Abstract
Introduction
A common bottleneck during ontology evaluation is knowledge acquisition from domain experts for gold standard creation. This paper contributes a novel semi-automated method for evaluating the concept coverage and accuracy of biomedical ontologies by complementing expert knowledge with knowledge automatically extracted from clinical practice guidelines and electronic health records, which minimizes reliance on expensive domain expertise for gold standards generation.
Methods
We developed a bacterial clinical infectious diseases ontology (BCIDO) to assist clinical infectious disease treatment decision support. Using a semi-automated method we integrated diverse knowledge sources, including publically available infectious disease guidelines from international repositories, electronic health records, and expert-generated infectious disease case scenarios, to generate a compendium of infectious disease knowledge and use it to evaluate the accuracy and coverage of BCIDO.
Results
BCIDO has three classes (i.e., infectious disease, antibiotic, bacteria) containing 593 distinct concepts and 2345 distinct concept relationships. Our semi-automated method generated an ID knowledge compendium consisting of 637 concepts and 1554 concept relationships. Overall, BCIDO covered 79% (504/637) of the concepts and 89% (1378/1554) of the concept relationships in the ID compendium. BCIDO coverage of ID compendium concepts was 92% (121/131) for antibiotic, 80% (205/257) for infectious disease, and 72% (178/249) for bacteria. The low coverage of bacterial concepts in BCIDO was due to a difference in concept granularity between BCIDO and infectious disease guidelines. Guidelines and expert generated scenarios were the richest source of ID concpets and relationships while patient records provided relatively fewer concepts and relationships.
Conclusions
Our semi-automated method was cost-effective for generating a useful knowledge compendium with minimal reliance on domain experts. This method can be useful for continued development and evaluation of biomedical ontologies for better accuracy and coverage.
Keywords: Infectious disease, bacteria, antibiotic, ontology, evaluation, knowledge acquisition
Graphical abstract
1. INTRODUCTION
Ontologies enable formal representation and sharing of domain knowledge [1] and can augment clinical decision support systems by providing a standard vocabulary for biomedical entities to help standardize and integrate heterogeneous data resources [2–4]. Ontologies are now pervasive in biomedicine and function to address multiple requirements including knowledge management, data integration, exchange and semantic interoperability, and decision support and reasoning [2]. However, ontology evaluation remains difficult [5]. Common methods for the evaluation of biomedical ontologies include conformance to a philosophical principle [6], application or task-based evaluation [7], user-based evaluation [8], data-driven evaluation [9] and gold standard-based evaluation [10]. Evaluation of a large clinical knowledge base often centers on example applications and involves comparing ontologies against pre-defined gold standards [11]. This can be problematic for domain-specific ontologies since there may be no available gold standard for comparison [11]. The development of a new gold standard requires extensive domain expertise through a process that can have poor cost-effectiveness and cause long time delays.
Evaluation of the accuracy and comprehensiveness of a large domain-specific ontology typically relies on domain experts to manually develop a gold standard reference. This method faces several challenges. First, knowledge about a domain is constantly evolving, but knowledge acquisition directly from domain experts cannot happen as frequently as needed and often lags behind knowledge generation in any domain. Static gold standards can soon become outdated. Modern ontology design and evaluation requires an iterative and dynamic process so that newly emerging knowledge can be incorporated in frequent evaluations. Second, domain experts may not possess comprehensive knowledge about a domain all the time; therefore, relying on the single source of expert knowledge can lead to bias or limitations in the resulting gold standard.
In this paper, we presented a new semi-automated method for combining multiple knowledge sources to evaluate biomedical ontologies, which minimize the involvement of domain experts and augment them with knowledge automatically acquired from public electronic data sources, and applied it to evaluate a bacteria clinical infectious disease ontology. We explain how we utilized automated extraction of concepts and properties in conjunction with manual methods to integrate multiple diverse knowledge sources into a comprehensive compendium of infectious disease (ID) knowledge, and then compared BCIDO to this knowledge compendium. This method is superior to existing static methods for ontology development in that it can run anytime and multiple times so that emerging new domain knowledge can be incorporated in gold standard generation as often as preferred. On this basis, we discuss how this method can be used for the evaluation of other biomedical ontologies. Another contribution of this work is a validated bacteria clinical infectious disease ontology that provides comprehensive concept and concept relationships that are useful for portable decision support for antimicrobial prescription.
2. MATERIALS AND METHODS
2.1. Combining Expert Knowledge and Electronic Data for BCIDO Ontology Development
Antimicrobial resistance is an increasing problem worldwide and is often caused by inappropriate antimicrobial prescribing. Antibiotic resistance is now a major threat to public health and has the potential to affect anyone, of any age, in any country [12]. Incorporating an antibiotic decision support system (ADSS) into clinical decision-making has been shown to be effective at reducing inappropriate antibiotic prescribing and lowering local antimicrobial resistance [13–15]. However, despite their apparent benefits, ADSSs are infrequently used in the hospital in-patient setting [16]. The barriers to widespread adoption and implementation of successful ADSSs include standalone systems that are independent from the electronic health record (EHR) and require interruption of the clinical workflow to use [14, 17], a single infectious disease focus (i.e. acute bronchitis)[18, 19] or single clinical location (i.e. intensive care or primary care)[14, 20–24], and ADSSs that use their own terminology and cannot be transferred to other EHR systems [15, 18, 25, 26]. To improve the interoperability of future portable ADSSs, we developed and published a bacterial clinical infectious diseases ontology (BCIDO)[16].
BCIDO defines common concept definitions for clinical infectious diseases along with domain knowledge commonly used in the hospital in-patient setting for the diagnosis of these diseases. BCIDO encompasses concept definitions for common clinical presentations of infections, patient-specific factors that influence differential diagnoses and treatment options, the organisms themselves, and the antimicrobial agents used to treat infections. The design of BCIDO has been described previously [16]. In brief, the ontology covers factors relevant to making an antimicrobial decision in the hospital setting, including patient factors and microbiology results, such as gram stain and culture results. Specific antimicrobial treatment recommendations are not defined in BCIDO because they vary widely among clinicians, institutions and countries and are therefore not “universal truths”. However, the factors required for making an antimicrobial treatment decision are included so that treatment decisions in an ADSS can be tailored to local preferences. BCIDO is limited to bacterial infections. However, it has been designed to be easily extended to include antimicrobial treatments for mycobacterial, viral, and fungal infections. The concept granularity of the ontology is often chosen to ensure a diagnosis or treatment recommendation can be made at this granularity level.
When designing BCIDO, the Infectious Disease Ontology (IDO)[28] (http://infectiousdiseaseontology.org/) was selected as the upper ontology. IDO is a suite of interoperable ontology modules that together aim to cover the entire infectious disease domain. The suite consists of the core IDO, covering terms and relations generally relevant to the infectious disease domain, and a set of domain-specific ontologies developed as extensions from the core [29]. To date, disease and pathogen specific extension ontologies have been developed for malaria [30], dengue fever [31], brucellosis[32], and Staphylococcus aureus [33, 34]. The primary purpose of the core IDO is to maximize interoperability between IDO extensions as well as with ontologies outside the IDO suite. To accomplish this, IDO is developed within the framework of the OBO Foundry [29] (http://obofoundry.org/) and adheres to the Foundry’s ontology development guidelines. BCIDO was developed using the core IDO as an upper ontology, and thus the Basic Formal Ontology and Ontology of General Medical Science, which serve as upper ontologies for the IDO suite. BCIDO adheres to the Foundry’s ontology development guidelines and to Cimino’s Desiderata for terminologies [1]. Together these include: (1) using Aristotelian definitions with a single mode of classification, (2) using single inheritance hierarchies, (3) using relations with formal, logical definitions based on a distinction between types and instances, and (4) writing definitions and ontology assertions as compositions of ontology terms and relations.
To help standardize and integrate data resources, clinical infectious disease concepts and antibiotics in BCIDO were mapped to the reference resource, the Unified Medical Language System (UMLS) [35] concept unique identifiers (CUIs), where possible. UMLS integrates many terminologies and coding standards. Mapping BCIDO to UMLS CUIs enables BCIDO to be linked to many other relevant biomedical resources such as SNOMED-CT and ICD version 9 or 10[36]. Bacterial terms were imported from the National Center for Biotechnology Information Organismal Classification (NCBITaxon). Anatomical terms were imported from The Foundational Model of Anatomy (http://sig.biostr.washington.edu/projects/fm/index.html) (FMA) [11] and were used to define the location of infectious processes (i.e. osteomyelitis located_in some bone).
