T-cell acute lymphoblastic leukemia (T-ALL) is a malignant disorder resulting from the leukemic transformation of T-cell precursors and is one of the most common forms of cancer in children and is also found in adults.1–3 The most frequent genetic abnormality in T-ALL is the dysregulation of transcription factor genes, including aberrant expression of TAL1/SCL.1–3 TAL1 is ectopically expressed in 40–60% of T-ALL cases due to chromosomal translocation, intrachromosomal rearrangement (“SIL-TAL deletion”) or a somatic mutation in a non-coding intergenic element.1, 3–5 We previously reported that TAL1 forms a positive interconnected auto-regulatory loop with its regulatory partners GATA3, RUNX1 and MYB (“core regulatory circuits”).3, 6 To identify critical downstream targets that contribute to T-cell leukemogenesis, we have also performed a loss-of-function RNA interference screen in the TAL1-positive T-ALL cell lines in our previous study.6 Among the high-confidence TAL1 targets, only four genes, including the Tribbles homolog 2 (TRIB2), were selected by this screen. TRIB2 is a pseudo-kinase protein and has been implicated as an oncogene that contributes to acute myeloid leukemia (AML) by negatively regulating the C/EBPα tumor suppressor protein.7, 8 TRIB2 is also a transcriptional target of the oncogene NOTCH1 in T-ALL.9 However, the oncogenic role of TRIB2 in T-ALL pathogenesis, in particular, as a downstream consequence of the core regulatory circuits, has not been elucidated. In this study, we identified molecular pathways regulated by TRIB2 in T-ALL cells.
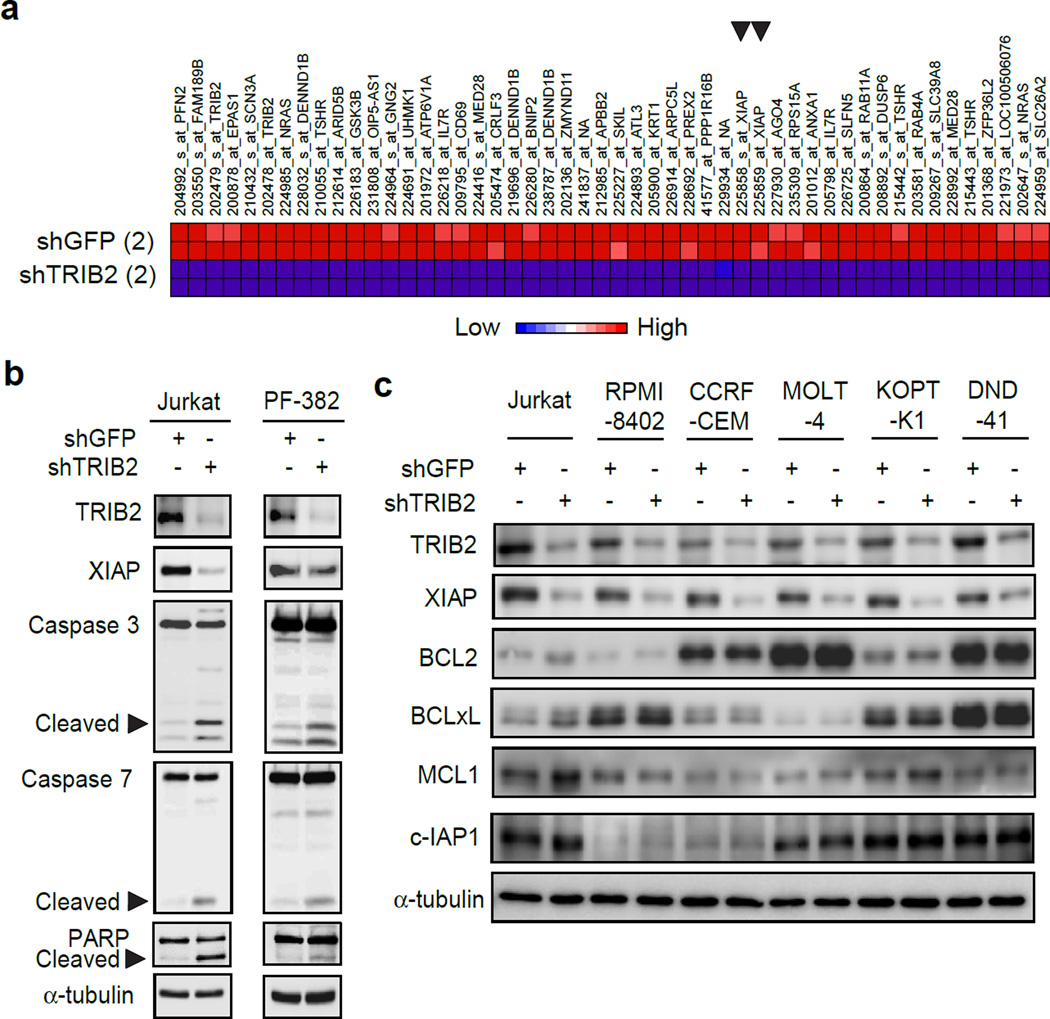
Western blot analysis showed that TRIB2 was expressed in all TAL1-positive T-ALL cell lines examined (Supplementary Figure 1a). We performed microarray gene expression profiling after TRIB2 knockdown in Jurkat cells that expresses high levels of TAL1 and TRIB2 proteins. This analysis revealed that 123 and 77 gene probes were significantly down- and upregulated, respectively, after TRIB2 knockdown (Supplementary Figures 1b and 1c; Supplementary Table 1). A striking observation was the markedly decreased expression of the XIAP gene, which was found among the top 50 downregulated genes (Figure 1a, indicated by arrowheads), suggesting that TRIB2 positively regulates this gene. In fact, a significant positive correlation in mRNA expression between these two genes (p<0.05 by Pearson's r) was observed