CtIP, which promotes an initial resection step by the MRN (Mre11/Rad50/Nbs1) complex (reviewed in (Symington and Gautier, 2011)). Further resection is mediated by the Exo1 exonuclease or the DNA2 nuclease acting with the BLM and WRN helicase (Sturzenegger et al., 2014). This processing results in the extended 3’ overhangs required for Rad51-mediated HDR. Whereas these events closely follow the paradigm for DSB resection established in budding yeast (Mimitou and Symington, 2008; Zhu et al., 2008), the control of resection in mammalian cells additionally involves a negative regulator, Rif1, which associates with 53BP1 at sites of DNA damage (Chapman et al., 2013; Di Virgilio et al., 2013; Feng et al., 2013; Escribano-Diaz et al., 2013; Zimmermann et al., 2013). In G1, Rif1 promotes non-homologous end joining (NHEJ) by limiting DSB resection through its interaction with Rev7/MAD2L2 (Xu et al., 2015; Boersma et al., 2015). In S/G2, BRCA1 prevents Rif1 from acting at DSBs, allowing the formation of 3’ overhangs and promoting HDR on sister chromatids (reviewed in (Panier and Boulton, 2014; Panier and Durocher, 2013; Zimmermann and de Lange, 2014)).
At fully functional telomeres, 5’ end resection is an important post-replicative step to generate the 3’ overhang needed for telomere function. However, excessive 5’ resection is a threat to genome integrity because it can lead to telomere shortening. The telomere specific shelterin complex functions to protect chromosome ends from this and other detrimental outcomes of the DDR (reviewed in (Palm and de Lange, 2008)), ensuring that chromosome ends do not activate the ATM and ATR signaling pathways and do not succumb to inappropriate resection and DSB repair. Shelterin is compartmentalized such that its TRF2 component is dedicated to the repression of the ATM kinase whereas the TPP1/POT1 heterodimers (TPP1/POT1a and TPP1/POT1b in the mouse) block the activation of the ATR kinase.
Telomere hyper-resection is repressed by shelterin as well as by 53BP1/Rif1. In the absence of Rif1 or 53BP1, the removal of the whole shelterin complex through the simultaneous deletion of TRF1 and TRF2 results in extensive 5’ end resection and shortening of telomeres (Sfeir et al., 2009; Zimmermann et al., 2013). Previous data showed that the repression of ATM/CtIP-controlled resection at telomeres depends on TRF2 (Lottersberger et al., 2013). Here we establish that in addition to TRF2, the TPP1/POT1 heterodimers repress inappropriate 5’ end resection. The TPP1/POT1-controlled pathway is distinct from that controlled by TRF2 since it is stimulated by the ATR kinase and appears to be independent of CtIP. 53BP1-bound Rif1 is critical for preventing inappropriate resection by both pathways.
RESULTS
The role of TRF2 in the control of 5’ end resection at telomeres
To study the regulation of 5’ end resection at telomeres, we took advantage of immortalized TRF1F/FTRF2F/F53BP1−/− conditional triple knockout (TKO) mouse embryo fibroblasts (MEFs), which show extensive hyper-resection after shelterin is removed from telomeres through the simultaneous deletion of TRF1 and TRF2 (Sfeir and de Lange, 2012). Since absence of TRF2 alone cannot explain the nucleolytic processing of the dysfunctional telomeres, we first determined to what extent the removal of TRF1 contributed to hyper-resection. Expression of exogenous TRF2 in TRF1F/FTRF2F/F53BP1−/− TKO cells can mediate the telomere binding of all expressed shelterin components (Rap1, TIN2, TPP1, and POT1a/b), such that the resulting telomeric complexes will only lack TRF1. As expected, reintroduction of TRF2 protected telomeres from much of the resection, resulting in a modest 3–5 fold increase in telomeric overhang signal rather than the 20–25 fold increase observed in TRF1F/FTRF2F/F53BP1−/− TKO cells lacking exogenous TRF2 (Fig. 1A–D). Thus, consistent with the minor resection previously reported for TRF1 deletion from 53BP1-deficient cells (Sfeir and de Lange, 2012), the absence of TRF1 from the telomeres leads to a moderate resection phenotype, whereas absence of both TRF1 and TRF2, and thus the whole shelterin complex, unleashes maximal resection.
Figure 1. Domains in TRF2 required for the repression of telomere hyper-resection.
(A) In-gel overhang assay to monitor 5’ end resection at telomeres in TRF1F/FTRF2F/F 53BP1−/−p53−/− Cre-ERT2 MEFs expressing the indicated TRF2 mutants or wild type TRF2 analyzed at 96 h after 4-hydroxytamoxifen (4-OHT) induction of Cre. Dashed lines indicate the quantified area.
(B) In-gel overhang assay to monitor the effect of ATM inhibition on telomere hyper-resection in TRF1F/FTRF2F/F53BP1−/−p53−/− Cre-ERT2 MEFs expressing wild type TRF2 or TRF2ΔTIN2. Cells were analyzed at 96 h after 4-OHT. 2.5 µM KU55933 was added 12 hr before 4-OHT.
(C) Quantification of the relative normalized overhang signals in the indicated cells before and after Cre. Cells and treatments as in (A) and (B). *, P < 0.05 **, P < 0.01 ***, P < 0.001 (two-tailed Student’s t test). Bars represent means of 3–4 independent experiments ±SDs. For each condition, the relative normalized overhang signal without Cre treatment was set to 1.0 and the +Cre value was expressed relative to this reference.
(D) Schematic representation of the shelterin proteins present at telomeres under the conditions examined in (A–C) and the extent of resection in each condition. See also Figure S1.
Complementation of the TRF1F/FTRF2F/F53BP1−/− TKO MEFs with TRF2 mutants that lacked either the Rap1 binding site (TRF2ΔRap1 (Sfeir et al., 2010)) or the iDDR region (TRF2ΔiDDR lacking aa 403–427 (Okamoto et al., 2013)) (Fig. 1A–D; Fig. S1) showed that these two domains of TRF2 are not required for the repression of resection. In contrast, a TRF2 mutant lacking the TIN2 binding site (TRF2ΔTIN2; (Takai et al., 2011)) was unable to provide the same repression of resection as wild type TRF2 (Fig. 1A–D). In TKO cells expressing TRF2ΔTIN2, the telomeres are predicted to contain TRF2 and Rap1 but not TRF1, TIN2, TPP1, and POT1a/b. The TRF2ΔTIN2 mutant was expressed at the same level as wild type TRF2, could be detected at telomeres by ChIP (albeit at slightly reduced levels), and largely restored the protection of telomeres from NHEJ (Fig. S1A–C). Nonetheless, the TRF2ΔTIN2 cells showed a 7–10 fold increase in the overhang signals (Fig. 1A–D).
Whereas the resection at telomere lacking all shelterin components showed the expected contribution of ATM signaling (Fig. 1B and C; Fig. S1D) (Sfeir and de Lange, 2012), the increase in the overhang signal at telomeres containing the TRF2ΔTIN2 mutant, examined in parallel, was not affected by inhibition of ATM (Fig. 1B and C; Fig. S1D). Thus, an ATM-independent pathway might be involved in the hyper-resection at telomeres containing only TRF2 and Rap1.