The ontology was represented in the OWL 2 EL Web Ontology Language (OWL) as a single hierarchical structure using the Protégé-OWL editor (http://protege.stanford.edu). The entire IDO core ontology was imported as the upper ontology (http://purl.obolibrary.org/obo/ido.owl). The Basic Formal Ontology was used to assist in designing the structure of our ontology and defining additional ontology classes and properties. Clinical infectious disease concepts and antibiotics were mapped to UMLS using the “identifier” annotation property, and synonyms or related terms were recorded using the “has_related_term”, “has_exact_synonom” or “has_broad_synonom” annotation properties, as defined by the Dublin core. Bacterial terms were imported from the NCBITaxon and anatomical terms were imported from the FMA using the minimal information to reference an external ontology term (MERIOT) principle using the web-based OntoFox application [37]. Domain knowledge was obtained from the first author’s experience as an ID physician and supplemented by common clinical ID textbooks and guidelines [38, 39]. In addition antibiotic and bacteria concepts were manually extracted from RxNorm and LOINC and included in BCIDO.
BCIDO focuses on three areas “infectious disease”, “antibiotic” and “bacteria” and includes 599 distinct concepts and 2355 class attributes. Figure 1 shows an example from the infectious disease area demonstrating classes and class attribute definitions. The “infectious disease” hierarchy contains 255 classes, of which 90% nearly all were mapped to UMLS CUI. The “antibiotic” hierarchy contains 98 classes, of which 90% were mapped to UMLS CUI on March 10th 2013. The “bacteria” hierarchy contains 255 classes, of which all were imported from NCBITaxon on May 16th, 2013. These high-level BCIDO classes are shown in Table 1 along with descriptions, their BFO class type, the corresponding IDO term or parent type in IDO core, and the source ontology for imported terms, as previously reported [16].
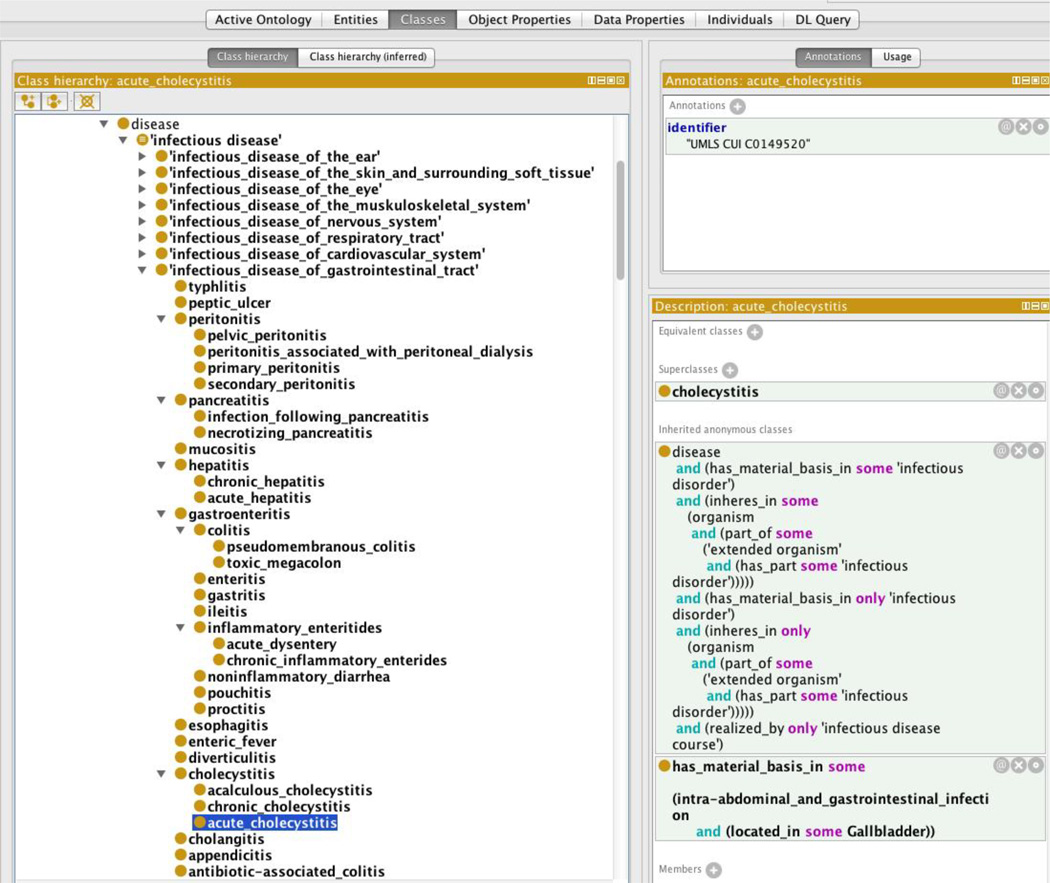
Figure 1.
Domain class of infectious disease showing “acute cholecystitis” as an example
Table 1.
Ontology classes, descriptions, BFO and IDO class types and source ontology
| Class | Description | BFO class type | IDO class type |
Source ontology |
|---|---|---|---|---|
| Bacteria | Bacteria are a large group of single-celled prokaryotic organisms, which may have a variety of shapes ranging from spherical, rod- like, comma-shaped to spiral. | Independent Continuant: object | Bacteria | Bacteria is a term in the IDO core imported from NCBITaxon. |
| Infectious disease | A disease whose physical basis is an infectious disorder. | Dependent Continuant: realizable entity | Disease | “Infectious disease” is a term in the IDO core and is a subtype of “disease” which is imported to IDO core from Ontology for General Medical Science. |
| Antibiotic | A chemotherapeutic agent or substance that kills (microbicidal) or inhibits (microbiostatic) the growth of bacteria and treats bacterial infections. | Independent Continuant: object | Antibiotic | “Antibiotic” is a term in IDO core which is linked to the term “antibiotic” in ChEBI. |
| Bacterial quality | The properties of bacteria that allow bacteria to be classified according to phenotypic or morphologic features. These properties assist with narrowing the differential diagnosis before definitive culture results are available. | Dependent Continuant: quality | Quality | “Bacterial quality” is a subtype of “quality” imported to IDO core from BFO. |
The object properties defined in BCIDO are shown in Table 2, as previously reported[16]. There were 571 object properties between the “bacteria” and “infectious disease” hierarchy; 522 object properties between the “antibiotics” and “bacteria” hierarchies; and 48 object properties between the “bacteria” and “bacterial quality” hierarchies. The object property causes asserts the relation between a “bacteria” and an “infectious disease” and is defined by the existence of a known causative link between the bacteria and the infectious disease. For example, “Neisseria meningitidis causes some meningitis”. The object property is_antimicrobial_coverage_for asserts the relation between an “antibiotic” and a “bacteria” and is asserted when at least some strains of the bacterial type are susceptible to the antimicrobial. For example, “penicillin is_antimicrobial_coverage_for some Treponema pallidum”. The object property has_shape asserts the relation between a “bacteria” and a “bacterial shape” and is defined by the typical shape of the bacteria. For example, “Staphylococcus has_shape spherical”.
Table 2.
Ontology object properties
| Domain class | Object property | Range class |
|---|---|---|
| Bacteria | Causes | Infectious disease |
| Antibiotic | Is_antibiotic_coverage_for | Bacteria |
| Bacteria | Has_characteristic_stain result | Bacterial quality |
| Bacteria | Has_shape | Bacterial quality |
| Infectious disease | Can_be_associated_with | Infectious disease |
| Infectious disease | Is_located_in | Anatomical entity |
2.2. Combining Expert Knowledge and Electronic Data for Ontology Evaluation
The metrics for evaluation of a clinically based knowledge resource include adherence to standard ontology development practices, internal consistency, accuracy and comprehensiveness of knowledge, generalizability and usefulness [11]. We have, thus far, focused our evaluation of BCIDO on the first four evaluation metrics and anticipate that the evaluation of usability and generalizability will occur as BCIDO is used in real-world clinical settings later. The evaluation of the accuracy and comprehensiveness of an earlier version of BCIDO has been reported previously [16]. Two domain experts evaluated the accuracy of the earlier version of BCIDO using the laddering technique [40] and visual review. The domain experts recommended the addition of 16 concepts and 110 relationships between concepts.
