Abstract
This Viewpoint article provides a brief and selective summary of research on the chemical ecology underlying symbioses between bacteria and animals. Animals engage in multiple highly specialized interactions with bacteria that reflect their long coevolutionary history. The article focuses on a few illustrative but hardly exhaustive examples in which bacterially produced small molecules initiate a developmental step with important implications for the evolution of animals, provide signals for the maturation of mammalian immune systems, and furnish chemical defenses against microbial pathogens.
1. Introduction
All animals originated and evolved on a planet already teeming with bacteria, and the two have been competing, co-existing, and cooperating ever since. Most research on the relations between animals and bacteria has focused on pathological interactions - the ways bacteria cause disease. Natural products chemistry has played a decisive role in these studies through defining bacterial virulence factors and discovering naturally occurring antibacterial agents. The pioneering studies leading to penicillin and streptomycin ushered in the antibiotic era, and even in the current era (1981-2010) the number of new small molecule antibacterial agents developed from natural sources outnumbered those developed from synthetic molecules by 2:1.1,2 New technological and bioinformatic approaches to natural product discovery will likely increase their contributions to new drugs.3-6 The biological motivations for these studies have been almost exclusively medical, not ecological, and the roles of these antibiotics in the lives of their producers is even today very imperfectly understood.7
In the last few years studies on the non-pathogenic interactions between animals and bacteria have become increasingly frequent as biologists have begun to pose and answer questions dealing with the ways in which bacteria facilitated the origin, evolution, and development of animals.8 As bacteria largely sense and respond to the world around them with molecules, a complete answer to these questions requires a full description of the chemical ecology underlying bacteria-animal interactions, and providing this description creates greatly expanded opportunities for natural products chemists to deploy their skills on a fresh set of significant questions.
A chemical ecology approach to natural products has several noteworthy features. It inverts an increasingly common procedure in natural products chemistry by putting biological function ahead of chemical identification. Many current studies begin by identifying a molecule through metabolomic and/or bioinformatic analyses and then searching for a biological function. In contrast, an ecological approach begins with a function, and then identifies the responsible molecule(s) - an approach that reprises the procedure that led to many of our most useful drugs. An ecological approach also studies molecules in the physiological and ecological contexts in which they evolved, and knowing the relevant context enables approaches such as identifying inducers for triggering cryptic metabolite production, unraveling the evolutionary history of biosynthetic pathways, and suggesting medically relevant assays for further exploration and possible exploitation. This Viewpoint will highlight some recent studies that illustrate how bacterially produced small molecules affected the evolution, influence developmental decisions, and provide chemical defenses for animals.
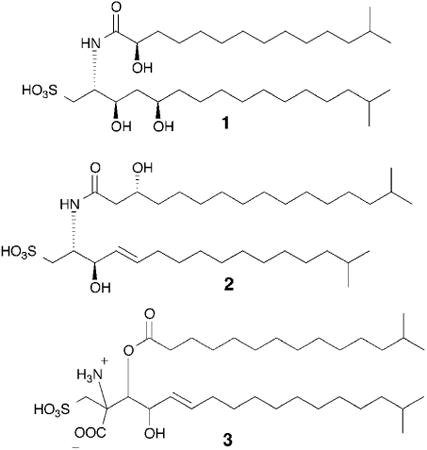
2. Evolution
Animals are multicellular, and the development of multicellularity was a major evolutionary step in the animal lineage.9 Multicellularity has evolved at least 25 times on Earth, but only once in animals. Since the 19th century, choanoflagellates have been considered a fitting candidate for understanding the transition to multicellularity, as phylogenetically they represent the last branch of unicellular organisms before multicellular animals emerged.10 Choanoflagellates, which subsist on bacteria, are found in fresh, brackish and marine environments. Some, most notably Salpingoeca rosetta, occur in both single-cell and colonial forms; the colonial form, which is called a rosette after its shape, is formed by incomplete cell division from a single founding cell.11 Surprisingly, the transition from the unicellular to colonial phenotype is induced by a bacterially produced signal. Using rosette formation as an assay, the inducing molecule, rosette-inducing factor 1 (RIF-1, 1) was identified as a sulfonolipid, a rare class of lipids that resemble sphingolipids.12 The complete stereostructure of RIF-1 had to be defined through total synthesis as its extraordinary potency – femtomolar! – made isolation of significant quantities problematic. The modular synthesis coupled with further isolation studies produced roughly a dozen RIF analogs, none of which had any discernible biological activity. This remarkably tight structure-activity relationship suggests a very restricted set of interactions between RIF-1 and its receptor.13 Characterizing the mechanism of action of RIF-1 could reveal homologous signaling pathways in other multicellular organisms, and the mechanism of action may even be general enough that examples could be found throughout the animal lineage. Additionally, while sulfonolipids are not well-studied molecules, they are produced by a number of different marine organisms (see examples 2 and 3).14,15 Investigation of these other sulfonolipid producers and their associations with marine eukaryotes, especially sponges, may reveal additional functions.