TPP1-bound POT1a/b and TRF2 are the main inhibitors of resection at telomeres
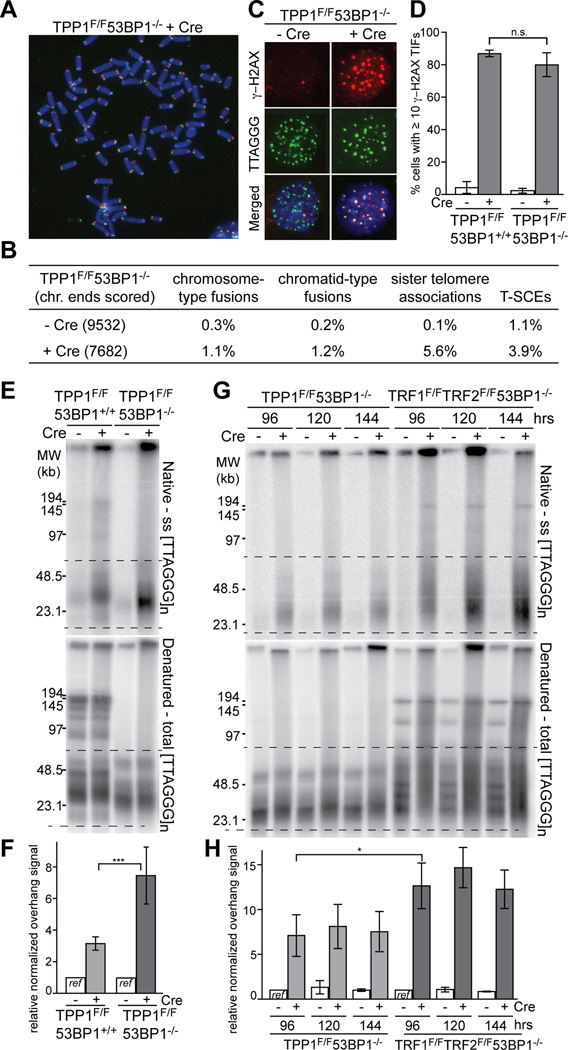
The results obtained with the TRF2 mutants suggested that either TIN2 itself or the TIN2-bound TPP1/POT1 heterodimers (or both) are involved in the repression of resection (Fig. 1D). As TIN2 deletion results in destabilization of the shelterin complex and thus can yield confounding results (Takai et al., 2011; Frescas and de Lange, 2014), we focused on the TPP1 and POT1 components of shelterin. We generated SV40LT-immortalized TPP1F/F53BP1−/− MEFs, which lose TPP1, POT1a, and POT1b from telomeres upon expression of Cre but retain the other shelterin components (TRF1, TRF2, TIN2, and Rap1 (Kibe et al., 2010)). Consistent with previous data, metaphase spreads from TPP1-deficient cells lacked telomere fusions, which often confound the analysis of telomere end resection (Fig. 2A and B). As expected, the absence of 53BP1 did not affect the induction of a telomere damage response upon loss of TPP1, as measured based on the appearance of γ-H2AX in Telomere dysfunction Induced Foci (TIFs; (Takai et al., 2003)) (Fig. 2C and D).
Figure 2. Loss of TPP1/POT1 leads to telomere hyper-resection.
(A–D) Characterization of telomeric phenotypes induced by Cre in SV40 large T immortalized TPP1F/F53BP1−/− MEFs at 96 h.
(A) Metaphase spread with telomeres detected with CO-FISH (green, TelC probe for G-rich lagging-strand template; red, TelG probe for the C-rich leading-strand template). DNA was stained with DAPI (blue).
(B) Quantification of telomere aberrations in TPP1/53BP1 DKO cells analyzed as in (A). Sister telomere associations were scored only at the long arm telomeres. Values are averages from three independent experiments.
(C) Induction of Telomere dysfunction-Induced Foci (TIF) upon deletion of TPP1 from 53BP1 knockout (KO) cells. γ-H2AX was detected by indirect immunofluorescence (IF) (red) in combination with FISH for telomeres (green).
(D) Quantification of γ-H2AX TIFs detected as in (C). Cells with ≥10 telomeric γ-H2AX foci were scored. Bars show means of ≥3 experiments ± SDs (>100 cells per experiment).
(E) In-gel overhang assay to detect resection at telomeres in TPP1F/F53BP1+/+ and TPP1F/F53BP1−/− MEFs at 96 h after Cre.
(F) Quantification of changes in 3’ overhang signals at telomeres of the indicated cells analyzed as in (E). Area between dashed lines was quantified and compared among lanes in the same gel. Bars represent means of four independent experiments and SDs. Values from the each cell line without Cre treatment were set at 1.0. ***, P < 0.001 (two-tailed Student’s t test).
(G and H) Comparison of 5’ end resection in TPP1/53BP1 DKO and TRF1/TRF2/53BP1 triple knockout cells. Cells were analyzed in parallel at indicated time points after infection with Hit & Run Cre. Bars show means of three independent experiments and SDs. Values for each cell line without Cre at 96 h were set to 1.0. *, P < 0.05 (two-tailed Student’s t test).
See also Figures S2, S3, and S4.
The removal of TPP1 and concomitant loss both POT1 proteins from the telomeres in 53BP1-deficient MEFs resulted in a 7–10 fold increase in the single-stranded telomeric DNA signal (Fig. 2E and F), which suggested a level of resection similar to that at telomeres containing the TRF2ΔTIN2 mutant and Rap1 but lacking all other shelterin proteins (Fig. 1). The increase in the single-stranded TTAGGG repeat signal upon deletion of TPP1 from 53BP1-deficient cells was due to 5’ end resection since it was removed by treatment of the DNA by the E.coli 3’ exonuclease ExoI (Fig. S2). Deletion of TPP1 from 53BP1-proficient cells resulted in a more modest increase in the telomeric overhang signal (Fig. 2E and F), consistent with the requirement for TPP1-tethered POT1b in restricting telomere resection after DNA replication (Kibe et al., 2010; Wu et al., 2012). Also consistent with prior data (Wu et al., 2012), the absence of telomerase had a minor effect on the overhang signals after TPP1 deletion, indicating that most of the increase in ssDNA is due to hyper-resection rather than 3’ end extension (Fig. S3). Thus, telomeres lacking TPP1 (and POT1a/b) undergo extensive resection when 53BP1 is absent. Side-by-side comparison showed that removal of the whole shelterin complex results in more extensive resection than in absence of TPP1, consistent with the role of TRF2 in blocking resection (Fig. 2G and H). To determine whether the effect of TPP1 deletion is primarily due to loss of the POT1 proteins, we identified a mutant of TPP1 that fails to interact with the POT1 proteins while retaining its TIN2 interaction (TPP1ΔPOT1; Fig. S4). Although wild type TPP1 repressed the hyper-resection in TPP1/53BP1 DKO cells, the TPP1ΔPOT1 mutant failed to protect the telomeres, consistent with resection being controlled by POT1a/b (Fig. S4).