To evaluate the comprehensiveness of BCIDO, ten clinical case notes from patients with infectious diseases were reviewed and BCIDO’s coverage of antibiotic, clinical infectious disease and bacterial concepts in the case notes was determined. Although the coverage of antibiotic and bacteria concepts was excellent (100% and 94%, respectively), the coverage of infections disease concepts was lower (78%). Taken together, our early evaluation highlighted the need for a larger scale evaluation of the accuracy and comprehensiveness of the knowledge contained in BCIDO. The results of our early evaluation provided the rationale for our approach described here.
Evaluation of the accuracy and comprehensiveness of a large knowledge resource typically relies on domain experts to manually develop a gold standard reference. However, the time required for the domain experts to evaluate BCIDO even on a very small scale was not trivial, about 6 hours per domain expert. It was anticipated that the exclusive use of domain expertise to manually develop a new gold standard to evaluate BCIDO against was neither cost-effective nor feasible. To overcome this challenge, we used a novel semi-automated method to integrate multiple diverse knowledge sources into a comprehensive compendium of ID knowledge (Figure 2), which served as a gold-standard reference to compare BCIDO against. In our evaluation, firstly we describe our evaluation of the internal consistency and adherence to standard ontology practices of BCIDO. Secondly, we describe the use of a semi-automated method to create a compendium of ID knowledge from three sources of ID knowledge. Finally, we evaluate the accuracy and comprehensiveness of BCIDO using the compendium of ID knowledge as the gold standard reference.
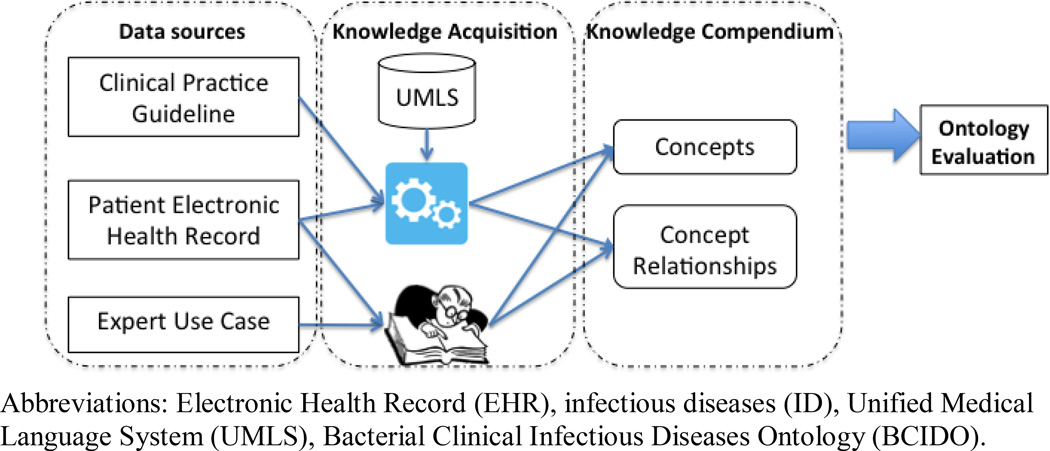
Figure 2.
Infectious disease knowledge acquisition and integration from multiple knowledge sources for comparison to the Bacterial Clinical Infectious Disease Ontology
2.2.1 Internal consistency and adherence to standard ontology practices
Adherence to standard ontology practices was evaluated by checking for adherence to the OBO Foundry’s ontology development guidelines and to Cimino’s Desiderata listed above (see 2.1)[1]. Four ID experts (two Physicians and two Fellows) were recruited via email to participate in the evaluation of an ID knowledge resource and were compensated with a $50 gift card. All ID experts were practicing physicians in the U.S., and one ID expert had also previously practiced in Australia. The four ID experts reviewed randomly selected sections of BCIDO to evaluate internal consistency. The same four ID experts generated the ID case scenarios as described below (see 2.2.2.3).
2.2.2 Knowledge accuracy and comprehensiveness
To complement domain experts in the large-scale evaluation of knowledge accuracy and comprehensiveness of BCDIO, we used a semi-automated method to integrate multiple diverse knowledge sources into a comprehensive ID compendium. This approach increased the volume and velocity of knowledge available for serving as the gold standard for evaluations and reduced the cost of manual labeling by experts. Moreover, the automated method can be called as often as preferred for knowledge generation. Knowledge sources with unique and complementary content relevant to the goal of BCIDO for supporting ADSSs were selected and included to create the ID knowledge compendium. The diversity in the knowledge sources improved the depth and breadth of the ID knowledge compendium.
2.2.2.1 Knowledge source #1: Infectious disease guidelines
Published ID guidelines contain knowledge that is frequently consulted by healthcare practitioners to guide diagnostic and management decisions. Publically available ID guidelines were downloaded on October 1st 2014 from three major ID societies and one national guideline repository from around the world; the Infectious Disease Society of America (IDSA; http://www.idsociety.org/idsa_practice_guidelines/), the European Society of Clinical Microbiology and Infectious Diseases (ESCMID; https://www.escmid.org/escmid_library/medical_guidelines/escmid_guidelines/), the Australasian Society for Infectious Diseases (ASID; http://www.asid.net.au/resources/clinical-guidelines) and National Institute for Health and Care Excellence, United Kingdom (NICE; https://www.nice.org.uk/guidance). Those guidelines that were exclusively related to non-bacterial infectious diseases such as viral, parasitic, mycobacterial or fungal infections were excluded because BCIDO focuses solely on bacterial infection decision support.
2.2.2.2 Knowledge source #2: Patient electronic medical records
Patient medical records contain the knowledge required to practice medicine in a real life setting and contain the most common ID conditions. In addition, electronic health records (EHRs) contain the knowledge required to be in an ID knowledge representation that will support an EHR-based ADSS. To meet this knowledge requirement, we used the publically available Multi-parameter Intelligent Monitoring in Intensive Care II (MIMIC II) database[41] (http://physionet.org/mimic2). MIMIC II is a diverse EHR database of 32,535 critically ill patients admitted to Beth Israel Deaconess Medical Center intensive care units between 2001 and 2007. The MIMIC II database contains patients admitted to medical units who typically have a high incidence of infections, as well as patients admitted to neonatal, post-surgical and cardiothoracic intensive care units who have a lower incidence and a more limited range of infections. Twenty patient records from non-medical units and 20 patient records from medical unites were randomly selected for inclusion in the ID knowledge compendium. Random numbers corresponding to the ICU admission identifier number were generated using the web application random.org (https://www.random.org/).
2.2.2.3 Knowledge source #3: Infectious diseases expert generated case scenarios
The four ID experts who evaluated the internal consistency of BCIDO (see 2.2.1) also generated scenarios of common infectious disease conditions seen in the hospital. The purpose of using ID expert generated case scenarios was to identify the most frequent and/or important, infectious disease conditions. ID experts complemented the previous knowledge sources by providing a perspective of relevance and importance that may be missed if guidelines and medical records are solely used as knowledge sources. The ID experts were instructed to independently develop ten bacterial infectious disease scenarios that are commonly seen in the hospital setting. The components of the scenarios included the infectious disease condition, the bacteria that cause the infectious disease condition and the antibiotics commonly used to treat the infectious disease condition. An example of an ID case scenario was given to the ID experts to provide guidance on the required depth of scenarios and the required format (Table 3). To cover the full diversity of antibiotic options, BCIDO aims to include all possible antibiotic choices. To ensure that a wide variety of antibiotic options were provided by the ID experts, experts were instructed to disregard antibiotic stewardship concerns and other nuances of antibiotic prescribing. As an example of an antibiotic stewardship concern; meropenem is an option for treating a urinary tract infection but would not be recommended if a narrower-spectrum antibiotic were just as effective. As an example of a nuance of antibiotic prescribing; gentamicin is an option for treating a urinary tract infection but is contra-indicated in a patient with renal failure. ID experts generated their own scenarios without further supervision and submitted their scenarios via email. When more than one expert developed the same ID case scenario, antibiotic options and causative bacteria from both experts were combined.
Table 3.
