Abstract
OBJECTIVE
Levels of insulin-like growth factor (IGF) proteins are associated with risk of cancer and mortality. IGF assays produced by Diagnostics Systems Laboratories (DSL) were widely used in epidemiological studies, were not calibrated against recommended standards and are no longer commercially available.
DESIGN
In a split sample study among 1471 adults participating in the Cardiovascular Health Study, we compared values obtained using DSL assays with alternative assays for serum IGF-I (Immunodiagnostic Systems, IDS), IGFBP-1 (American Laboratory Products Company, ALPCO) and IGFBP-3 (IDS).
RESULTS
Results were compared using kernel density estimation plots, quartile analysis with weighted kappa statistics and linear regression models to assess the concordance of data from the different assays. Participants had mean age of 77 years. Results between alternative assays were strongly correlated (IGF-I, r=0.93 for DSL versus IDS; log-IGFBP-1, r= 0.90 for DSL versus ALPCO; IGFBP-3, r= 0.92 for DSL versus IDS). Cross tabulations showed that participants were usually in the same quartile categories regardless of the assay used (overall agreement, 74% for IGF-I, 64% for IGFBP-1, 71% for IGFBP-3). Weighted kappa also showed substantial agreement between assays (kw, 0.78 for IGF-I, 0.69 for IGFBP-1, 0.76 for IGFBP-3). Regression of levels obtained with DSL assays (denoted X) to alternative assays were, IGF-I: 0.52X +15.2 ng/ml, log-IGFBP-1: 1.01X – 1.73 ng/ml IGFBP-3: 0.87X + 791.1 ng/ml. Serum values of IGF-I, IGFBP-1 and IGFBP-3 measured using alternative assays are moderately correlated.
CONCLUSIONS
Care is needed in interpretation of data sets involving IGF analytes if assay methodologies are not uniform.
Keywords: insulin-like growth factor, diabetes, glycemic status
Introduction
The insulin-like growth factor (IGF) axis is an evolutionarily conserved system with important biologic roles during embryonic development, growth and adulthood [1]. IGF-I has mitogenic and antiapoptotic activity and acts as the primary mediator of the effects of growth hormone. IGF-I also has insulin-like activity with direct effects on glucose and free fatty acid metabolism. IGF-I circulates in blood bound to six binding proteins (IGFBP-1 to IGFBP-6), and the acid-labile subunit, which together prolong IGF-I half-life and regulate its bioavailability [2, 3]. Only approximately 1% of IGF-I circulates unbound to IGFBPs. IGFBPs also may have IGF-I independent functions that may include cell growth, apoptosis, as well as metabolism.
IGF-I and IGFBPs have been extensively studied in epidemiologic investigations. Several studies have associated circulating levels of these proteins with incident diabetes, heart failure, stroke, coronary heart disease, several types of cancer and overall mortality [4–12]. In addition, measures of IGF-I are used clinically to diagnose and assess response to treatment in those with growth hormone deficiency and acromegaly [13, 14]. Several commercial assays for IGF-I, IGFBP-1, and IGFBP-3 are available.
Differences in assay performance may have contributed to some of the conflicting results from prior population studies investigating associations between IGF-related blood analytes and disease risk. Many, but not all, large-scale studies of IGF-related serum analytes used assays produced by one manufacturer, Diagnostics Systems Laboratory (DSL, Webster, TX)[15]. However, these assays have become unavailable. Moreover, they do not adhere to the consensus statement regarding standards in IGF assays [16, 17]. In order to facilitate our own ongoing research, we compared values measured by currently available assays that follow the recommended calibration standards [17] to levels previously obtained with the DSL assays. It cannot be assumed a priori that different assay methods would yield comparable results. Comparisons were completed using linear and deming regression models as well as quantile analysis, since epidemiological studies often use quantiles for grouping individuals into categories in risk assessment studies.
In the Cardiovascular Health Study [18], we previously obtained measurements of circulating IGF-I, IGFBP-1 and IGFBP-3 levels using assays produced by DSL. Subsequently, other commercial assays have been used to measure IGF proteins in a subset of the CHS cohort. In a validation study conducted among over 1000 participants, we sought to evaluate the comparability of measurements of serum IGF-I, IGFBP-1 and IGFBP-3 levels.
Materials and Methods
Study Population
The CHS is a population-based, prospective cohort study of community-dwelling older adults aged ≥65 years [19]. The cohort consists of 5888 participants who were recruited from four U.S. communities (Washington County, MD; Allegheny County, PA; Forsyth County, NC; and Sacramento County, CA), using a randomly generated sampling frame derived from Medicare eligibility lists of the Health Care Financing Administration (HCFA). Standardized clinic examinations and questionnaires were administered annually. Informed consent was provided by participants in accordance with the institutional review board guidelines at their clinic site.