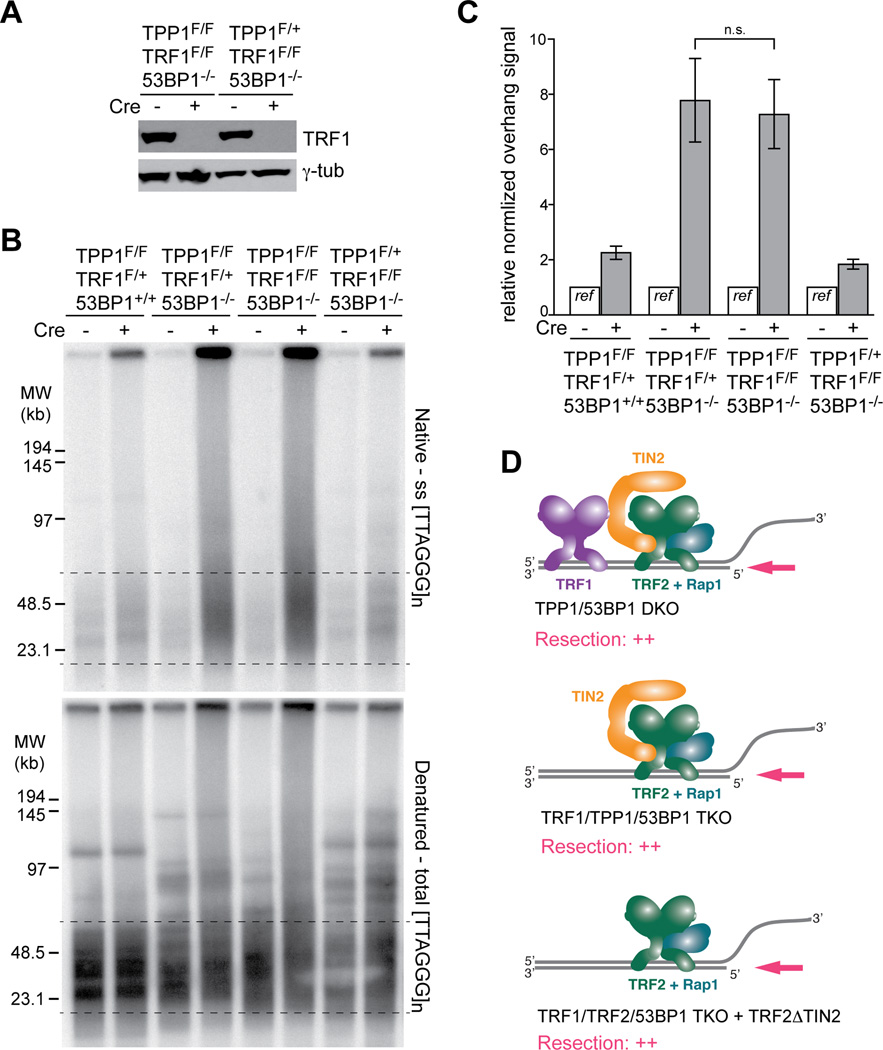
To confirm that the maximal resection in the shelterin-free setting was not due to the simultaneous absence of TRF1 and TPP1, we generated immortalized TRF1F/FTPP1F/F53BP1−/− MEFs and control MEFs expressing either TRF1 or TPP1. Co-deletion of TRF1 and TPP1 is predicted to generate telomeres containing only TRF2, TIN2, and Rap1. As expected, the resection at the dysfunctional telomeres lacking TRF1 and TPP1 was not significantly increased compared to deletion of TPP1 alone (Fig. 3A–D). This result is consistent with TRF2, not TRF1, being one of the main repressors of resection at telomeres. We therefore conclude that resection at telomeres is primarily blocked by TRF2 and the TPP1/POT1 heterodimers.
Figure 3. Deletion of TRF1 does not exacerbate the resection in absence of TPP1.
(A) Immunblot verifying the deletion of TRF1 from the indicated cells.
(B) In-gel overhang assay showing that co-deletion of TRF1 and TPP1 from 53BP1-deficient cells does not exacerbate the resection phenotype. The first three MEF lines were isolated from littermates. The fourth (control) MEF line (TPP1F/+TRF1F/F53BP1−/−) carries one copy of the TRF2 floxed allele (TRF2F/+). As TRF2 is not haploinsufficient the presence of the floxed allele is unlikely to affect the results. Cells were analyzed at 96 h after the second Cre infection.
(C) Quantification of overhang signals of the indicated cells analyzed as in (B). Bars show means of three independent experiments with SDs. Values from the each cell line without Cre treatment set at 1.0 and the plus Cre value was expressed relative to this reference. P value from two-tailed Student’s t test.
(D) Schematic summarizing the similar resection at telomeres lacking TPP1/POT1 but retaining TRF2 and various other shelterin components.
ATR stimulates telomere hyper-resection in absence of TPP1
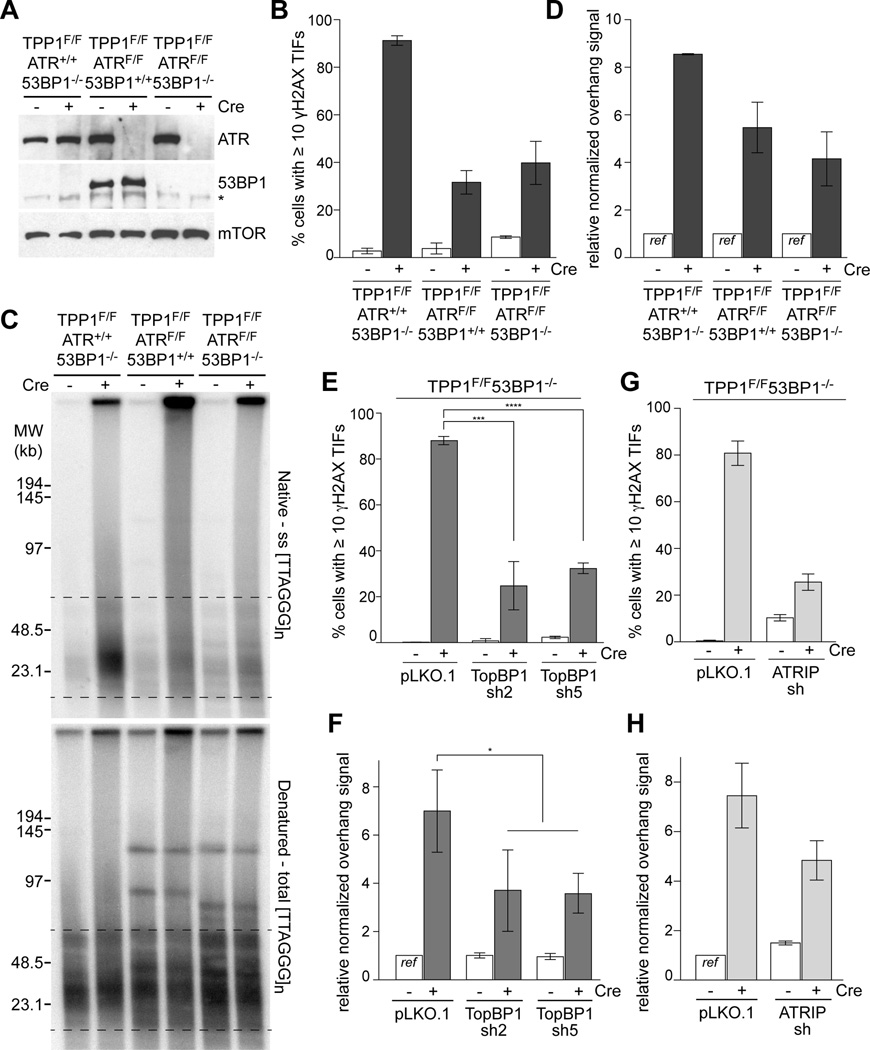
Since the ATM kinase is not activated at telomeres lacking TPP1 (Kibe et al., 2010) and inhibition of ATM did not affect the resection in TRF1/2 double knockout (DKO) cells complemented with TRF2ΔTIN2 (Fig. 1C), we queried the role of the ATR kinase in this setting. We generated conditional TPP1F/FATRF/F MEFs and then used CRISPR to remove 53BP1 (Fig. 4A; Fig. S5A–C). As expected, absence of ATR strongly diminished the formation of γ-H2AX foci at telomeres lacking TPP1 (Fig. 4B) but had no significant effect on cell cycle progression at the time point used (Fig. S5C). Although ATR signaling had not previously been implicated in promoting resection, the absence of ATR severely diminished the telomere hyper-resection in cells lacking TPP1 and 53BP1 (Fig. 4C,D). The residual 4-fold increase in overhang signals at telomeres in the TPP1/ATR/53BP1 TKO cells is consistent with a deficiency in CST-mediated fill-in after replication of the telomeric DNA. For instance, in the TPP1/53BP1 KO cell line used here the absence of POT1a/b at telomeres leads to a 3.5-fold increase in the overhang signal (Fig. S4) and in POT1b KO cell lines where CST-mediated fill-in is abrogated, the telomeric overhang signals increases 2–4 fold (Wu et al., 2012).