The example of an infectious disease (ID) case scenario provided to ID experts
| ID scenario | Bacteria that cause ID scenario |
Antibiotics used to treat ID scenarior |
|---|---|---|
| Hospital-acquired pneumonia | Staphylococcus aureu s | Vancomycin |
| Enterobacter species | Ceftriaxone | |
| Klebsiella species | Piperacillin-tazobactam | |
| Escherichia coli | Ticlarcillin-clavulanate | |
| Serratia species | Cefepime | |
| Proteus species | Ceftazidime | |
| Citrobacter species | Meropenem | |
| Pseudomonas aeruginosa | Gentamicin | |
| Acinetobacter baumannii | Tobramycin | |
| Polymixin B | ||
| Colistin |
2.2.3 Procedure for integrating knowledge and comparison to BCIDO
BCIDO encompasses terms and knowledge about clinical presentations of infection, the causative bacteria of infection and the antibiotics used for treatment, and has three major class hierarchies (infectious disease conditions, bacteria, antibiotics) are linked by two relationships (“Antibiotic is_antimicrobial_coverage_for Bacteria”, “Bacteria causes Infectious disease relationships”,) and a third inferred relationship (“Infectious disease is_treated_with Antibiotic”) [16]. Therefore, the evaluation of the accuracy and comprehensiveness of BCIDO needed to occur in two components: evaluation of the definitions of the concepts themselves as well as the relationships between concepts. The ID expert generated scenarios were formatted such that these two components were readily identified. The procedure for integrating concepts and relationships between concepts for the other two knowledge sources, guidelines and patient records, is described below.
2.2.3.1 Concepts
Concepts and their UMLS semantic types were extracted automatically from guidelines and text from patient records using a UMLS concept extraction algorithm as previously described [42]. In summary: first, each line of text is automatically annotated with a part-of-speech tagger to identify the grammatical role of each word. In this application, the grammatical role of a word was used only for noise reduction. The text is then processed to remove special characters and punctuation and to build all the possible n-grams (i.e., continuous subsequences of n words). N-grams composed of only English stop words or irrelevant grammatical structures are removed. Each n-gram is matched against the UMLS Metathesaurus and retained only if at least one substring of it is a recognizable UMLS concept. Each n-gram term found in the UMLS lexicon is also normalized according to its preferred CUI in order to reduce the sparseness of the concepts. Using the CUIs also enables the handling of synonyms, since similar concepts are aligned to the same preferred term because of the UMLS specification (e.g., “atrial fibrillation” and “auricular fibrillation” are both mapped to “atrial fibrillation”).
The process for integrating a list of relevant ID concepts occurred in several stages. Firstly, each resource or repository from the knowledge source was combined into a single text document (i.e. all IDSA guidelines were contained in a single text document) and UMLS concepts were automatically extracted as described above. An excel file of extracted UMLS concepts was generated for each knowledge resource or repository (see Appendix A). A subset of 5 guidelines and 15 patient records were manually reviewed to determine the accuracy of the UMLS concept extraction process. While the UMLS extraction process was very accurate for the guidelines (two additional infectious disease concepts and no additional antibiotic or bacteria concepts were identified), many concepts were missed in the patient records. In the patient records, 16 concepts were not identified by the UMLS extraction process; 19% (7/32) of infectious disease concepts, 20% (5/25) of antibiotic concepts and 18% (4/22) of bacterial concepts were missed. Subsequently, all patient records were manually reviewed after UMLS concept extraction to detect concepts that were missed by the UMLS extraction process. Non-medical ICU patient records were reviewed manually due to the paucity of concepts contained in the text.
Secondly, relevant concepts in the UMLS extract were identified using two methods. In the first method, concepts labeled with the semantic types “antibiotic”, “bacterium” or “disease or syndrome” were selected. Concepts were subsequently manually excluded if they were not relevant to an infection (i.e. myocardial infarction), or more specifically, not relevant to a bacterial infection (i.e. herpes encephalitis was excluded because it is exclusively caused by a virus). To ensure that no ID-related concepts were missed by this approach, concepts that were not labeled with the semantic types “antibiotic”, “bacterium” or “disease or syndrome” were manually reviewed in the UMLS extract from the IDSA guidelines. Two thousand and eight hundred UMLS concepts extracted from IDSA guidelines that were not labeled with the semantic types “antibiotic”, “bacterium” or “disease or syndrome” were reviewed and revealed 32 missing terms (1.2% of terms reviewed). Eleven of 32 missing concepts were assigned an incorrect semantic type (i.e. lyme disease classed as a pharmacologic substance) and the rest were assigned a less specific semantic type (i.e. doripenem was labeled the semantic type “pharmacologic substance” but should been labeled “antibiotic”). This assessment led to the second method, in which terms that were not labeled with the semantic types “antibiotic”, “bacterium” or “disease or syndrome” were manually reviewed to detect ID-related concepts that may not have been identified by the semantic types.
Thirdly, synonyms or related terms were identified and documented for comparison to BCIDO annotation properties and then removed. Examples include “skin infection” is a related term for “cutaneous infection”, and “nosocomial pneumonia” is a synonym for “health-care associated pneumonia”.
Fourthly, commercial names for antibiotics were converted to the generic form (i.e. “Zosyn” was converted to “piperacillin-tazobactam”) and commonly used forms of the antibiotic were used (i.e. “cefuroxime” was used instead of “cefuroxime axetil”).
Fifthly, bacterial concepts were processed into the formal taxonomical form (i.e. “Bacillus species” became “Bacillus”).
Sixthly, some concepts were a combination of causative bacteria and infectious disease (i.e. “meningococcal meningitis”). Concepts that contained both causative bacteria and infectious disease were manually identified and split into two distinct concepts (i.e. Bacteria, Infectious disease) and the connecting relationship added (i.e. “Bacteria causes Infectious disease”). For example, the concept “pneumococcal pneumonia” became the two concepts “Streptococcus pneumoniae” and “pneumonia” and the relationship “Streptococcus pneumoniae causes pneumonia”. This process resulted in a list of distinct concepts related to bacterial infections that were divided into the following classes: infectious disease, bacteria and antibiotic (see Appendix B).
2.2.3.2 Relationships among concepts
Separate methods were used to identify the relationships among concepts in guidelines and in patient records. For guidelines, the UMLS extraction process was used to identify the location in the text of the relationships among concepts. In addition to an excel sheet of all extracted UMLS concepts, the UMLS extraction process generated a separate text document identifying the location in the text of where a concept was detected (see Appendix C). The ID-related concepts identified above (and related terms or synonyms) were identified in the text document and any relationship between concepts in the text was manually recorded. For the patient records, the relationships between concepts were manually extracted at the same time that the record was reviewed for concepts missed by the UMLS extraction process. Concepts identified by the UMLS extraction process were also searched for within the patient record at the time of manual review.
2.2.3.3 Integrating concepts and relationships among concepts across knowledge sources into an ID knowledge compendium
Concepts and relationships between concepts identified in each knowledge source were combined into an ID knowledge compendium to compare BCIDO against. The ID knowledge compendium was created by integrating the concepts and concept relationships from all three knowledge sources into a single excel file (see Appendix D). Duplicate concepts and relationships between concepts were removed. The class concepts and relationships between class concepts in the ID knowledge compendium are the same as those in BCIDO and are shown in Table 4. To compare the effectiveness of each knowledge source for contributing concepts and relationships to the ID compendium, an effectiveness index was calculated by dividing the number of concepts or relationships contributed, by the number of resources used from a particular knowledge source. For example, the effectiveness index of 15 antibiotic concepts extracted from 20 patient records is 0.75 (15/20).
Table 4.