Specimen Collection
After an 8–10 hour fast, blood samples were collected from participants by trained phlebotomists. The samples were allowed to stand in room temperature for 30 minutes. They were then centrifuged at 3000g at 4°C temperature for 10 minutes. After centrifugation, the samples were put into −70°C storage. Frozen samples packaged with frozen CO2 in insulated styrofoam boxes were shipped weekly to the specimen repository at the Central Blood Analysis Laboratory (CBAL) at the University of Vermont (Burlington, VT). All refrigerators, −70°C freezers and refrigerated centrifuges were monitored daily. Systemic errors in phlebotomy procedures, processing, shipping and storage were monitored by the CHS Coordinating Center [20]. Measurements of IGF-I, IGFBP-1 and IGFBP-3 were performed using stored fasting venous blood samples obtained during the 1996–1997 (year 9) examination cycle. These measurements were completed at the Cancer Prevention Research Unit, Lady Davis Research Institute of Jewish General Hospital [8] using the DSL, ALPCO and IDS assays described below.
Assays
Using stored blood specimens collected at the Year 9 CHS clinic visit (1996/1997), we performed IGF assays at the Cancer Prevention Research Unit, McGill University, Montreal [7]. [8].
Diagnostic Systems Laboratories assays
IGF-I: This assay involved acid –ethanol extraction to separate IGF-I from its binding proteins, followed by a sandwich ELISA assay. Intra-assay coefficient of variation was 1.49%. IGFBP-1: This ELISA method had an intra-assay coefficient of variation of 1.88%. IGFBP-3: This ELISA method had an intra-assay coefficient of variation of 1.77%. In 249 samples of specimens, repeat measures of IGF-I, IGFBP-1 and IGFBP-3 were obtained over the 2–3 year period of testing to assess the within-individual variability over time (Pearson Correlation coefficient (r) = 0.74 – 0.86)) [9]. These assays were produced prior to the publication of recommended standards and thus, these assays were not calibrated against the WHO International Standard 02/254 [17].
Comparison Assays
IGF-I
Measures were obtained using the IDS-iSYS Insulin like Growth Factor-I Assay (IDS-iSYS IGF-I), an automated chemiluminescence immunoassay provided by Immunodiagnostic Systems Ltd (IDS, Boldon Business Park, Boldon, Tyne & Wear, England). Samples are incubated in an acidic solution to dissociate IGF-I from its binding proteins and the addition of excess of IGF-II prevents re-aggregation. After neutralization, the solution is further incubated with a biotinylated anti-IGF-I monoclonal antibody and an acridinium labeled anti-IGF-I monoclonal antibody. Magnetic particles labeled with streptavidin are added to the solution, incubated, and then captured using a magnet. IGF-I concentration is then determined by measuring the amount of light emitted by the acridinium label. This IGF assay is calibrated against the WHO International Standard 02/254 [17] and has no interference from IGF-II, insulin, proinsulin, and any of the IGFBPs (IGFBP-1 to IGFBP-6). The intra-assay coefficient of variation is 2.2% and the assay detects IGF-I levels in the range of 10–1,200 ng/mL.
IGFBP-3
Measures were obtained using IDS-iSYS Insulin-Like Growth Factor Binding Protein -3 (IGFBP-3), an automated chemiluminescence immunoassay provided by Immunodiagnostic Systems Ltd (IDS, Boldon Business Park, Boldon, Tyne & Wear, England)[21]. A diluent is used to dilute the original specimen sample. The solution is then incubated with a biotinylated anti-IGFBP-3 monoclonal antibody and an acridinium labeled anti-IGFBP-3 antibody. Magnetic particles labeled with streptavidin are added to the solution, incubated, and then captured using a magnet. IGFBP-3 concentration is then determined by measuring the amount of light emitted by the acridinium label. This IGFBP-3 assay if calibrated to the Reference Material: Insulin-Like Growth Factor Binding Protein-3 NIBSC code: 93/560 and has no cross-reactivity with IGF-I, IGF-II, proinsulin, insulin and any of the other IGFBPs. The intra-assay coefficient of variation is 1.94%. The assay detects IGFBP-3 levels in the range of 80–10,000 ng/mL and has no cross-reactivity with other IGFBPs.
IGFBP-1
Measures were obtained using a two-step sandwich ELISA assay method from American Laboratory Products Company (ALPCO, Keewaydin Drive, Salem, NH). The two antibodies in the two-step sandwich assay are a monoclonal IGFBP-1 antibody and a horseradish peroxidase tagged monoclonal IGFBP-1 antibody with specificity for a different region of IGFBP-1. The Intra-assay coefficient of variation is 2.25% and this assay does not cross-react with any of the other IGFBPs. The assay detects IGFBP-1 levels in the range of 1–250 ng/mL and has no cross-reactivity with other IGFBPs.