Figure 4. Resection at telomeres lacking TPP1 is stimulated by ATR.
(A) Verification of ATR and 53BP1 deletion in TPP1F/F53BP1−/− cells and clones derived from TPP1F/FATRF/F MEFs at 96 h after Cre. mTOR is used as loading control. Asterisk: non-specific band.
(B, E, and G) Quantifications of cells with ≥10 γ-H2AX foci at telomeres in the indicated cell lines at 96 h after Cre treatment. Bars in (B, and G) show means and SEMs of two independent experiments (>100 cells per experiment). Bars in (E) show means and SDs from three independent experiments. ***, P < 0.001 (two-tailed Student’s t test).
(C) In-gel overhang assay on the TPP1/53BP1 DKO, TPP1/ATR DKO, TPP1/ATR/53BP1 TKO cells at 96 h after Cre treatment.
(D) Quantification of overhang signals analyzed as in (C). Bars represent means of two independent experiments ± SEMs.
(F, and H) Quantification of in-gel overhang assays (Fig. S5E, G) on TPP1F/F53BP1−/− MEFs treated with TopBP1, and ATRIP shRNAs at 96 h after Cre treatment. Bars in (F) show means of three independent experiments and SDs. *, P < 0.05 (two-tailed Student’s t test). Bars in (H) show means of two independent experiments and SEMs. See also Figure S5.
The involvement of ATR kinase signaling was corroborated with shRNA mediated depletion of TopBP1 and ATRIP, two factors required for the activation of the ATR kinase (reviewed in (Ciccia and Elledge, 2010)). Both methods showed the expected reduction in γ-H2AX TIFs induced by deletion of TPP1 (Fig. 4) but did not affect the S phase index of the cells (Fig. S5). Consistent with the involvement of ATR signaling in resection, two shRNAs to TopBP1 and one shRNA to ATRIP significantly reduced the resection at telomeres lacking TPP1 (Fig. 4E–H; Fig. S5). In contrast, ATM inhibition had no effect on the resection at telomeres lacking TPP1/POT1 (Fig. S5I), similar to the result obtained in TRF1/TRF2/53BP1 TKO cells expressing TRF2ΔTIN2 (Fig. 1). These results are expected, since the ATM kinase is not activated at telomeres that retain TRF2.
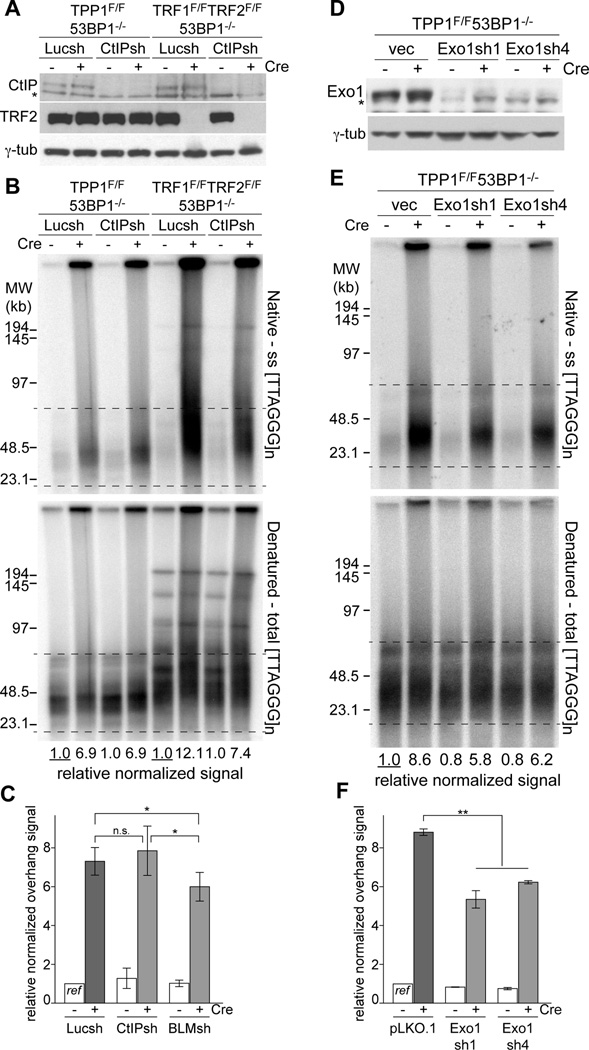
Contributions of Exo1 and BLM but not CtIP to resection
To identify the nucleolytic factors involved in the ATR-regulated resection pathway, we inhibited CtIP and BLM with shRNAs that were previously used to show that resection at shelterin-free telomeres is mediated by these factors (Sfeir and de Lange, 2012). The data indicated that knockdown of CtIP had no effect in the TPP1/53BP1 DKO setting whereas the CtIP shRNA showed the expected effect on resection in TRF1/TRF2/53BP1 TKO cells treated in parallel (Fig. 5A–C). In contrast, the shRNA knockdown experiments implicated both BLM and Exo1 in the ATR-stimulated resection (Fig. 5C–F; Fig. S6). Thus, the ATR-stimulated resection at telomeres lacking TPP1/POT1 is in part mediated by the Exo1 nuclease and the BLM helicase. Neither Exo1 nor BLM are solely responsible for resection, which is expected based on the redundancy of these resection pathways. The minor effect of BLM depletion could be due to the DNA2 nuclease being assisted by both the WRN and BLM helicases (Sturzenegger et al., 2014). We have not been able to test whether the DNA2 nuclease because its depletion impedes S phase progression in our MEFs.
Figure 5. Exo1 and BLM, but not CtIP-dependent resection in TPP1/53BP1 DKO.
(A) Immunoblotting showing equal knockdown of CtIP in TPP1F/F53BP1−/− and TRF1F/FTRF2F/F53BP1−/−p53−/− MEFs treated with Cre. Cells were analyzed at 96 h after the second Cre infection. Asterisk: non-specific band.
(B) In-gel overhang assay of shCtIP treated TPP1/53BP1 DKO and TRF1/TRF2/53BP1 TKO cells as in (A). For each cell line, the relative normalized overhang signal obtained from Luciferase sh treated cells without Cre were set as 1.0 and the other values were expressed relative to this reference.
(C) Quantification of the overhang signals as in (B). Bars show means ± SDs from ≥3 independent experiments. *, P < 0.05 (two-tailed Student’s t test).