Number of Bacterial Clinical Infectious Disease Concepts and Relationships Between Concepts Identified From Multiple Knowledge Sources
| Class Concepts | Relationship between Class Concepts | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Knowledge Source | No. UMLS concepts extracted |
ID | Antibiotic | Bacteria | Total | Bacteria causesID |
Antibioticis_ antimicrobial_coverag e_forBacteria |
ID is_treated_with Antibiotic |
Total |
| Patient records | |||||||||
| MIMIC II | |||||||||
| Non- medical | - | 18 | 12 | 8 | 38 | 21 | 7 | 29 | 57 |
| Medical | 3067 | 29 | 23 | 20 | 72 | 28 | 10 | 58 | 96 |
| Guidelines | |||||||||
| IDSA | 1148 | 205 | 120 | 213 | 555 | 485 | 298 | 436 | 1219 |
| ESCMID | 5409 | 113 | 55 | 54 | 225 | 156 | 125 | 85 | 366 |
| NICE | 2349 | 34 | 12 | 18 | 74 | 26 | 9 | 19 | 54 |
| ASID | 4403 | 74 | 56 | 27 | 161 | 52 | 43 | 33 | 128 |
| Expert ID scenarios | - | 30 | 52 | 104 | 206 | 256 | 222 | 267 | 745 |
Abbreviations: Unified Medical Language System (UMLS), Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC II), Infectious Disease Society of America (IDSA), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), the Australasian Society for Infectious Diseases (ASID), National Institute for Health and Care Excellence (NICE), infectious disease (ID)
2.2.4 Comparison to BCIDO and error analysis
The concepts and relationships between concepts in the ID knowledge compendium were manually compared to BCIDO to identify semantically related terms and synonyms, especially in the infectious disease class, and add them to BCIDO under the annotation properties “has_related_term”, “has_exact_synonom” or “has_broad_synonom” as appropriate. Errors were corrected in BCIDO and included spelling errors and use of outdated terms for concepts. Missing knowledge in BCIDO was corrected in an iterative manner. For example, a missing concept was entered into BCIDO and relationships immediately added before moving onto the next concept in the ID knowledge compendium.
3. RESULTS
3.1. Internal consistency and adherence to standard ontology practices
BCIDO adheres to all four features of standard ontology practice listed above (see 2.1). In addition, BCIDO adheres to six of the eight relevant Desiderata characteristics[1]: concept orientation, formal definitions, multiple granularities, reject “not elsewhere classified”, recognize redundancy and context representation. Two characteristics (non-semantic concept identifiers, multiple consistent views) were not satisfied. However, many of the concepts were mapped to UMLS CUIs and therefore fulfill the requirement for non-semantic concept identifiers. Four ID experts evaluated random sections of BCIDO for internal consistency and no errors in the bacterial, antibiotic and infectious disease class heirarcheis were found. In addition, 92% (289/313) of “Antibiotic is_antimicrobial_coverage_for Bacteria” relationships and 100% (60/60) of “Bacteria causes Infectious disease” relationships were correct. After reviewing BCIDO, the ID experts recommended 11 new “Bacteria causes Infectious disease” and 7 new “Antibiotic is_antimicrobial_coverage_for Bacteria” relationships be added to BCIDO.
3.2. ID knowledge compendium
A summary of number of overall UMLS concepts and ID concepts, and relationships between ID concepts identified from each knowledge source is shown in Table 4. In addition, 236 related terms and synonyms were identified and added to BCIDO under the annotation properties “has_related_term”, “has_exact_synonom” or “has_broad_synonom” (111 infectious disease terms, 86 bacteria terms and 39 antibiotic terms).
3.2.1 Infectious disease guidelines
Thirty-seven ISDA guidelines were obtained and 11,148 UMLS concepts extracted (Table 4). Nine ESCMID guidelines were obtained and 5409 UMLS concepts extracted. Eight ASID guidelines were obtained and 4403 UMLS concepts extracted. Thirteen NICE guidelines were obtained and 2349 UMLS concepts extracted. Manual extraction of relevant concepts from all 54 guidelines would have taken an expert approximately 18 hours based on the assumption that it would take 20 minutes to evaluate a single guideline. In contrast, automated concept extraction took a few minutes for all 54 guidelines. Each guideline repository added more concepts to the ID knowledge compendium, and there was extensive overlap of concepts between the four-guideline sources.
Figure 3 demonstrates the overlap of infectious disease concepts between the three guidelines repositories contributing the most ID concepts (IDSA, ESCMID, and ASID guidelines). The effectiveness of each guideline repository for providing ID related concepts and relationships differed. Compared to the other guidelines repositories, the NICE guidelines provided fewer concepts and relationships per guideline (Table 5). In contrast to the other guidelines, the NICE guidelines focus on general management principles (i.e. what tests to perform, when to refer to hospital) rather than providing definitive lists of differential bacterial diagnoses and antibiotic treatment options. This difference may account for the lower contribution of concepts and relationships to the ID compendium. After combining the concepts obtained from all guideline sources, 238 infectious disease concepts, 127 antibiotic concepts and 229 bacterial concepts were identified (see Appendix D). Similarly, 819 “Bacteria causes Infectious disease”, 468 “Antibiotic is_antimicrobial_coverage_for Bacteria”, 630 “Infectious disease is_treated_with Antibiotic” relationships were identified (see Appendix D).
Figure 3.
Overlap of infectious disease (ID) concepts obtained from three guideline repositories
Table 5.
Effectiveness of knowledge sources for identifying bacterial clinical infectious disease concepts and relationships among concepts
| Effectiveness Index* | ||||||
|---|---|---|---|---|---|---|
| Class Concepts | Relationship between Class Concepts | |||||
| Knowledge Source (no. of resources) |
ID | Antibiotic | Bacteria | Bacteria causesID |
Antibioticis_ antimicrobial_cover age_forBacteria |
IDis_treated_with Antibiotic |
| Patient records | ||||||
| MIMIC II | ||||||
| Non- medical (20) | 0.9 | 0.6 | 0.4 | 1.05 | 0.35 | 1.45 |
| Medical (20) | 1.45 | 1.15 | 1.0 | 1.4 | 0.5 | 2.9 |
| Guidelines | ||||||
| IDSA (37) | 5.54 | 3.24 | 5.76 | 13.1 | 8.05 | 11.78 |
| ESCMID (9) | 12.56 | 6.1 | 6.0 | 17.3 | 13.89 | 9.44 |
| NICE (13) | 2.62 | 0.92 | 1.38 | 2.0 | 0.69 | 1.46 |
| ASID (8) | 9.25 | 7.0 | 3.38 | 6.5 | 5.38 | 4.13 |
| Expert ID scenarios (30) | 1.0 | 1.9 | 5.3 | 8.53 | 7.4 | 8.97 |
Effectiveness index was calculated by dividing the number concepts of relationships by the number of knowledge resources contained in the knowledge source.
Abbreviations: Unified Medical Language System (UMLS), Multiparameter Intelligent Monitoring in Intensive Care II (MIMIC II), Infectious Disease Society of America (IDSA), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), the Australasian Society for Infectious Diseases (ASID), National Institute for Health and Care Excellence (NICE), infectious disease (ID)
3.2.2 Patient electronic medical records
Twenty medical records from patients admitted to non-medical ICUs and 20 medical records from patients admitted to a medical ICUs were included. The number of concepts and relationships among concepts identified are shown in Table 4. As anticipated, more concepts and relationships among concepts were identified from patients who were admitted to a medical ICU compared to patients who were admitted to a non-medical ICU, especially since sick patients have more data in electronic medical records [43, 44]. However, even patients admitted to a medical ICU contributed fewer concepts and relationships per patient medical record compared to other knowledge sources (ranges: antibiotics 0–6, infectious diseases 0–4, bacteria 0–3; Figure 4). In addition, there was extensive overlap of concepts and relationships among concepts between the patients (i.e. “neonatal sepsis is_treated_with ampicillin and gentamicin” occurred in nearly all neonatal ICU patients), and only common infectious diseases, bacteria and antibiotics were present in the 40 medical records reviewed (i.e. “nosocomial pneumonia is_treated_with piperacillin/tazobactam” occurred in many adult ICU records). As a result, the effectiveness of medical records of ICU patients for providing ID-related concepts and relationships to the ID compendium was very low (Table 5).
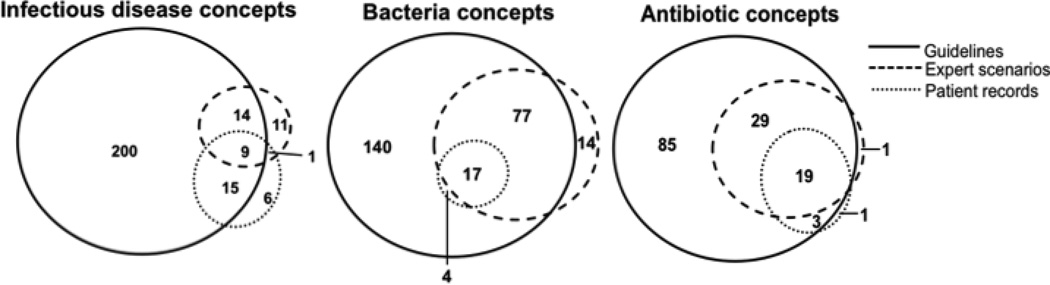
Figure 4.