Statistical Analysis
Measures of IGF-I and IGFBP-1 were log-transformed to normalize the distribution and log-transformed values were used for all analysis. Descriptive statistics of IGF-I, IGFBP-1 and IGFBP-3 concentrations are presented for the entire analytic population and then separately for men and women and for diabetics and non-diabetics. For each biomarker, student’s t-tests were used to compare differences in means of each IGF protein in men and women and in diabetics and non-diabetics. Bland Altman plots, which do not designate one assay as the gold standard, were used to visually assess the correlation between measures from the two assays. To evaluate the linear correlation between the values produced by the two assays, we calculated the Pearson correlation coefficient.
Epidemiological studies often follow the practice of using quantiles from the empirical distribution of biomarkers for grouping individuals into categories. Therefore, we also compared for each IGF-axis protein the quartile classifications obtained when using quartiles of measurements from the old and new assays. To evaluate the degree of agreement between these classifications we calculated a weighted Cohen’s kappa coefficient, which ranges from 0 (maximum discordance) to 1 (perfect concordance) [22]. The following standard criteria were used to interpret the strength of agreement between measures: slight (κw = 0.01 – 0.20), fair (κw = 0.21 – 0.40), moderate (κw = 0.41 – 0.60), substantial (κw = 0.61 – 0.80), and almost perfect (κw = 0.81 – 1.0) [23]. Further, to evaluate the linear relationship between measures from the two assays, we fitted an Ordinary Least Squares (OLS) Regression with the measurements obtained with the old assay as predictor of the measurements obtained from the new assay. To account for variation in both assays, a deming regression model was fit to further evaluate the relationship between measures from the two assays.
Results
The individuals studied had a mean age of 76.6 years, mean BMI of 27.2 and 12.8% were African American, 68% were female, and 9.8% had self-reported diabetes. Log-IGF-I, log-IGFBP-1 and IGFBP-3 levels were not significantly different among the diabetics and non-diabetics using measures from either assays (Table 1). Correlations between old and new assay levels were similar in the non-diabetic and diabetic populations for all three IGF proteins (non-diabetics: r= 89 – 93, diabetics r= 87 – 94). Men and women had statistically significant differences in both assay measures for all IGF proteins (p<0.0001).
Table I.
IGF-I, IGFBP-1 and IGFBP-3 measurements obtained with old assay (All-Stars study) and new assay (Kaplan Study) stratified by gender and diabetes status
| DSL Assay IGF Measures
|
|||||||
|---|---|---|---|---|---|---|---|
| Entire Cohort | Men | Women | p-value | Non-Diabetics | Diabetics | p-value | |
|
|
|
|
|||||
| log-IGF-I, ng/ml | |||||||
| N | 1158 | 371 | 787 | 1041 | 113 | ||
| Range | 3.37 – 6.19 | 3.95 – 5.97 | 3.37 – 6.19 | 3.37 – 6.19 | 3.78 – 5.97 | ||
| Mean ± SD | 5.05 ± 0.39 | 5.15 ± 0.34 | 5 ± 0.41 | <0.0001 | 5.05 ± 0.39 | 5.07 ± 0.43 | 0.537 |
| log-IGFBP-1, ng/ml | |||||||
| N | 1384* | 473 | 911 | 1241 | 138 | ||
| Range | 0.03 – 4.3 | 1.59 – 5.58 | 1.16 – 5.51 | 1.16 – 5.51 | 1.44 – 5.58 | ||
| Mean ± SD | 1.80 ± 0.79 | 3.38 ± 0.66 | 3.55 ± 0.69 | <0.0001 | 3.48 ± 0.68 | 3.61 ± 0.76 | 0.059 |
| IGFBP-3, ng/ml | |||||||
| N | 1334 | 448 | 886 | 1188 | 141 | ||
| Range | 868 – 7230 | 893 – 5742 | 1025 – 6446 | 893 – 6446 | 1562 – 6374 | ||
| Mean ± SD | 3719 ± 837 | 3049 ± 751 | 3551 ± 880 | <0.0001 | 3376 ± 852 | 3446 ± 1025 | 0.438 |
|
|
|||||||
|---|---|---|---|---|---|---|---|
| IDS and ALPCO IGF Measures
| |||||||
| Entire Cohort | Men | Women | p-value | Non-Diabetics | Diabetics | p-value | |
|
|
|
|
|||||
| log-IGF-I, ng/ml | |||||||
| N | 1158 | 371 | 787 | 1041 | 113 | ||
| Range | 3.03 – 5.71 | 3.54 – 5.45 | 3.03 – 5.71 | 3.03 – 5.71 | 3.7 – 5.45 | ||
| Mean ± SD | 4.56 ± 0.35 | 4.67 ± 0.3 | 4.5 ± 0.36 | <0.0001 | 4.55 ± 0.35 | 4.59 ± 0.35 | 0.258 |
| log-IGFBP-1, ng/ml | |||||||
| N | 1384* | 473 | 911 | 1241 | 138 | ||
| Range | 1.16 – 5.58 | 0.06 – 4.3 | 0.03 – 4.18 | 0.03 – 4.18 | 0.06 – 4.3 | ||
| Mean ± SD | 3.49 ± 0.69 | 1.64 ± 0.75 | 1.88 ± 0.8 | <0.0001 | 1.8 ± 0.78 | 1.82 ± 0.89 | 0.719 |
| IGFBP-3, ng/ml | |||||||
| N | 1334 | 448 | 886 | 1188 | 141 | ||
| Range | 893 – 7178 | 1475 – 5891 | 868 – 6689 | 868 – 6575 | 2012 – 6689 | ||
| Mean ± SD | 3385 ± 891 | 3474 ± 716 | 3849 ± 845 | <0.0001 | 3715 ± 804 | 3803 ± 966 | 0.3 |
Includes IGF levels 52 individuals with IGFBP-1 levels that were below the assay detectability threshold for the ALPCO assay. Values of half the lower limit of detection were assigned for these individuals.