Not all signals produced by bacteria that play roles in the influencing the evolutionary steps along the animal lineage are small molecules. It has been known for several decades that bacterially produced signals induce larval settling and the initiation of cell differentiation in the marine invertebrate Hydroides elegans – a process that has fascinated developmental biologists and is also implicated in biofouling.16 Recently, the larval settlement inducer was identified as phage tail-like bacteriocins, which are contractile proteinaceous structures.17 While these sorts of molecules had previously been shown to have antibacterial, insecticidal, and anti-feeding activity, they had never been associated with an essential morphological change in an organism's life history.18-20 Further study of how H. elegans came to depend on a bacterial signal, while other closely related cnidarians settle in the absence of bacterial biofilms, will greatly enhance our understanding of the first steps in the evolution of the animal lineage.
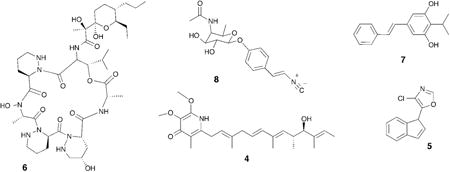
3. Defense
As noted in the introduction, bacteria produce a staggering array of antibiotics, and humans are not the only animals that have benefited from their biosynthetic fecundity.21 Beewolf digger wasps host symbiotic Streptomyces bacteria in specialized female glands, and they provide these bacteria to their larvae as they spin their protective cocoons.22,23 Examination of the cocoons revealed that the incorporated Streptomyces sp. produce a diverse set of antibiotics that serve to protect the cocoon, and more importantly its inhabitant, from a variety of microbial predators. While each of the antibiotics alone had moderate activity against a range of predators, the antibiotic cocktail produced by multiple bacterial species (piericidin 4 and streptochlorin 5 as examples), created a potent broad spectrum antibiotic activity, which argues that insect-bacterial systems evolved not only the use of bacterially produced antibiotics but also combination therapy long before humans.
Fungus-farming ants provide a variation on the beewolf system. Fungus-farming ants, as the name implies, cultivate a fungal food source that provides all of their nutrition. These cultivated fungi, which are grown in underground gardens by the ants, are plagued by a specialized pathogenic fungus that can overwhelm the fungal gardens and destroy the colony.24 The ants host a single strain of Actinobacteria, which are often housed and fed in highly derived anatomical features called crypts, which provide chemical defenses against the fungal pathogen. In an early study on this system, dentigerumycin (6), was isolated from the bacterial symbionts (Pseudonocardia sp.) of the ant Apterostigma dentigerum, and dentigerumycin selectively killed the Escovopsis sp. pathogen rather than the ants' fungal cultivar.25
The ants and wasps in the first two examples benefited from symbiotic bacteria, but other animals, like insectivorous Heterorhabditis nematode worms, use symbiotic bacteria (Photorhabdus luminescens) to prey upon insects.26 The bacteria live peacefully within their nematode host while it searches for insect larvae in the soil, but when the worm enters an insect larva, the bacteria emerge and begin producing insect toxins, an array of degrading proteases and esterases, antibiotics, and developmental signals to initiate feeding and reproduction in the worms. These nematodes are used as agricultural control agents, and the system attracted the attention of both biologists and chemists.27-29 One barrier to discovering the antibiotic (and other) molecules being produced was the differential lifestyles of the P. luminescens symbionts. While the pathogenic bacteria in the insect produced interesting molecules, the quiescent ones living in the worms were not nearly as prolific. In laboratory culture, the bacteria displayed little of their biosynthetic potential. The productive lifestyle could be triggered by a factor in insect hemolymph (L-proline), which could be used to induce the production of formerly cryptic metabolites in laboratory cultures. L-proline induction led to the identification of several upregulated metabolites - including stillbene-3 (7), an antibiotic and inhibitor of the insect innate immune system, the antibacterial nematophin, as well as a number of cryptic metabolites, such as the isocyanide rhabduscin (8), which disables a key enzyme in the insect's innate immune response.30,31 Cryptic metabolites – metabolites that are not produced in standard laboratory settings – are typically cryptic because their production is tightly regulated. In some cases, an environmental trigger like L-proline in the above example is sufficient to upregulate production, but in other cases the regulation have additional layers of repression that need to be lifted.32 Investigation of these types of interactions can not only give us access to novel natural products, but can further our understanding of how these molecules are regulated in the environment.