(D) Immunoblot for Exo1 knockdown in the TPP1/53BP1 DKO cells at 96 h after Cre treatment. Asterisk: non-specific band.
(E) In-gel overhang assays on indicated MEFs (as in (D)) at 96 h after Cre infection.
(F) Quantification of the overhang signals as in (E). Graph shows means ± SEMs from two independent experiments. **, P < 0.01 from two-tailed Student’s t test on the combined results of the two Exo1 shRNAs. See also Figure S6.
Rif1 inhibits ATM/CtIP-independent resection
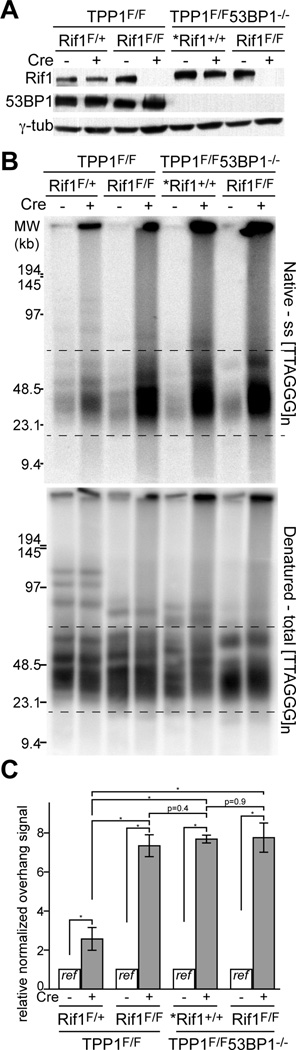
As the ATR-stimulated resection pathway has not previously been studied, we asked whether it is controlled by 53BP1 through its interaction partner Rif1, as is the case for ATM/CtIP-dependent resection. To this end, we generated SV40LT-immortalized MEFs from which TPP1 could be deleted together with Rif1 and compared the level of telomere resection after TPP1 deletion in the absence of 53BP1, in the absence of Rif1, and in the absence of both (Fig. 6). The results indicated that the same resection occurred in each of these settings (Fig. 6C). Thus, Rif1 is the major factor acting downstream of 53BP1 to block the ATR/BLM/Exo1-promoted resection at telomeres lacking TPP1/POT1 (Fig. 6D).
Figure 6. Rif1 is the main factor that mediates inhibition of resection by 53BP1.
(A) Immunoblotting showing loss of Rif1 and 53BP1 from the indicated Cre-treated MEFs. Cells were analyzed at 96 h after Cre infection. The asterisk indicates MEFs that are heterozygous for the floxed allele of TRF1 in addition to the indicated genotype.
(B) In-gel overhang assay on indicated MEFs (as in (A)) at 96 h after Cre infection.
(C) Quantification of the overhang signals from three independent experiments as in (B). Bars show means ± SDs. *, P < 0.05 (two-tailed Student’s t test).
DISCUSSION
Using telomeres lacking TPP1 and the POT1 proteins, we uncovered a previously unknown 5’ end resection pathway that involves signaling by ATR rather than ATM and does not require CtIP/MRN. Compared to genome-wide DSBs, telomeres lacking TPP1 provide a unique opportunity to identify this pathway because two specific circumstances obviate the need for the nicking step that is thought to create the initial 3’ overhang needed for further 5’ resection (Cannavo and Cejka, 2014; Garcia et al., 2011). First, the telomere termini originating from lagging-strand DNA synthesis are predicted to have a natural 3’ overhang due to the removal of the RNA primer for DNA synthesis. Second, at the presumably blunt-ended telomeres generated by leading-strand DNA synthesis, the TRF2-bound Apollo/SNMB1 nuclease is thought to mediate an initial 5’ resection step that circumvents the need for CtIP/MRN mediated cleavage (Wu et al., 2012; Lam et al., 2010; Wu et al., 2010). In contrast, at most DSBs, the ATM-dependent processing by CtIP/MRN can obscure the role of ATR in further resection.
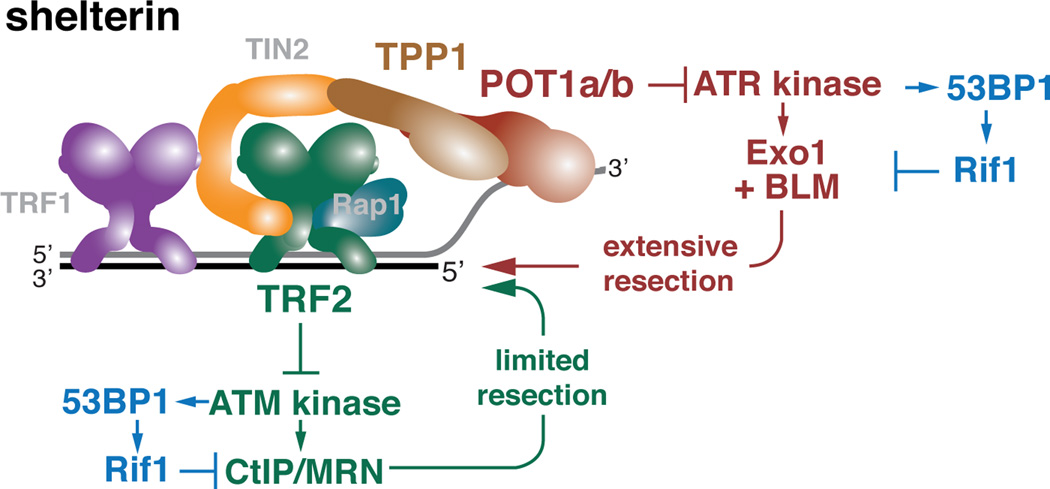
A similar architecture for the control of two 5’ end resection pathways
The pathways that can lead to 5’ end resection at deprotected telomeres differ in the primary processing step (CtIP/MRN versus BLM and Exo1) and the DDR kinase involved (ATM versus ATR) but have a similar overall architecture (Fig. 7). ATM signaling promotes resection through CtIP/MRN and inhibits resection through Rif1. Similarly, the regulation of resection by ATR, as proposed here, includes a negative regulation that is exerted through Rif1 and positive regulation at the level of BLM- and/or Exo1-mediated resection. We have no information on the target of ATR in the positive regulation. In non-replicating Xenopus extracts, ATR can stimulate DSB resection by DNA2 through phosphorylation of CtIP (Peterson et al., 2013). However, CtIP appears not to play a role in the ATR-promoted resection described here. In terms of negative regulation, we consider it likely that ATR affects Rif1 in the same manner as ATM, acting at the level of its recruitment by 53BP1.
Figure 7. Two distinct resection pathways are repressed at telomeres.
Schematic depicting the repression of two independent resection pathways by distinct shelterin proteins and the regulatory role of the DDR kinases and 53BP1/Rif1. The ATM-regulated resection involves CtIP/MRN and is inhibited by 53BP1/Rif1. Similarly, ATR regulated resection involves positive regulation through Exo1 and/or BLM and is inhibited through 53BP1/Rif1. 53BP1 and Rif1 are epistatic in both pathways. At telomeres, shelterin can prevent hyper-resection through repression of the DDR kinases. ATM is repressed by TRF2, most likely through the formation of the t-loop structure (not shown). ATR is blocked by POT1a (and to a lesser extent POT1b), which are tethered to TRF1 and TRF2 by TPP1 and TIN2. POT1 proteins are thought to prevent ATR activation by excluding RPA from the single-stranded telomeric DNA.