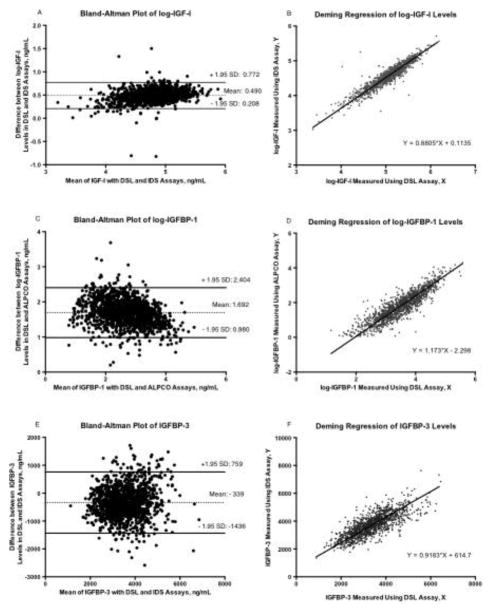
The range of log-IGF-I levels obtained with the DSL and IDS assays are similar (Table 1), although the measurements are highly correlated (Table 3: r = 0.93, 95% CI: 0.925 – 0.94). The Bland Altman plot shows a bias of 0.49 ng/ml, with a relatively wide range as indicated by the ± 1.95 SD (Figure 1A). The difference between measures from the two assays is greater at higher measures of log-IGF-I using both assays. Cross tabulations of IGF-I quartiles determined using the two assays showed that 74% of the participants were in exact concordant categories (Table 2), while all others varied by only one quartile, as reflected in the strong weighted kappa statistic (kw = 0.78, “substantial” agreement). Deming regression analysis shows that the regression line do not deviate significantly from the regression lines (Table 3: Y=0.88*X + 0.11). Results from the linear regression model show that the estimated intercept and slope are 0.38 ng/ml (95%CI: 0.29, 0.48) and 0.83 ng/ml (95% CI: 0.81, 0.85), respectively, with this model explaining 87% of the variability observed in the measurements from the IDS assay.
Table III.
Results of Regressions for IGF-1, IGFBP-1 and IGFBP-3 new assay measurements on old assay measurements.
| Estimate ± SE | 95% IC | R-squared | Pearson correlation coefficient | |
|---|---|---|---|---|
| Log-IGF-I | ||||
| OLS regression | ||||
| Intercept | 0.383 ± 0.048 | (0.288, 0.477) | 0.87 | 0.933 |
| Slope | 0.827 ± 0.010 | (0.808, 0.846) | ||
| Deming Regression | ||||
| Intercept | 0.114 ± 0.051 | (0.013, 0.214) | ||
| Slope | 0.881 ± 0.010 | (0.861, 0.900) | ||
| Log-IGFBP-1 | ||||
| OLS regression | ||||
| Intercept | −1.730 ± 0.045 | (−1.818, −1.643) | 0.815 | 0.903 |
| Slope | 1.011 ± 0.013 | (0.986, 1.036) | ||
| Deming regression | ||||
| Intercept | −2.298 ± 0.05798 | (−2.412 to −2.184) | ||
| Slope | 1.173 ± 0.01629 | (1.142 to 1.205) | ||
| IGFBP3-1 | ||||
| OLS regression | ||||
| Intercept | 791.1 ± 33.6 | (725.2, 857.1) | ||
| Slope | 0.865 ± 0.010 | (0.846,0.883) | 0.846 | 0.92 |
| Deming regression | ||||
| Intercept | 614.7 ± 72.4 | (472.7 to 756.7) | ||
| Slope | 0.918 ± 0.021 | (0.878 to 0.959) | ||
Figure 1. Bland-Altman Plots and Scatter Plot with Fitted Deming Regression Lines.