Overlap of infectious disease, bacteria and antibiotic concepts between three types of knowledge sources
3.2.3 Infectious disease expert generated case scenarios
Four ID experts contributed ten common ID clinical cases independently of each other. An example of an ID scenario generated by an ID expert is shown in Table 6. There was overlap in the ID cases submitted and 30 unique ID case scenarios were ultimately identified. Overlap of submitted clinical cases reassured us that the final set of cases reflected the most common ID scenarios seen in the hospital setting. Many of the non-overlapping ID scenarios were scenarios which occur more frequently outside of the hospital setting (i.e. non-gonococcal urethritis). Overall, 186 ID-related concepts and 745 relationships between concepts were contained in the ID expert generated case scenarios (Table 4).
Table 6.
An example of an expert generated infectious disease case scenario
| ID scenario | Bacteria that cause ID scenario |
Antibiotics used to treat ID scenario |
|---|---|---|
| Cellulitis | Staphylococcus aureus | Cephazolin |
| β-hemolytic streptococci | Cephalexin | |
| Clostridium perf ringens | Amoxycillin-clavulanate | |
| Pasteurella multocida | Clarithromycin | |
| Pasteurella canis | Doxycycline | |
| Capnocytophaga canimorsu s | Trimethoprim-suxamethoxazole | |
| Vibrio vulnificus | Clindamycin | |
| Erysipelothrix rhusiopathiae | Vancomycin | |
| Linezolid | ||
| Telavancin |
3.3 ID knowledge compendium
After combining concepts and relationships obtained from all knowledge sources, 256 infectious disease concepts, 138 antibiotic concepts, 252 bacterial concepts, 613 “Bacteria causes Infecious disease relationships”, 438 “Antibiotic is_antimicrobial_coverage_for Bacteria” relationships, and 503 “Infectious disease is_treated_with Antibiotic” relationships were identified. The guideline knowledge source contributed the most infectious disease, bacterial and antibiotic concepts to the ID compendium (Figure 4). The addition of concepts identified in patient records and ID expert generated scenarios contributed relatively few additional concepts to the ID compendium (Figure 4). In contrast to the ID expert generated scenarios, the cost of using guideline repositiories as a source on knowledge was small. Using the guideline repository, the combination of automated UMLS concept extration and manual identification of relationships among concepts at a highlighted section of text generated 238 infectious disease concepts, 127 antibiotic concepts and 229 bacteria concepts, 819 “Bacteria causes Infectious disease”, 468 “Antibiotic is_antimicrobial_coverage_for Bacteria”, 630 “Infectious disease is_treated_with Antibiotic”. In comparison, four ID experts generated 30 infectious disease concepts, 52 antibiotic concepts and 104 bacteria concepts, 256 “Bacteria causes Infectious disease”, 222 “Antibiotic is_antimicrobial_coverage_for Bacteria” and 267 “Infectious disease is_treated_with Antibiotic”. The personnel-time saving or efficiency improvement from having four ID experts to generate the case scenarios to having one ID expert to integrate ID knowledge from the guidelines was around 80% (4 experts 3 hours each for 186 concepts and 745 relationships, which is 15.5 concepts per person per hour, vs. 1 expert 8 hours for 594 concepts and 1916 relationships, which is 74.25 concepts per person per hour). Furthermore, the effectiveness of providing concepts or relationships among concepts was higher per guideline compared to per expert generated case scenario (Table 5).
3.4 Comparison to BCIDO and error analysis
Overall, BCIDO covered 79% (504/637) of concepts in the ID compendium and 89% (1378/1554) of the relationships between concepts in the ID compendium. BCIDO performed well on coverage of antibiotic concepts with 92% (121/131) of antibiotic concepts in ID knowledge compendium represented in BCIDO. BCIDO had good coverage of infectious disease concepts with 80% (205/257) of terms in the knowedge compendium represented in BCIDO, while bacterial concepts were the least well represented in BCIDO (72%; 178/249). Low coverage of infectious disease concepts may reflect the difficulty in capturing nuanced clinical knowledge into a structured formal knowledge representation, however this is an improvement from an early evaulation in which 72% coverage was observed [16]. Nearly all of the bacterial concepts missing from BCIDO were obtained from guidelines and were bacterial species or subspecies that infrequently cause infection in clinial practice. This difference is consistent with a difference in granularity between BCIDO and published guidelines. The list of bacteria that can cause human infection is incredibly long. Bacteria were initially included in BCIDO if they had been reported to cause a human infection in several individuals (i.e. had been reported in a case series). In comparison, the guidelines contained bacterial species that had been reported to cause infection in only one individual. Nearly all missing bacterial concepts were added to BCIDO on the premise that they had been judged by the expert guideline authors to cause clinically important infections, albeit rarely. Six additional bacterial concepts were not added to BCIDO because they were judge to be too granular defined granularity of BCIDO (i.e. Bacteroides bivus). All concept relationships contained in the ID compendium were well represented in BCIDO: 91% (558/613) of “Bacteria causes Infectious disease”, 92% (327/438) of “Antibiotic is_antimicrobial_coverage_for Bacteria” and 98% (493/503) “Infectious disease is_treated_with Antibiotic” relationships in the ID compendium were also in BCIDO.
All three knowledge sources tended to group bacteria together by clinical or micrbiological features in a manner that is common in clinical medicine. For example, the terms “oral anaerobes”, “enteric gram negatives”, “group C streptococcus”, “beta-hemolytic streptococci”, “non-typhoidal salmonella”, “coagulase-negative staphylococci” and “enteric gram negative bacilli” were frequently used. While BCIDO had attempted to incorporate some groupings based on microbial features (ie. “Staphylococcus” has_shape “spherical”), many of these common groupings were missing. We now include clinical and microbial groupings using the ‘alternative term’ annotation property.
Both ID experts and guidelines divided infectious disease concepts into more subclasses than was present in BCIDO. For example, “acute sinusitis” was divided into “community-acquired acute sinusitis”, “hospital-acquired sinusitis” and “post-surgical sinusitis”. This extra degree of granularity of infectious disease concpets was added to BCIDO. ID experts also sometimes grouped clinical diseases together (i.e. vertebral osteomyelitis, spondylodisckitis and epidural abscess was considered to be one case scenario). However, to preserve the detail of infectious disease concepts, this approach was not adopted in BCIDO.
4. DISCUSSION
As antimicrobial resistance continues to rise at an alarming rate, solutions such as appropriate antimicrobial prescribing become increasingly important. Although ADSSs have been shown to reduce both antimicrobial prescribing and antibacterial drug resistance, the use of successful ADSS is not widespread. With the exception of the Evans et al. system [13], ADSSs address a single infectious disease or a narrow range of clinical syndromes represented in clinical guidelines and do not comprehensively cover the broad domain of clinical infectious disease. One of the main barriers to dissemination of successful ADSSs is that they tend to use their own terminology. Thus, often ADSSs cannot be easily transferred among different EHR systems. Ontologies can improve the portability of such ADSSs by providing a standard vocabulary for biomedical entities, helping to standardize and integrate data resources. BCIDO serves as an application ontology capturing a controlled terminology for clinical infectious diseases along with domain knowledge commonly used in the hospital in-patient setting with the aim of improving the interoperability of portable ADSSs. BCIDO captures much of the knowledge necessary to make clinical decisions about treatment and diagnosis across a broad scope of clinical infectious diseases.
The use of ontologies in decision support is increasing. Members of the Infectious Disease Ontology Consortium have developed and are developing a number of ontologies related to specific infectious diseases such as Brucellosis and Malaria[32, 45]. The antibiotic prescribing ontology provides the “proof of concept” that an ontology in the infectious diseases domain can successfully enable decision support [46]. In comparison to existing infectious disease ontologies, BCIDO includes the clinical infectious diseases knowledge required to make clinical decisions before microbiological information is known or in the absence of positive microbial results. This approach accurately reflects the knowledge management tasks of hospital antimicrobial prescribers. BCIDO extends existing ontologies by using the core IDO as the upper ontology, re-using terms and mapping to UMLS. Although not publically available, the antibiotic prescribing ontology could be integrated with BCIDO. Currently, the antibiotic terms in BCIDO are not re-used from an existing ontology. However, the Drug Ontology (DrOn) [47] mediates resources such as Chemical Entities of Biological Interest (ChEBI) and RxNorm and ultimately terms from DrOn will be imported as DrON coverage expands.