Potential roles for ATR-controlled resection
We suspect that the role for ATR in resection discovered at dysfunctional telomeres is relevant to what happens at S phase DSBs and sites of replication stress. After the initial ATM-dependent processing by CtIP/MRN, the activation of ATR by the initially short region of ssDNA could ensure that resection continues in order for HDR to take place. At sites of replication stress, ATR regulated resection by Exo1 and DNA2 might also be advantageous, for instance to prevent fork regression (Zeman and Cimprich, 2014; Hu et al., 2012; Neelsen and Lopes, 2015). At the same time, the ability of ATR to use 53BP1/Rif1 to avoid the formation of extensive single-stranded regions could be important to avert deleterious events, such as inappropriate recombination between repeat elements. Interestingly, in vitro experiments have shown that the level of ATR signaling depends on the length of the ssDNA (MacDougall et al., 2007). This feature could provide ATR with an adjustable output to either stimulate or inhibit of resection depending on the length of the ssDNA. For instance, if 53BP1/Rif1 are resistant to low levels of ATR activation, resection might not be blocked until sufficient ssDNA has been generated.
Consistent with a role for Rif1 in the control of resection at sites of replication stress, Rif1 deficient cells are hypersensitive to replication inhibitors, such as aphidicolin and HU, but not to other DNA damage agents, including as TopII inhibitors, MMC, or IR (Buonomo et al., 2009). Furthermore, Rif1 loss leads to an increase in chromatid breaks upon treatment with aphidicolin, further indicating a problem in the management of replication stress (Buonomo et al., 2009). These phenotypes are unlikely to be related to the role of Rif1 in controlling replication timing (reviewed in (Yamazaki et al., 2013)) but could be explained if Rif1 is needed to block excessive resection when replication has stalled or when the fork has collapsed.
The data also argue that the ability of Rif1 to block resection extends beyond G1 to S and/or G2 phase. In S/G2, BRCA1 is thought to ensure that Rif1 does not block resection at DSBs (Escribano-Diaz et al., 2013). Indeed, BRCA1 has been documented to prevent the accumulation of Rif1 at DSBs in S phase but not in G1. Yet, Rif1 can be observed at sites of replication stress (Buonomo et al., 2009), indicating that BRCA1 is incapable of (fully) repressing Rif1 in this setting. Furthermore, the 53BP1/Rif1-controlled resection at telomeres lacking TRF2 takes places after telomeric DNA replication but before mitosis, suggesting that Rif1 can act in S/G2 (Zimmermann et al., 2013; Lottersberger et al., 2013). Thus, the interplay between BRCA1 and Rif1 at DSBs, dysfunctional telomeres, and sites of replication stress in S/G2 merits further attention.
The inhibition of Exo1 and BLM-mediated resection by 53BP1/Rif1 is reminiscent of the role of Saccharomyces cerevisiae Rad9 checkpoint protein at DSBs. Rad9 is phosphorylated by the ATR ortholog Mec1 and is often referred to as a 53BP1 ortholog, although 53BP1 is dispensable for checkpoint signaling. Nonetheless, like 53BP1, Rad9 limits resection through a mechanism that involves Sgs1 (the only BLM/WRN-like helicase in yeast) and Exo1 (Lydall and Weinert, 1995; Lazzaro et al., 2008; Ngo et al., 2014; Bonetti et al., 2015; Clerici et al., 2014). However, the inhibition of resection by Rad9 does not involve Rif1. Instead, Rif1 has been reported to promote resection (Martina et al., 2014). Thus, the control of resection by Mec1 and Rad9 may differ considerably from that by ATR and 53BP1/Rif1.
How shelterin guards against telomere hyper-resection
Resection is carefully controlled at telomeres. After DNA replication, regulated resection and fill-in, both governed by shelterin, regenerates the unique 3’ overhang structure of telomeres (reviewed in (Doksani and de Lange, 2014)). However, in fungi and in mammals, telomeres have evolved mechanisms to prevent inappropriate resection and the accompanying loss of telomeric DNA. A major player in fungi is the CST complex that is thought to counteract resection by mediating polymerase α/primase fill in (reviewed in (Price et al., 2010)). In mammals, shelterin provides the first line of defense against resection with TRF2 dedicated to blocking ATM/CtIP-dependent resection and TPP1/POT1 assigned to the ATR-stimulated pathway. In both cases, the resection pathways are likely to be primarily blocked at the level of DNA damage signaling (Fig. 7). TRF2 has long been known to block MRN-activated ATM signaling and recent data indicate that it acts through its ability to alter telomere structure into the t-loop configuration which has been proposed to block the MRN complex from accessing the telomere end (Doksani et al., 2013). Apart from repressing ATM kinase signaling, the t-loop structure itself may also be protective against resection (Doksani and de Lange, 2014). On the other hand, TPP1-tethered POT1a (and to lesser extent POT1b) are known to prevent ATR kinase activation through excluding RPA from the ss telomeric DNA (Denchi and de Lange, 2007; Gong and de Lange, 2010; Takai et al., 2011; Kibe et al., 2010; Flynn et al., 2011). We propose that TPP1/POT1-dependent repression of resection is important at telomeres regardless of their DNA configuration. Given the similarities of the presumed structures at the base of the t-loop and at a stalled replication forks, telomeres are likely to be threatened by the ATR-stimulated resection pathway identified here, even when in they are in the t-loop configuration.
EXPERIMENTAL PROCEDURES
Cell culture, retroviral infections, and inhibitors
TPP1F/F SV40-LT, TRF1F/FTRF2F/F53BP1−/−p53−/− Cre-ERT2 mouse embryonic fibroblasts (MEFs) have been described previously (Kibe et al., 2010; Sfeir and de Lange, 2012). TPP1F/F53BP1−/− Cre-ERT2, TPP1F/FmTR−/−, TPP1F/FATRF/F, TPP1F/FTRF1F/+, TPP1F/FTRF1F/+53BP1−/−, TPP1F/FTRF1F/F53BP1−/−, TPP1F/+TRF1F/FTRF2F/+53BP1−/−, TPP1F/FRif1F/+, TPP1F/FRif1F/F, TPP1F/FRif1F/F53BP1−/− MEFs were obtained by standard mouse crosses. Primary MEFs isolated from E12.5 or E13.5 embryos were cultured in DMEM (Cellgro) with 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 1 mM sodium-pyruvate (Sigma-Aldrich), 100 U/ml penicillin (GIBCO), 0.1 µg/ml streptomycin (GIBCO), 0.2 mM L-glutamine (GIBCO), 0.1 mM non-essential amino acids (GIBCO), and 15% fetal bovine serum (FBS) (GIBCO). Genotyping was carried out by Transnetyx Inc. MEFs were immortalized at passage 2 using infection with pBabe-SV40-LT (a gift from Greg Hannon) and maintained in the same media without β-mercaptoethanol, and sodium-pyruvate as described (Celli et al., 2006). Cre recombinase was introduced by two retroviral infections with Hit and Run-Cre in pMMP at 12 h interval (Celli et al., 2006; Sfeir and de Lange, 2012). To delete TPP1 gene in TPP1F/F53BP1−/− Cre-ERT2 SV40-LT MEFs, Hit and Run-Cre was induced because Cre-ERT2 did not induce properly in the cell line. For the Cre-ERT2 system, Cre was induced with 0.5 µM 4-OH tamoxifen (4-OHT, Sigma-Aldrich) for 6 h; cells were washed with PBS twice and the media was exchanged to fresh media without 4-OHT. t=0 was set at 12 h after first the infection or at the time of addition of media without 4-OHT. TRF2 wt, TRF2 mutants, TPP1 wt, and TPP1ΔPOT1 (deletion of aa 181–195) in pLPC-N-Myc were expressed by three infections at 12 h intervals. Retroviral shRNAs (Denchi and de Lange, 2007; Sfeir and de Lange, 2012) and Luciferase shRNA in pSuper as a control were introduced with five infections (three infections at 4 h intervals followed by the fourth and last infection at 12 h intervals). For TopBP1 knockdown, lentiviruses derived from pLKO.1 vector were introduced by two infections for 4 h each (Gong and de Lange, 2010). Lentiviral ATRIP shRNA and (TRCN000012418, Sigma-Aldrich) and Exo1 shRNAs (sh1: TRCN0000238466, sh4: TRCN0000218614, Sigma-Aldrich) were introduced by two infections for 6 h each. The retroviral infected MEF cells were selected in media with puromycin for 2–3 days. ATM was inhibited with 2.5 µM KU55933 for 108 h. DMSO was used as the negative control.