in primary T-ALL patient samples reported by Winter et al10 (Supplementary Figure 1d). XIAP is a member of the inhibitor of apoptosis family of proteins (IAP) and blocks apoptosis.11 Western blot analysis showed that TRIB2 knockdown induced downregulation of the XIAP protein and cleavage of PARP and caspase-3 and -7 (markers of the intrinsic apoptosis pathway) in T-ALL cells (Figure 1b). The similar result was observed with an independent shRNA targeting TRIB2 (Supplementary Figure 1e). Other genes involved in the intrinsic pathway were not significantly altered after TRIB2 knockdown (Supplementary Table 2). Downregulation of the XIAP but not of other anti-apoptotic proteins, such as BCL2, BCLxL, MCL1 or c-IAP1, was observed in multiple T-ALL cell lines after TRIB2 knockdown (Figure 1c) where cell growth was significantly inhibited (Supplementary Figure 1f). Overexpression of XIAP or BCL2 was able to partially rescue the cells from apoptosis (Supplementary Figure 1g) and cell growth inhibition (Supplementary Figure 1h) after TRIB2 knockdown. These results suggest that T-ALL cells has been already “primed” to cell death so that the cells undergo apoptosis when XIAP was downregulated. In contrast, the T-ALL cells overexpressing TRIB2 were more resistant to treatment with cytotoxic drugs (etoposide and doxorubicin) compared to the control cells transduced with an empty vector (Supplementary Figure 1i). These results indicate that TRIB2 overexpression contributes to cell survival.
Figure 1. TRIB2 contributes to cell survival by regulating the XIAP anti-apoptotic protein in T-ALL cells.
(a) Global gene expression in Jurkat cells transduced with TRIB2 shRNA (shTRIB2) or control GFP shRNA (shGFP) were measured by microarray analysis. The experiment was performed with biological duplicates. The top 50 downregulated gene probes were indicated. Arrowheads represent the XIAP genes. (b) The TRIB2 shRNA and control GFP shRNA were transduced by lentivirus infection into T-ALL cell lines (Jurkat and PF-382). Whole cell extracts were subjected to immunoblot analysis with antibodies indicated. Cleaved forms of the caspases and PARP were shown by arrowheads. (c) The TRIB2 shRNA and control GFP shRNA were transduced by lentivirus infection into T-ALL cell lines (Jurkat, RPMI-8402, CCRF-CEM, MOLT-4, KOPT-K1 and DND-41). Whole cell extracts were subjected to immunoblot analysis with antibodies indicated.