for log-IGF-I (A and B), log-IGFBP-1 (C and D) and IGFBP-3 (E and F). SD stands for standard deviation. Bland Altman Plots (A, C, E) show plots of the mean difference between the two assay measures for each IGF protein on the X-axis and the difference between the two assays for each IGF protein on the Y axis. The dotted lines are the mean of the difference between the two assays, while the solid lines represent the 1.96 SD range of these differences. Deming regression graphs (B, D, F) show plots of a scatter of measures from the two assays superimposed with the regression lines. Measures from the older assays are plotted on the X-axis and the new assays on the Y-axis. The regression equation is also shown.
Table II.
Cross-tabulation of quartiles of IGF protein levels measured using the old DSL assays and alternative assays (IDS for IGF-I and IGFBP-3, ALPCO for IGFBP-1).
Values are row percentages. Kw indicates weighted kappa statistics
| DSL Assay Levels | Alternative Assay Levels | ||||||
|---|---|---|---|---|---|---|---|
| Quartiles | Quartiles, (ng/ml) | ||||||
| Log-IGF-I, (ng/ml) (n=1158) | % | 1 | 2 | 3 | 4 | Total | Kw |
|
| |||||||
| 1 | 86 | 13 | 0 | 1 | 100 | ||
| 2 | 10 | 68 | 21 | 1 | 100 | ||
| 3 | 1 | 17 | 66 | 15 | 100 | ||
| 4 | 0 | 1 | 23 | 75 | 100 | 0.78 | |
|
| |||||||
| Log-IGFBP-1, (ng/ml) (n=1436) | % | 1* | 2 | 3 | 4 | ||
|
| |||||||
| 1 | 72 | 26 | 2 | 0 | 100 | ||
| 2 | 13 | 52 | 32 | 4 | 100 | ||
| 3 | 1 | 13 | 53 | 33 | 100 | ||
| 4 | 0 | 1 | 12 | 87 | 100 | 0.69 | |
|
| |||||||
| IGFBP-3, (ng/ml) (n=1471) | % | 1 | 2 | 3 | 4 | ||
|
| |||||||
| 1 | 88 | 10 | 2 | 0 | 100 | ||
| 2 | 25 | 62 | 12 | 1 | 100 | ||
| 3 | 0 | 28 | 59 | 13 | 100 | ||
| 4 | 1 | 1 | 20 | 78 | 100 | 0.76 | |
Includes IGF levels 52 individuals with IGFBP-1 levels that were below the assay detectability threshold for the ALPCO assay. Values of half the lower limit of detection were assigned for these individuals.
The range of Log-IGFBP-1 measures obtained from the DSL and ALPCO assays were very different; 3.4 ng/ml – 6.2 ng/ml for the DSL assay and 1.2 ng/ml – 5.6 ng/ml for the ALPCO assay (Table 1). Measures reported by both assays were highly correlated (Table 3: r = 0.90, 95% CI: 0.893, 0.912). Bland Altman plots show a bias of 1.69 ng/ml and the difference between measures from the two assays is higher at lower analytic values of log-IGFBP-1 (Figure 1C). 52 individuals had IGFBP-1 results below the threshold for detection in the ALPCO assay while having available IGFBP-1 measurements from the DSL assay. They were included in the lowest ALPCO quartile in cross-tabulation analyses. Cross tabulations of log-IGFBP-1 quartiles of measures from the two assays show that 64% of the participants were in concordant categories (Table 2). Overall, the weighted kappa showed substantial agreement (kw = 0.69). Deming regression analysis shows that the regression line do not deviate significantly from the regression lines (Table 3: Y= 1.17*X – 2.30). Results from linear regression of log-transformed values were: estimated intercept: −1.73 ng/ml (95%CI: −1.82, −1.64), slope: 1.011 ng/ml (95% CI: 1.986, 1.036). The linear regression model explained approximately 82% of the variation observed in the ALPCO assay measures.
The range of values reported by the DSL and IDS IGFBP-3 assays were similar and levels were highly correlated (Table 3*********: r = 0.92, 95% CI: 0.912, 0.927). Bland-Altman analysis showed a large bias of −339 with a wide range, as indicated by the ± 1.95 SD (Figure 1E). The difference between measures from the DSL and IDS assays did not vary at different levels of IGFBP-3. Additionally, cross tabulations of IGFBP-3 quartiles of measures from the two assays show that 71.1% of the participants were in concordant quartiles, (Table 2) with discordant values differing by no more than one level, as reflected in the weighted kappa statistic (kw = 0.76). Deming regression analysis shows that the regression line do not deviate significantly from the regression lines (Table 3: Y= 0.92*X 615). Results from the linear regression model show that the estimated intercept and slope are 791.1 (95%CI: 725.2, 857.1) and 0.86 (95%CI: 0.85, 0.88), respectively, with this model explaining 85% of the variability observed in the measurements from the IDS assay.