CRISPR/Cas9-mediated 53BP1 knockout
The guide sequence was determined by ZiFit (http://zifit.partners.org): sg53BP1-3, 5’-GCATCTGCAGATTAGGA-(PAM)-3’. Oligonucleotides were obtained from Sigma-Aldrich and introduced into an AflII-digested gRNA cloning vector (Addgene) by Gibson Assembly (New England Biolabs). The gRNA and hCas9 expression vectors were induced by electroporation using MEF 2 Nucleofector Kit (Lonza). Single cell clones isolated with limiting dilution were screened by immunoblotting. T7 endonuclease1 (T7E1) assay and DNA sequencing were performed to verify the gene modification. PCR product was amplified by nested PCR with following primers (fw for 1st PCR: GGGAGCAGATGGACC, rev for 1st PCR: GTACCCAAATGAGAAGACTCC, fw for 2nd PCR: GTCAATTGGATTCAGATTTCTCT, rev for 2nd PCR: CACAGAAGACATTTGCCATCA). For T7E1 assays, re-annealed PCR product was digested with T7 Endonuclease 1 (New England Biolabs) for 20 min at 37°C and then analyzed on 2.5% agarose/0.5× TAE. GenScript USA Inc carried out DNA sequencing of sub-cloned PCR product.
Immunoblotting
Immunoblotting was performed as described previously (Celli et al., 2006; Kibe et al., 2010). The following primary antibodies were used: TRF1 (1449, de Lange lab); TRF2 (1254 or 1255, de Lange lab); Rif1 (1240, de Lange lab); TPP1 (ab104297, Abcam); 53BP1 (NB100–305, Novus Biological); ATR (N-19, Santa Cruz); CtIP (H-300, Santa Cruz); Exo1 (A302–640A, Bethyl Laboratories, Inc.); BLM (ab2179, Abcam); TopBP1 (ab2402, Abcam); mTOR (#2972, Cell Signaling Technology); γ-tubulin (GTU-88, Sigma-Aldrich); FLAG (M2, Sigma-Aldrich). The chemiluminescent signals were detected using ECL Western Blotting Detection Reagents (GE healthcare) and BioMax MR film or XAR film (Kodak) according to the manufacturer’s protocol.
Telomere overhang assay
Telomere overhangs were analyzed as described (Celli et al., 2006). To verify whether single-stranded telomeric DNA detected in native gels were derived from overhangs at telomere ends, DNA in plugs were treated with E.coli Exonuclease I in vitro prior to the digestion with MboI. After five washes with TE (10 mM Tris-HCl pH 7.5, 1 mM EDTA), DNA in plugs were washed with water for 1 h, equilibrated with ExoI buffer (67 mM glycine-NaOH, pH 9.5, 6.7 mM MgCl2, 10 mM β-mercaptoethanol) twice for 3 h each, and incubated with 1,000 U E.coli ExoI (NEB) overnight at 37°C. Overhang signals were obtained by hybridizing a labeled C-strand oligo to native DNA in gel. Overhang signals were normalized to the total telomeric DNA signals in the same lane after re-hybridization of the telomeric oligo to DNA that was denatured in situ. Normalized overhang signals were compared between samples to determine changes in the telomere resection. For each condition at least three biological replicates (e.g. independent Cre medited deletion experiments) were analyzed.
IF-FISH
IF-FISH was performed as described previously (Dimitrova and de Lange, 2006). Images were captured with a Zeiss Axioplan II or Zeiss Axioimager microscope using a Hamamatsu C4742-95 camera and Improvision OpenLab or Volocity software.
Chromosome Orientation (CO)-FISH and ChIP (Chromatin IP) assay
Metaphase spreads were formed on glass slides in the Cytogenic Drying Chamber (Thermotron) (20°C, 50% humidity) and processed for CO-FISH as described (Celli et al., 2006). The ChIP assay was performed as previously described (Loayza and de Lange, 2003). Telomeric DNA bound with TRF2 was immunoprecipitated with anti-mTRF2 (1254) and Protein G magnetic beads (#9006, Cell Signaling).
FACS analysis
Cell cycle profile was analyzed as previously described (Takai et al., 2007). Briefly, cells were labeled for 2 h with 10 µM BrdU and fixed with cold 70% ethanol. BrdU incorporated DNA were denatured with 2N HCl and 0.5% TritonX-100 for 30 min at room temperature. After neutralizing with 0.1 M Na2B4O[H2O]10, pH 8.5, cells were incubated with FITC-conjugated anti-BrdU antibody (BD Biosciences) in PBS with 0.5% Tween 20 and 0.5% BSA for 30 min at room temperature. The cells were re-suspended with 5 µg/ml propidium iodide, 0.5% BSA, 0.2 mg/ml RNase A in PBS prior to analysis with an AccuriC6 (BD Biosciences). Data were analyzed by FlowJo software.