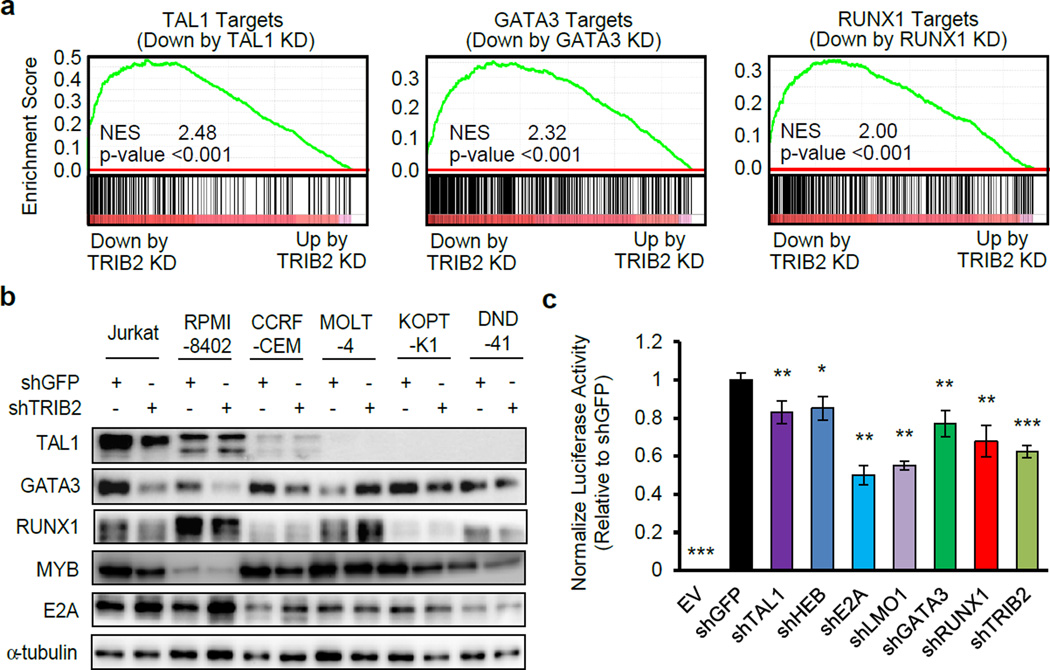
In addition to the apoptosis pathway, we found that genes regulated by TRIB2 included a number of genes directly regulated by the TAL1 complex that have been identified earlier by our ChIP-seq and microarray analyses6 (Supplementary Figure 2a). Gene set enrichment analysis (GSEA) showed that many of the genes downregulated after knockdown of TAL1, GATA3 or RUNX1 were also significantly downregulated after TRIB2 knockdown (Figure 2a). The result was validated by qRT-PCR analysis for representative TAL1 target genes (ALDH1A2 and LIG4; Supplementary Figure 2b). These results indicate that TRIB2 selectively regulates expression of the direct targets of the TAL1 complex.
Figure 2. TRIB2 coordinately regulates the expression of target genes of the TAL1 complex in T-ALL cells.
(a) The high-confident target genes of TAL1, GATA3 and RUNX1 determined previously6 were used as gene sets. A gene set enrichment analysis (GSEA) was performed to analyze the expression profile of these genes after TRIB2 knockdown (KD) in Jurkat cells. GSEA plots indicate the degree to which genes are overrepresented at the extreme left (downregulated by TRIB2 KD) or right (upregulated by TRIB2 KD) of the entire ranked list. Solid bars represent genes. Normalized enrichment score (NES) and P-values are indicated. (b) The TRIB2 shRNA and control GFP shRNA were transduced by lentivirus infection into T-ALL cell lines. Whole cell extracts were subjected to immunoblot analysis with antibodies indicated. (c) Jurkat cells stably expressing the ALDH1A2 enhancer reporter construct were established and transduced with the respective shRNA targeting a transcription factor by lentivirus infection. The luciferase activity and cell viability were measured after 3 days of virus infection. The luciferase activity was normalized by cell viability and is shown as fold changes compared to the control sample, which was transduced with shGFP. The values are the mean ± SD of biological triplicates; *p<0.05, **p<0.01, ***p<0.001 by two sample, two-tailed t-test.
We next examined the mechanism of regulation of the TAL1 targets by TRIB2. Analysis of mRNA expression revealed that four major components of the TAL1 complex, namely TAL1, GATA3, RUNX1 and MYB, were downregulated after TRIB2 knockdown in Jurkat cells (Supplementary Figure 2b). This trend was more significant at the protein level in the same cell line (Figure 2b, left), presumably due to a high protein turn-over of these factors. The similar result was observed with an independent shRNA targeting TRIB2 (Supplementary Figure 1e). We previously reported that these genes positively regulate each other through an auto-regulatory loop in this cell line.4, 6 Indeed, overexpression of TRIB2 upregulated expression of all of these proteins in the same cell line both at the mRNA (Supplementary Figure 2c) and protein levels (Supplementary Figure 2d). Notably, TAL1 was not downregulated after TRIB2 knockdown in two TAL1-positive cell lines (RPMI-8402 and CCRF-CEM; Figure 2b) where TAL1 is driven by the SIL gene promoter, which is activated independently of the TAL1 complex.12 Additionally, downregulation of GATA3, RUNX1 and MYB was not significant in the TAL1-negative cell lines (Figure 2b). Although additional study is needed to clarify the mechanism of regulation of the TAL1 complex by TRIB2, these results suggest that TRIB2 upregulates the expression of the TAL1 complex members by affecting the positive auto-regulatory loop. Importantly, the decrease in protein expression of these transcription factors was also observed after TRIB2 knockdown in the XIAP or BCL2-overexpressing cells (Supplementary Figures 2e and 2f) where apoptosis was blocked (Supplementary Figure 1g; and see PARP cleavage in Supplementary Figures 2e and 2f), demonstrating that the regulation of transcription factor proteins is not a result of cell death.