Discussion
In this study, we evaluated the comparability of levels of serum IGF-I, IGFBP-1 and IGFBP-3 measured using the recently discontinued DSL assays to those of selected commercially assays (IDS for IGF-I and IGFBP-3 and ALPCO for IGFBP-1) that adhere to the recent recommendations for IGF standards [17].
The results showed that although each of the three sets of paired assays were highly correlated with one another, the magnitude of the values varied. These differences were greatest for IGFBP-1 and least for IGFBP-3. For all analytes, the IDS and ALPCO assays procedures were performed on the serum obtained at the same venipuncture as the samples used for the DSL assays, albeit the DSL aliquots had been frozen for fewer years at the time the assays were performed. However, studies have shown that IGF analytes are stable over long periods in sera frozen at −70°C or lower [24]. Moreover, repeat measures of IGF-I, IGFBP-1 and IGFBP-3 were obtained over the 2–3 year period of testing in our laboratory to assess the within-individual variability over time (Pearson Correlation coefficient (r) = 0.74 – 0.86) [9]. Differences between measures from the two assays may be due to random variations or differences in assay sensitivity and antibody specificities for IGF-I and IGFBPs.
In all three cases, analysis of the IGF-axis data by quartile was shown to produce considerable agreement between paired assays. It has been common in published analyses of associations of IGF-related analytes with health and disease endpoints for the analytes to be analyzed based on categorical data (eg, quartiles or quintiles). Several other studies have reported high correlation and agreement between measures of IGF-I from different assays [13, 14, 24, 25], though the majority of these studies were in populations with growth hormone disorders. Some IGF-I assays only correlate at either low or high levels of IGF-I; however our data shows that the DSL and IDS IGF-I assays have relatively similar bias at varying levels of IGF-I. Assays for IGFBP-3 had the least bias while IGFBP-1 had the most bias at varying levels of each analyte. Correlation between results from different assays may depend on health status. For example, adequate correlation of results across alternative IGF-I assays has been observed in healthy subjects but not in those with diabetes [26]. However, this finding was not supported by a previous study [24]. In our population-based cohort, assay measure correlations were similar in the non-diabetic and diabetic populations for all three IGF proteins (non-diabetics: r= 89 – 93, diabetics r= 87 – 94). Finally, we report linear and deming regression models that build upon the prior literature addressing this methodological problem [14, 25, 26]. The regression models were well-fit as indicated by a high r-square value.
Our study was completed using a cohort of individuals 65 years and older who have lower IGF-I levels, thus, our results should not be generalized to cohorts of varying age groups because assays may correlate at lower ranges but not in higher ranges of IGF protein levels. In addition, we recommend that whenever possible, all samples of a study should be run using a single method. This is particularly important for studies of population cohorts that identify disease associations associated with modest between-person differences within the normal ranges of IGF-related analytes.
Highlights.
IGF assays commonly used in epidemiological research do not always produce perfectly comparable levels of IGF-I, IGFBP-1 and IGFBP-3.
There is substantial agreement between measures of IGF proteins obtained using different assays (kw, 0.78 for IGF-I, 0.69 for IGFBP-1, 0.76 for IGFBP-3).
Use of regression equations determined in this study can be used to obtain comparable IGF measures for the assays mentioned in this study.
Such conversions should only be used when absolutely necessary in epidemiological studies and should not be used when assay measures are used to diagnose or monitor treatment in the clinical setting.
Acknowledgments
Funding Source: The project described was supported by the MSTP Training Grant, T32-GM007288 and by the National Center for Advancing Translational Sciences (NCATS), a component of the National Institutes of Health (NIH), through CTSA grant numbers UL1TR000086, TL1RR000087, and KL2TR000088. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. Additional support by contracts HHSN268201200036C, N01HC85239, N01 HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, and grant HL080295 from the National Heart, Lung, and Blood Institute (NHLBI), with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided by AG023629 from the National Institute on Aging [27]. A full list of principal CHS investigators and institutions can be found at http://www.chs-nhlbi.org.
Footnotes
DISCLOSURE STATEMENT: The authors have nothing to disclose.
Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Le Roith D. Seminars in medicine of the beth israel deaconess medical center. Insulin-like growth factors. N Engl J Med. 1997;336:633–640. doi: 10.1056/NEJM199702273360907. [DOI] [PubMed] [Google Scholar]
- 2.Binkert C, Landwehr J, Mary JL, Schwander J, Heinrich G. Cloning, sequence analysis and expression of a cdna encoding a novel insulin-like growth factor binding protein (igfbp-2) EMBO J. 1989;8:2497–2502. doi: 10.1002/j.1460-2075.1989.tb08386.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimasaki S, Shimonaka M, Zhang HP, Ling N. Identification of five different insulin-like growth factor binding proteins (igfbps) from adult rat serum and molecular cloning of a novel igfbp-5 in rat and human. J Biol Chem. 1991;266:10646–10653. [PubMed] [Google Scholar]
- 4.Chan JM, Stampfer MJ, Giovannucci E, Gann PH, Ma J, Wilkinson P, Hennekens CH, Pollak M. Plasma insulin-like growth factor-i and prostate cancer risk: A prospective study. Science. 1998;279:563–566. doi: 10.1126/science.279.5350.563. [DOI] [PubMed] [Google Scholar]
- 5.Gunter MJ, Hoover DR, Yu H, Wassertheil-Smoller S, Rohan TE, Manson JE, Howard BV, Wylie-Rosett J, Anderson GL, Ho GY, Kaplan RC, Li J, Xue X, Harris TG, Burk RD, Strickler HD. Insulin, insulin-like growth factor-i, endogenous estradiol, and risk of colorectal cancer in postmenopausal women. Cancer Res. 2008;68:329–337. doi: 10.1158/0008-5472.CAN-07-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan RC, Buzkova P, Cappola AR, Strickler HD, McGinn AP, Mercer LD, Arnold AM, Pollak MN, Newman AB. Decline in circulating insulin-like growth factors and mortality in older adults: Cardiovascular health study all-stars study. J Clin Endocrinol Metab. 2012;97:1970–1976. doi: 10.1210/jc.2011-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaplan RC, McGinn AP, Pollak MN, Kuller L, Strickler HD, Rohan TE, Cappola AR, Xue X, Psaty BM. High insulinlike growth factor binding protein 1 level predicts incident congestive heart failure in the elderly. Am Heart J. 2008;155:1006–1012. doi: 10.1016/j.ahj.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan RC, McGinn AP, Pollak MN, Kuller L, Strickler HD, Rohan TE, Xue X, Kritchevsky SB, Newman AB, Psaty BM. Total insulinlike growth factor 1 and insulinlike growth factor binding protein levels, functional status, and mortality in older adults. J Am Geriatr Soc. 2008;56:652–660. doi: 10.1111/j.1532-5415.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan RC, McGinn AP, Pollak MN, Kuller LH, Strickler HD, Rohan TE, Cappola AR, Xue X, Psaty BM. Association of total insulin-like growth factor-i, insulin-like growth factor binding protein-1 (igfbp-1), and igfbp-3 levels with incident coronary events and ischemic stroke. J Clin Endocrinol Metab. 2007;92:1319–1325. doi: 10.1210/jc.2006-1631. [DOI] [PubMed] [Google Scholar]
- 10.Rajpathak SN, He M, Sun Q, Kaplan RC, Muzumdar R, Rohan TE, Gunter MJ, Pollak M, Kim M, Pessin JE, Beasley J, Wylie-Rosett J, Hu FB, Strickler HD. Insulin-like growth factor axis and risk of type 2 diabetes in women. Diabetes. 2012;61:2248–2254. doi: 10.2337/db11-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rinaldi S, Peeters PH, Berrino F, Dossus L, Biessy C, Olsen A, Tjonneland A, Overvad K, Clavel-Chapelon F, Boutron-Ruault MC, Tehard B, Nagel G, Linseisen J, Boeing H, Lahmann PH, Trichopoulou A, Trichopoulos D, Koliva M, Palli D, Panico S, Tumino R, Sacerdote C, van Gils CH, van Noord P, Grobbee DE, Bueno-de-Mesquita HB, Gonzalez CA, Agudo A, Chirlaque MD, Barricarte A, Larranaga N, Quiros JR, Bingham S, Khaw KT, Key T, Allen NE, Lukanova A, Slimani N, Saracci R, Riboli E, Kaaks R. Igf-i, igfbp-3 and breast cancer risk in women: The european prospective investigation into cancer and nutrition (epic) Endocr Relat Cancer. 2006;13:593–605. doi: 10.1677/erc.1.01150. [DOI] [PubMed] [Google Scholar]
- 12.Yu H, Rohan T. Role of the insulin-like growth factor family in cancer development and progression. J Natl Cancer Inst. 2000;92:1472–1489. doi: 10.1093/jnci/92.18.1472. [DOI] [PubMed] [Google Scholar]