To evaluate the correctness and accuracy of BCIDO without relying on expensive domain expert knowledge, we developed a scalable, reusable semi-automated method for generating a comprehensive ID knowledge compendium from multiple diverse knowledge sources. Each knowledge source made a unique contribution to the ID knowledge compendium, emphasizing the importance of using diverse knowledge sources for evaluating ontologies. Guidelines offered the most diversity of concepts and relationships among concepts compared to patient records and expert generated scenarios. This reflects the intention of guidelines to cover the vast majority of possibilities for that particular infection or bacteria. In addition, the automated UMLS concept extraction process was very accurate for the guidelines, which made the development of a list of relevant concepts and relationships among concepts relatively easy. However, guidelines can only provide knowledge about a particular topic if a guideline on that topic exists. For example, none of the available guidelines contained knowledge about eye infections. Patient medical records had the least diversity of concepts and relationships among concepts, which is likely to be contributed to by the small sample size of patient notes reviewed, the low number of infectious disease conditions which occur during a hospital admission for each patient (range 0–4 infectious disease conditions per patient) and the lower diversity of infectious disease conditions occurring in patients admitted to ICU compared to the general hospital ward. ID expert generated scenarios focused on ID conditions commonly seen in clinical practice and used expert experience to determine the most relevant and important infectious disease conditions. The list of bacterial causes and antibiotics used for treatment generated by the ID experts was extensive and covered uncommon concepts and relationships between concepts that were missed in the patient medical record knowledge source. The inclusion of multiple diverse knowledge sources in the development of the knowledge compendium allowed greater confidence in the reliability of our evaluation of BCIDO and its suitability for providing a knowledge representation for ADSSs in the hospital setting.
Overall, BCIDO performed well against the ID knowledge compendium and several areas for improvement were identified. BCIDO performed well on antibiotic concepts as well as all the relationships between these concepts (ie “Antibiotic is_antimicrobial_coverage_for Bacteria). Infectious disease concepts were less well represented in BCIDO (80% coverage), which was an improvement from an early evaluations (72% coverage) [16]. Low coverage of infectious disease terms may reflect the difficulty in capturing nuanced clinical knowledge into a structured formal knowledge representation. We have attempted to allow for varying descriptions of the same or similar clinical ID concepts by including the annotation properties “has_related_term”, “has_exact_synonom” or “has_broad_synonom”, however, ongoing evaluation of clinical terms is required to improve the coverage and accuracy of clinical ID terms. Bacterial concepts were the least well represented in BCIDO (72%). Nearly all of the bacterial terms missing from BCIDO were obtained from guidelines and were bacterial species or subspecies that infrequently cause infections in clinical practice. This difference in granularity of between BCIDO and guidelines explained nearly all of the missing bacterial concepts. Concepts that were contained in the ID compendium but not BCIDO were added to BCIDO before the relationship comparison was performed. Therefore, BCIDO’s coverage of the relationships between concepts also contained in the ID compendium was excellent with > 90% of relationships in the ID compendium also present in BCIDO. The discrepancies between BCIDO and the ID compendium suggest that the manual methods used to develop BCIDO may have been incomplete in building a comprehensive ontology. Semi-automated approaches combining manual work from domain experts and automatic extraction from multiple sources can be used when developing domain ontologies.
Our study has some limitations. First, ontology evaluation is a multi-faceted problem and may involve measures from different dimensions. The approach proposed here may minimize expert involvement for coverage and accuracy evaluation but certainly cannot replace human testers for usability evaluation. Therefore, the decision to use this method for ontology evaluation should first start with assessment of the evaluation goals. Second, The ID knowledge compendium used the MIMICII database of patients in intensive care and infections seen in patients admitted to an intensive care unit may not reflect infections seen in the rest of the hospital. In addition, a small number of medical records were reviewed and more knowledge may be obtained if larger numbers of medical records were reviewed. However, we believe that infections more commonly seen in non-ICU wards would have been identified in the other knowledge sources. The manual extraction and evaluation of relationships from the guidelines and patient records was performed by the person who developed and evaluated BCIDO (C.L.G), which may have introduced bias. A higher-level evaluation of the usefulness and generalizability of BCIDO to other projects in knowledge representation and applications will occur when BCIDO is used for these purposes.
5. CONCLUSIONS
BCIDO is a comprehensive application ontology capturing a controlled terminology of bacterial clinical infectious diseases along with domain knowledge commonly used in the hospital setting. To evaluate the accuracy and comprehensiveness of BCIDO, we developed a semi-automated method to generate an ID knowledge compendium integrated from the multiple diverse knowledge sources with minimal reliance on domain experts. This method can be used for continued evaluation of biomedical ontologies and be expanded to include other knowledge sources such as journal articles and textbooks.
Supplementary Material
Highlights.
A novel method is proposed to evaluate the accuracy and completeness of ontologies
This method combines expert input and public knowledge for distant supervision
This method minimizes reliance on gold standards created by domain experts
This method improves efficiency for concept evaluation by 80% over manual methods
ACKNOWLEDGEMENTS
We are very grateful to Lindsay Cowell and Albert Goldfain for assistance with designing BCIDO, and to Anil Yaman for performing extraction of UMLS concepts from involved textual documents. This work was conducted using the Protégé resource, which is supported by grant GM10331601 from the National Institute of General Medical Sciences of the United States National Institutes of Health.
FUNDING
This research was partially supported by National Library of Medicine grant R01LM009886 to Dr. Weng. CLG was supported by a Hutchins Family Fellowship in Transplant Infectious Diseases and a Fulbright Scholarship. The article’s contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest.