Interestingly, we found that expression of the E-protein E2A was upregulated by TRIB2 knockdown in Jurkat cells (Figure 2b) and conversely was downregulated by TRIB2 overexpression (Supplementary Figure 2d). The same trend was found in other TAL1-positive cell lines but not in the TAL1-negative cell lines (Figure 2b). E2A is a heterodimerization partner of TAL1 but can also form a homodimer or heterodimer with other E-proteins (e.g., HEB).13 E2A has been reported as a tumor suppressor in T-ALL.14 Our result indicates that E2A is negatively regulated by TRIB2 overexpression. The mRNA expression of E2A was not altered after knockdown or overexpression of TRIB2 (Supplementary Figures 2b and 2c), indicating that E2A is regulated at the protein level. To further investigate this mechanism, we have treated the cells with the MG-132 proteasome inhibitor. The result demonstrated that MG-132 treatment increased protein expression of E2A, indicating that this protein is degraded in a proteasome-dependent manner (Supplementary Figure 2g). MG-132 treatment was able to restore E2A expression close to the endogenous level after overexpression of TRIB2 protein without affecting the mRNA expression of E2A (Supplementary Figure 2g). Our results indicate that TRIB2 affects the stability of E2A protein.
Finally, we analyzed the functional contribution of TRIB2 to the transcriptional regulation of the TAL1 targets in T-ALL. We have mapped by ChIP-seq analysis the enhancer region for one of the high-confidence TAL1 targets, ALDH1A2, which is bound by TAL1 and its regulatory partners (HEB, E2A, LMO1, GATA3 and RUNX1) (Supplementary Figure 2h).6, 15 We cloned this sequence into a luciferase reporter construct (Supplementary Figure 2i). The introduction of the construct into a TAL1-positive T-ALL cell line (Jurkat) led to an increase in luciferase activity compared to a control transduced with an empty vector (Figure 2c; compare lanes 1 and 2), indicating that the enhancer is activated by transcription factors endogenously expressed in T-ALL cells. Indeed, knockdown of the TAL1 complex members (TAL1, HEB, E2A, LMO1, GATA3 or RUNX1) inhibited the luciferase activity (Figure 2c; compare lanes 2–8). Importantly, TRIB2 knockdown also significantly reduced the activity of this enhancer (Figure 2c; compare lanes 2 and 9). We have also confirmed that DNA bindings of TAL1, GATA3 and RUNX1 proteins at this locus and the LIG4 gene were significantly reduced after TRIB2 knockdown (Supplementary Figure 2j). These results indicate that TRIB2 coordinately and positively regulates expression of the TAL1 target genes.
Taken together, our study shows that TRIB2 contributes to cell survival by regulating the XIAP anti-apoptotic gene in T-ALL cells. It is worth noting that XIAP is not a direct transcriptional target of TAL1, and that downregulation of the XIAP protein could be observed in T-ALL cell lines regardless of the TAL1 expression (Figure 1c), indicating that TRIB2 may generally regulate XIAP expression independently of the TAL1 complex. According to the database by the Immunological Genome Project (http://www.immgen.org/), both Trib2 and Xiap genes are expressed in mouse T-cells, suggesting that Trib2 may regulate Xiap expression in normal T-cells. Detailed analysis with animal studies is needed to further elucidate the relationship between these two genes. Importantly, TRIB2 positively regulates the expression of TAL1, GATA3, RUNX1 and MYB and negatively regulates E2A. TRIB2 likely affects the stability of E2A protein. Notably, upregulation of E2A after TRIB2 knockdown was observed only in TAL1-positive cases. O’Neil and coworkers have previously reported that haploinsufficiency of E2a was able to accelerate tumor onset and the penetrance of T-ALL in a transgenic murine model overexpressing Tal1,14 demonstrating the tumor suppressive function of E2A in TAL1-positive T-ALL. Our result suggests that TRIB2 overexpression reinforces the oncogenic transcriptional program in TAL1-positive T-ALL cells by altering the balance between the oncogenic TAL1 complex and the E2A tumor suppressor (Supplementary Figure 2k).