- 13.Massart C, Poirier JY. Serum insulin-like growth factor-i measurement in the follow-up of treated acromegaly: Comparison of four immunoassays. Clin Chim Acta. 2006;373:176–179. doi: 10.1016/j.cca.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 14.Granada ML, Ulied A, Casanueva FF, Pico A, Lucas T, Torres E, Sanmarti A. Serum igf-i measured by four different immunoassays in patients with adult gh deficiency or acromegaly and in a control population. Clin Endocrinol (Oxf) 2008;68:942–950. doi: 10.1111/j.1365-2265.2007.03120.x. [DOI] [PubMed] [Google Scholar]
- 15.Renehan AG, Zwahlen M, Minder C, O’Dwyer ST, Shalet SM, Egger M. Insulin-like growth factor (igf)-i, igf binding protein-3, and cancer risk: Systematic review and meta-regression analysis. Lancet. 2004;363:1346–1353. doi: 10.1016/S0140-6736(04)16044-3. [DOI] [PubMed] [Google Scholar]
- 16.Clemmons DR. Consensus statement on the standardization and evaluation of growth hormone and insulin-like growth factor assays. Clin Chem. 2011;57:555–559. doi: 10.1373/clinchem.2010.150631. [DOI] [PubMed] [Google Scholar]
- 17.Bidlingmaier M, Friedrich N, Emeny RT, Spranger J, Wolthers OD, Roswall J, Korner A, Obermayer-Pietsch B, Hubener C, Dahlgren J, Frystyk J, Pfeiffer AF, Doering A, Bielohuby M, Wallaschofski H, Arafat AM. Reference intervals for insulin-like growth factor-1 (igf-i) from birth to senescence: Results from a multicenter study using a new automated chemiluminescence igf-i immunoassay conforming to recent international recommendations. J Clin Endocrinol Metab. 2014;99:1712–1721. doi: 10.1210/jc.2013-3059. [DOI] [PubMed] [Google Scholar]
- 18.Newman AB, Arnold AM, Sachs MC, Ives DG, Cushman M, Strotmeyer ES, Ding J, Kritchevsky SB, Chaves PH, Fried LP, Robbins J. Long-term function in an older cohort--the cardiovascular health study all stars study. J Am Geriatr Soc. 2009;57:432–440. doi: 10.1111/j.1532-5415.2008.02152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, et al. The cardiovascular health study: Design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 20.Cushman M, Cornell ES, Howard PR, Bovill EG, Tracy RP. Laboratory methods and quality assurance in the cardiovascular health study. Clin Chem. 1995;41:264–270. [PubMed] [Google Scholar]
- 21.Friedrich N, Wolthers OD, Arafat AM, Emeny RT, Spranger J, Roswall J, Kratzsch J, Grabe HJ, Hubener C, Pfeiffer AF, Doring A, Bielohuby M, Dahlgren J, Frystyk J, Wallaschofski H, Bidlingmaier M. Age- and sex-specific reference intervals across life span for insulin-like growth factor binding protein 3 (igfbp-3) and the igf-i to igfbp-3 ratio measured by new automated chemiluminescence assays. J Clin Endocrinol Metab. 2014;99:1675–1686. doi: 10.1210/jc.2013-3060. [DOI] [PubMed] [Google Scholar]
- 22.Warrens MJ. Chance-corrected measures for 2 x 2 tables that coincide with weighted kappa. Br J Math Stat Psychol. 2011;64:355–365. doi: 10.1348/2044-8317.002001. [DOI] [PubMed] [Google Scholar]
- 23.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 24.Khosravi J, Diamandi A, Bodani U, Khaja N, Krishna RG. Pitfalls of immunoassay and sample for igf-i: Comparison of different assay methodologies using various fresh and stored serum samples. Clin Biochem. 2005;38:659–666. doi: 10.1016/j.clinbiochem.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Ranke MB, Osterziel KJ, Schweizer R, Schuett B, Weber K, Robbel P, Vornwald A, Blumenstock G, Elmlinger MW. Reference levels of insulin-like growth factor i in the serum of healthy adults: Comparison of four immunoassays. Clin Chem Lab Med. 2003;41:1329–1334. doi: 10.1515/CCLM.2003.203. [DOI] [PubMed] [Google Scholar]
- 26.Chestnut RE, Quarmby V. Evaluation of total igf-i assay methods using samples from type i and type ii diabetic patients. J Immunol Methods. 2002;259:11–24. doi: 10.1016/s0022-1759(01)00478-1. [DOI] [PubMed] [Google Scholar]
- 27.Bacha F, Arslanian SA. Ghrelin suppression in overweight children: A manifestation of insulin resistance? J Clin Endocrinol Metab. 2005;90:2725–2730. doi: 10.1210/jc.2004-1582. [DOI] [PubMed] [Google Scholar]