Statistics analysis
Data were shown as means ± standard deviations unless otherwise indicated. Graphs were obtained from GraphPad Prism6 or MS Excel and re-generated using Adobe Illustrator. Two-tailed Student’s t test was performed with GraphPad Prism6. *, p < 0.05 **, p < 0.01 ***, p < 0.001
Supplementary Material
Acknowledgments
We thank Devon White for invaluable assistance with generating MEFs. Francisca Lottersberger is thanked for generating the TRF2ΔiDDR allele and Roos Karssemeijer for generating the 53BP1 CRISPR sgRNA. Members of the de Lange lab are thanked for their comments on this work. This work was supported by grants from the NIH to TdL (CA181090 and GM049046).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Boersma V, Moatti N, Segura-Bayona S, Peuscher MH, van der Torre J, Wevers BA, Orthwein A, Durocher D, Jacobs JJ. MAD2L2 controls DNA repair at telomeres and DNA breaks by inhibiting 5’ end resection. Nature. 2015;521:537–540. doi: 10.1038/nature14216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti D, Villa M, Gobbini E, Cassani C, Tedeschi G, Longhese MP. Escape of Sgs1 from Rad9 inhibition reduces the requirement for Sae2 and functional MRX in DNA end resection. EMBO Rep. 2015;16:351–361. doi: 10.15252/embr.201439764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo S, Wu Y, Ferguson D, de Lange T. Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol. 2009;187:385–398. doi: 10.1083/jcb.200902039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- Celli GB, Lazzerini Denchi E, de Lange T. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Barral P, Vannier JB, Borel V, Steger M, Tomas-Loba A, Sartori AA, Adams IR, Batista FD, Boulton SJ. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49:858–871. doi: 10.1016/j.molcel.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerici M, Trovesi C, Galbiati A, Lucchini G, Longhese MP. Mec1/ATR regulates the generation of single-stranded DNA that attenuates Tel1/ATM signaling at DNA ends. EMBO J. 2014;33:198–216. doi: 10.1002/embj.201386041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, de Lange T. Protection of telomeres through independent control of ATM and ATR by TRF2 and POT1. Nature. 2007;448:1068–1071. doi: 10.1038/nature06065. [DOI] [PubMed] [Google Scholar]
- Di Virgilio M, Callen E, Yamane A, Zhang W, Jankovic M, Gitlin AD, Feldhahn N, Resch W, Oliveira TY, Chait BT, Nussenzweig A, Casellas R, Robbiani DF, Nussenzweig MC. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339:711–715. doi: 10.1126/science.1230624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, de Lange T. MDC1 accelerates nonhomologous end-joining of dysfunctional telomeres. Genes Dev. 2006;20:3238–3243. doi: 10.1101/gad.1496606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, de Lange T. The Role of Double-Strand Break Repair Pathways at Functional and Dysfunctional Telomeres. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doksani Y, Wu JY, de Lange T, Zhuang X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent T-loop formation. Cell. 2013;155:345–356. doi: 10.1016/j.cell.2013.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C, Orthwein A, Fradet-Turcotte A, Xing M, Young JT, Tkac J, Cook MA, Rosebrock AP, Munro M, Canny MD, Xu D, Durocher D. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49:872–883. doi: 10.1016/j.molcel.2013.01.001. [DOI] [PubMed] [Google Scholar]
- Feng L, Fong KW, Wang J, Wang W, Chen J. RIF1 Counteracts BRCA1-mediated End Resection during DNA Repair. J Biol Chem. 2013;288:11135–11143. doi: 10.1074/jbc.M113.457440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn RL, Centore RC, O’Sullivan RJ, Rai R, Tse A, Songyang Z, Chang S, Karlseder J, Zou L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature. 2011;471:532–536. doi: 10.1038/nature09772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frescas D, de Lange T. TRF2-Tethered TIN2 Can Mediate Telomere Protection by TPP1/POT1. Mol Cell Biol. 2014;34:1349–1362. doi: 10.1128/MCB.01052-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, de Lange T. A Shld1-controlled POT1a provides support for repression of ATR signaling at telomeres through RPA exclusion. Mol Cell. 2010;40:377–387. doi: 10.1016/j.molcel.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Sun L, Shen F, Chen Y, Hua Y, Liu Y, Zhang M, Hu Y, Wang Q, Xu W, Sun F, Ji J, Murray JM, Carr AM, Kong D. The intra-S phase checkpoint targets Dna2 to prevent stalled replication forks from reversing. Cell. 2012;149:1221–1232. doi: 10.1016/j.cell.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Kibe T, Osawa GA, Keegan CE, de Lange T. Telomere Protection by TPP1 Is Mediated by POT1a and POT1b. Mol Cell Biol. 2010;30:1059–1066. doi: 10.1128/MCB.01498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YC, Akhter S, Gu P, Ye J, Poulet A, Giraud-Panis MJ, Bailey SM, Gilson E, Legerski RJ, Chang S. SNMIB/Apollo protects leading-strand telomeres against NHEJ-mediated repair. EMBO J. 2010;29:2230–2241. doi: 10.1038/emboj.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;424:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- Lottersberger F, Bothmer A, Robbiani DF, Nussenzweig MC, de Lange T. Role of 53BP1 oligomerization in regulating double-strand break repair. Proc Natl Acad Sci U S A. 2013;110:2146–2151. doi: 10.1073/pnas.1222617110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martina M, Bonetti D, Villa M, Lucchini G, Longhese MP. Saccharomyces cerevisiae Rif1 cooperates with MRX-Sae2 in promoting DNA-end resection. EMBO Rep. 2014;15:695–704. doi: 10.1002/embr.201338338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Lopes M. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat Rev Mol Cell Biol. 2015;16:207–220. doi: 10.1038/nrm3935. [DOI] [PubMed] [Google Scholar]
- Ngo GH, Balakrishnan L, Dubarry M, Campbell JL, Lydall D. The 9-1-1 checkpoint clamp stimulates DNA resection by Dna2-Sgs1 and Exo1. Nucleic Acids Res. 2014;42:10516–10528. doi: 10.1093/nar/gku746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K, Bartocci C, Ouzounov I, Diedrich JK, Yates JR, Denchi EL. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013;494:502–505. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Panier S, Boulton SJ. Double-strand break repair: 53BP1 comes into focus. Nat Rev Mol Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol. 2013;14:661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, Gottesman ME, Gautier J. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol Cell. 2013;49:657–667. doi: 10.1016/j.molcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle. 2010;9:3157–3165. doi: 10.4161/cc.9.16.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012;336:593–597. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kabir S, van Overbeek M, Celli GB, de Lange T. Loss of Rap1 induces telomere recombination in the absence of NHEJ or a DNA damage signal. Science. 2010;327:1657–1661. doi: 10.1126/science.1185100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturzenegger A, Burdova K, Kanagaraj R, Levikova M, Pinto C, Cejka P, Janscak P. DNA2 cooperates with the WRN and BLM RecQ helicases to mediate long-range DNA end resection in human cells. J Biol Chem. 2014;289:27314–27326. doi: 10.1074/jbc.M114.578823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- Takai H, Wang RC, Takai KK, Yang H, de Lange T. Tel2 regulates the stability of PI3K-related protein kinases. Cell. 2007;131:1248–1259. doi: 10.1016/j.cell.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Takai KK, Kibe T, Donigian JR, Frescas D, de Lange T. Telomere protection by TPP1/POT1 requires tethering to TIN2. Mol Cell. 2011;44:647–659. doi: 10.1016/j.molcel.2011.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Takai H, de Lange T. Telomeric 3’ Overhangs Derive from Resection by Exo1 and Apollo and Fill-In by POT1b-Associated CST. Cell. 2012;150:39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, van Overbeek M, Rooney S, de Lange T. Apollo Contributes to G Overhang Maintenance and Protects Leading-End Telomeres. Mol Cell. 2010;39:1–12. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Chapman JR, Brandsma I, Yuan J, Mistrik M, Bouwman P, Bartkova J, Gogola E, Warmerdam D, Barazas M, Jaspers JE, Watanabe K, Pieterse M, Kersbergen A, Sol W, Celie PH, Schouten PC, van den Broek B, Salman A, Nieuwland M, de Rink I, de Ronde J, Jalink K, Boulton SJ, Chen J, van Gent DC, Bartek J, Jonkers J, Borst P, Rottenberg S. REV7 counteracts DNA double-strand break resection and affects PARP inhibition. Nature. 2015;521:541–544. doi: 10.1038/nature14328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Hayano M, Masai H. Replication timing regulation of eukaryotic replicons: Rif1 as a global regulator of replication timing. Trends Genet. 2013;29:449–460. doi: 10.1016/j.tig.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24:108–117. doi: 10.1016/j.tcb.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Lottersberger F, Buonomo SB, Sfeir A, de Lange T. 53BP1 regulates DSB repair using Rif1 to control 5’ end resection. Science. 2013;339:700–704. doi: 10.1126/science.1231573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.