Supplementary Material
Acknowledgments
We thank members of the Sanda and Look laboratories for discussions. We thank the Nature Publishing Group Language Editing for editing the manuscript. This research is supported by the U.S. National Cancer Institute (1K99CA157951, 5P01CA109901) and the National Research Foundation (NRF), Prime Minister’s Office, Singapore under its Competitive Research Programme (Award No. NRF-NRFF2013-02).
Footnotes
Supplementary information is available on the Leukemia web site. (http://www.nature.com/leu)
Conflicts of Interest:
The authors declare no conflicts of interest.
References
- 1.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nature reviews Immunology. 2008 May;8(5):380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 2.Look AT. Oncogenic transcription factors in the human acute leukemias. Science. 1997 Nov 7;278(5340):1059–1064. doi: 10.1126/science.278.5340.1059. [DOI] [PubMed] [Google Scholar]
- 3.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. The Journal of clinical investigation. 2012 Oct;122(10):3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014 Dec 12;346(6215):1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navarro JM, Touzart A, Pradel LC, Loosveld M, Koubi M, Fenouil R, et al. Site- and allele-specific polycomb dysregulation in T-cell leukaemia. Nature communications. 2015;6:6094. doi: 10.1038/ncomms7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanda T, Lawton LN, Barrasa MI, Fan ZP, Kohlhammer H, Gutierrez A, et al. Core transcriptional regulatory circuit controlled by the TAL1 complex in human T cell acute lymphoblastic leukemia. Cancer cell. 2012 Aug 14;22(2):209–221. doi: 10.1016/j.ccr.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keeshan K, He Y, Wouters BJ, Shestova O, Xu L, Sai H, et al. Tribbles homolog 2 inactivates C/EBPalpha and causes acute myelogenous leukemia. Cancer cell. 2006 Nov;10(5):401–411. doi: 10.1016/j.ccr.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dedhia PH, Keeshan K, Uljon S, Xu L, Vega ME, Shestova O, et al. Differential ability of Tribbles family members to promote degradation of C/EBPalpha and induce acute myelogenous leukemia. Blood. 2010 Aug 26;116(8):1321–1328. doi: 10.1182/blood-2009-07-229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wouters BJ, Jorda MA, Keeshan K, Louwers I, Erpelinck-Verschueren CA, Tielemans D, et al. Distinct gene expression profiles of acute myeloid/T-lymphoid leukemia with silenced CEBPA and mutations in NOTCH1. Blood. 2007 Nov 15;110(10):3706–3714. doi: 10.1182/blood-2007-02-073486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winter SS, Jiang Z, Khawaja HM, Griffin T, Devidas M, Asselin BL, et al. Identification of genomic classifiers that distinguish induction failure in T-lineage acute lymphoblastic leukemia: a report from the Children's Oncology Group. Blood. 2007 Sep 1;110(5):1429–1438. doi: 10.1182/blood-2006-12-059790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivasula SM, Ashwell JD. IAPs: what's in a name? Molecular cell. 2008 Apr 25;30(2):123–135. doi: 10.1016/j.molcel.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breit TM, Mol EJ, Wolvers-Tettero IL, Ludwig WD, van Wering ER, van Dongen JJ. Site-specific deletions involving the tal-1 and sil genes are restricted to cells of the T cell receptor alpha/beta lineage: T cell receptor delta gene deletion mechanism affects multiple genes. The Journal of experimental medicine. 1993 Apr 1;177(4):965–977. doi: 10.1084/jem.177.4.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kee BL. E and ID proteins branch out. Nature reviews Immunology. 2009 Mar;9(3):175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- 14.O'Neil J, Shank J, Cusson N, Murre C, Kelliher M. TAL1/SCL induces leukemia by inhibiting the transcriptional activity of E47/HEB. Cancer cell. 2004 Jun;5(6):587–596. doi: 10.1016/j.ccr.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 15.Ono Y, Fukuhara N, Yoshie O. TAL1 and LIM-only proteins synergistically induce retinaldehyde dehydrogenase 2 expression in T-cell acute lymphoblastic leukemia by acting as cofactors for GATA3. Molecular and cellular biology. 1998 Dec;18(12):6939–6950. doi: 10.1128/mcb.18.12.6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.