REFERENCES
- 1.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Method Inform Med. 1998;37:394–403. [PMC free article] [PubMed] [Google Scholar]
- 2.Bodenreider O. Biomedical ontologies in action: role in knowledge management, data integration and decision support. Yearb Med Inform. 2008:67–79. [PMC free article] [PubMed] [Google Scholar]
- 3.Achour SL, Dojat M, Rieux C, Bierling P, Lepage E. A UMLS-based knowledge acquisition tool for rule-based clinical decision support system development. J Am Med Inform Assoc. 2001;8:351–360. doi: 10.1136/jamia.2001.0080351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kashyap V, Morales A, Hongsermeier T. On implementing clinical decision support: achieving scalability and maintainability by combining business rules and ontologies. AMIA Annu Symp Proc. 2006:414–418. [PMC free article] [PubMed] [Google Scholar]
- 5.Brank J, Grobelnik M, Mladenic D. A survey of ontology evaluation techniques. Proceedings of SiKDD 2005. Ljubljana, Slovenia. 2005 [Google Scholar]
- 6.Smith B, Ceusters W. Ontological realism: A methodology for coordinated evolution of scientific ontologies. Appl Ontol. 2010;5:139–188. doi: 10.3233/AO-2010-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke EL, Loguercio S, Good BM, Su AI. A task-based approach for Gene Ontology evaluation. J Biomed Semantics. 2013;4(Suppl 1):S4. doi: 10.1186/2041-1480-4-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liubo O, Beiji Z, Miaoxing Q, Chengming Z. A method of ontology evaluation based on coverage, cohesion and coupling; Eight International Conference on Fuzzy Systems and Knowledge Discovery (FSKD); 2011. pp. 2451–2455. [Google Scholar]
- 9.Brewster C, Alani H, Dasmahapatra S, Wilks Y. International Conference on Language Resources and Evaluation (LREC 2004) Portugal: Lisbon; 2004. Data driven ontology evaluation. [Google Scholar]
- 10.Maedche E, Staab S. Proceedings of the European Conference on Knowledge Acquisition and Management (EKAW) Spain: Sig enza; 2002. Measuring similarity between ontologies; pp. 251–263. [Google Scholar]
- 11.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J Biomed Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. Antimicrobial resistance: global report on surveillance 2014. 2014:257. [Google Scholar]
- 13.Evans RS, Pestotnik SL, Classen DC, Clemmer TP, Weaver LK, Orme JF, et al. A computer-assisted management program for antibiotics and other antiinfective agents. New Engl J Med. 1998;338:232–238. doi: 10.1056/NEJM199801223380406. [DOI] [PubMed] [Google Scholar]
- 14.Thursky KA, Mahemoff M. User-centered design techniques for a computerised antibiotic decision support system in an intensive care unit. Int J Med Inform. 2007;76:760–768. doi: 10.1016/j.ijmedinf.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 15.Paterson DL. The role of antimicrobial management programs in optimizing antibiotic prescribing within hospitals. Clin Infect Dis. 2006;42(Suppl 2):S90–S95. doi: 10.1086/499407. [DOI] [PubMed] [Google Scholar]
- 16.Gordon CL, Pouch S, Cowell LG, Boland MR, Platt HL, Goldfain A, et al. Design and evaluation of a bacterial clinical infectious diseases ontology. AMIA Annu Symp Proc. 2013;2013:502–511. [PMC free article] [PubMed] [Google Scholar]
- 17.Shortliffe EH, Davis R, Axline SG, Buchanan BG, Green CC, Cohen SN. Computer-based consultations in clinical therapeutics: explanation and rule acquisition capabilities of the MYCIN system. Comput Biomed Res. 1975;8:303–320. doi: 10.1016/0010-4809(75)90009-9. [DOI] [PubMed] [Google Scholar]
- 18.Gonzales R, Anderer T, McCulloch CE, Maselli JH, Bloom FJ, Graf TR, et al. A cluster randomized trial of decision support strategies for reducing antibiotic use in acute bronchitis. JAMA Intern Med. 2013;173:267–273. doi: 10.1001/jamainternmed.2013.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann D, Knaus M, McCullagh L, Sofianou A, Rosen L, McGinn T, et al. Measures of user experience in a streptococcal pharyngitis and pneumonia clinical decision support tools. Appl Clin Inform. 2014;5:824–835. doi: 10.4338/ACI-2014-04-RA-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gulliford MC, van Staa T, McDermott L, Dregan A, McCann G, Ashworth M, et al. Cluster randomised trial in the General Practice Research Database: 1 Electronic decision support to reduce antibiotic prescribing in primary care (eCRT study) Trials. 2011;12:115. doi: 10.1186/1745-6215-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans RS, Pestotnik SL, Classen DC, Burke JP. Evaluation of a computer-assisted antibiotic-dose monitor. Ann Pharmacother. 1999;33:1026–1031. doi: 10.1345/aph.18391. [DOI] [PubMed] [Google Scholar]
- 22.Nachtigall I, Tafelski S, Deja M, Halle E, Grebe MC, Tamarkin A, et al. Long-term effect of computer-assisted decision support for antibiotic treatment in critically ill patients: a prospective ‘before/after’ cohort study. BMJ Open. 2014;4:e005370. doi: 10.1136/bmjopen-2014-005370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez-Maresca M, Sorlozano A, Grau M, Rodriguez-Castano R, Ruiz-Valverde A, Gutierrez-Fernandez J. Implementation of a computerized decision support system to improve the appropriateness of antibiotic therapy using local microbiologic data. BioMed Res Int. 2014;2014:395434. doi: 10.1155/2014/395434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hum RS, Cato K, Sheehan B, Patel S, Duchon J, DeLaMora P, et al. Developing clinical decision support within a commercial electronic health record system to improve antimicrobial prescribing in the neonatal ICU. Appl Clin Inform. 2014;5:368–387. doi: 10.4338/ACI-2013-09-RA-0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linder JA, Schnipper JL, Tsurikova R, Yu T, Volk LA, Melnikas AJ, et al. Documentation-based clinical decision support to improve antibiotic prescribing for acute respiratory infections in primary care: a cluster randomised controlled trial. Inform Prim Care. 2009;17:231–240. doi: 10.14236/jhi.v17i4.742. [DOI] [PubMed] [Google Scholar]
- 26.Evans RS, Classen DC, Pestotnik SL, Clemmer TP, Weaver LK, Burke JP. A decision support tool for antibiotic therapy. Proc Annu Symp Comput Appl Med Care. 1995:651–655. [PMC free article] [PubMed] [Google Scholar]
- 27.Mintz M, Bills S, Snow R, Jurafsky D. Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP. Vol. 2. Suntec, Singapore: Association for Computational Linguistics; 2009. Distant supervision for relation extraction without labeled data; pp. 1003–1011. [Google Scholar]
- 28.Cowell L, Smith B. Infectious Diseases Ontology. In: Sintchenko V, editor. Infectious Disease Informatics. New York: Springer; 2010. pp. 373–395. [Google Scholar]
- 29.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topalis P, Mitraka E, Bujila I, Deligianni E, Dialynas E, Siden-Kiamos I, et al. IDOMAL: an ontology for malaria. Malar J. 2010;9:230. doi: 10.1186/1475-2875-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitraka E, Topalis P, Dialynas E, Dritsou V, Louis C. IDODEN: An Ontology for Dengue; Proceedings of the International Conference on Biomedical Ontology; 2012. p. 1. [Google Scholar]
- 32.Lin Y, Xiang Z, He Y. Brucellosis Ontology (IDOBRU) as an extension of the Infectious Disease Ontology. J Biomed Semantics. 2011;2:9. doi: 10.1186/2041-1480-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goldfain A, Smith B, Cowell LG. Towards an ontological representation of resistance: the case of MRSA. J Biomed Inform. 2011;44:35–41. doi: 10.1016/j.jbi.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldfain A, Smith B, Cowell LG. Constructing a Lattice of Infectious Disease Ontologies from a Staphylococcus aureus Isolate Repository; Proceedings of the 3rd International Conference on Biomedical Ontology (ICBO 2012), KR-MED Series; 2012. pp. 1–5. [Google Scholar]
- 35.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucl Acids Research. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. International Classification of Diseases (ICD) 2015 [Google Scholar]
- 37.Xiang Z, Courtot M, Brinkman RR, Ruttenberg A, He Y. OntoFox: web-based support for ontology reuse. BMC Res Notes. 2010;3:175. doi: 10.1186/1756-0500-3-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandell GL, Bennett JE, Dolin R. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 7th ed. Elsevier; 2010. [Google Scholar]
- 39.Gilbert DN, Moellering RC, Eliopoulos GM, Chambers HF, Saag MS. Antimicrobial Therapy, Inc. 42nd ed 2012. The Sanford Guide to Antimicrobial Therapy 2012. [Google Scholar]
- 40.Rugg G, McGeorge P. Laddering. Expert Syst. 1995:339–346. [Google Scholar]
- 41.Saeed M, Villarroel M, Reisner AT, Clifford G, Lehman LW, Moody G, et al. Multiparameter Intelligent Monitoring in Intensive Care II: a public-access intensive care unit database. Crit Care Med. 2011;39:952–960. doi: 10.1097/CCM.0b013e31820a92c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miotto R, Weng C. Unsupervised mining of frequent tags for clinical eligibility text indexing. J Biomed Inform. 2013;46:1145–1151. doi: 10.1016/j.jbi.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiskopf NG, Rusanov A, Weng C. Sick patients have more data: the non-random completeness of electronic health records. AMIA Annu Symp Proc. 2013;2013:1472–1477. [PMC free article] [PubMed] [Google Scholar]
- 44.Rusanov A, Weiskopf NG, Wang S, Weng C. Hidden in plain sight: bias towards sick patients when sampling patients with sufficient electronic health record data for research. BMC medical informatics and decision making. 2014;14:51. doi: 10.1186/1472-6947-14-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Topalis P, Mitraka E, Bujila I, Deligianni E, Dialynas E, Siden-Kiamos I, et al. IDOMAL: an ontology for malaria. Malar J. 2010;9:230. doi: 10.1186/1475-2875-9-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bright TJ, Yoko Furuya E, Kuperman GJ, Cimino JJ, Bakken S. Development and evaluation of an ontology for guiding appropriate antibiotic prescribing. J Biomed Inform. 2012;45:120–128. doi: 10.1016/j.jbi.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hogan WR, Hanna J, Joseph E, Brochhausen M. Towards a Consistent and Scientifically Accurate Drug Ontology; ICBO 2013 Conference Proceedings; 2013. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.