4. Development
Animal microbiomes, the microbial population living on or in an animal, have been largely studied using massive sequencing efforts. The data from the human microbiome project, for example, has already generated a million times more data than the initial human genome project, and these data have been useful in generating hypothesis free analyses including the discovery of new natural products. This approach is exemplified by a recent study on the biosynthetic potential of the microbiome, which used a bioinformatics driven approach to reveal that biosynthetic gene clusters encoding thiopeptide antibiotics are widely distributed in the human microbiota. Further, this study also reported a novel thiopeptide, lactocillin, that preferentially targets gram positive vaginal pathogens over commensal vaginal strains. While further investigation is required to determine the activity of lactocillin in vivo – this study suggests that the human microbiome could be a reservoir of novel therapeutics.33 Analyses springing from observational hypotheses are much rarer, but the sphingolipids that have been shown to mediate the interactions between Bacteroides, an abundant member of the human gut microbiome, and the human immune system form a very interesting exception. Multiple studies pointed to the ability of Bacteroides to antagonize invariant natural killer T-cells (iNKT), and later studies pinpointed bacterially produced sphingolipids as the relevant signal. Sphingolipids are important structural and signaling molecules in mammals, including humans – and ubiquitous sphingolipids such as sphingosine-1-phosphate (9) or ceramide (10) have been shown to regulate processes related to cell senescence, apoptosis, cell motility and inflammation.34 Sphingolipid diversity and function in bacteria, however, is largely unknown.35 These studies showed that bacterial sphingolipids regulate the iNKT cells through lipid-antigen presentation by the major histocompatibility complex protein, CD1d.36-38 This immunomodulatory activity can have important implications in the management, or exacerbation of conditions characterized by hyperactive immunological responses such as autoimmune disorders, or cell-mediated immunity against pathogens.
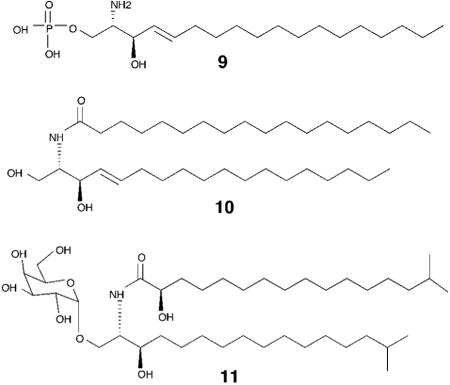
While the general scheme of iNKT regulation was understood, linking particular glycosphingolipids to specific immune responses had not been done. Recently, it was shown that a pervasive human (and mouse) gut microbiome member, Bacteroides fragilis, produces a glyocosphingolipid (α-GalCer) that protects against chemically induced colitis by restricting iNKT population size. This study also revealed that in mice, prenatal exposure to these bacterially produced glycosphingolipids is necessary for their full anti-proliferative effects, suggesting that exposure to certain bacterial species during early development is an important feature of the mammalian immune response.39,40 In a similar study also investigating the sphingolipid repertoire of B. fragilis, an α-GalCer was identified that acts as an agonist of iNKTs (11). While the net effect of these B. Fragilis glycosphingolipids on iNKT population size appears to vary between these studies, it is clear that these molecules are potent regulators of iNKT activation and that minor structural differences between these glycosphingolipids may lead to significant changes to their biological activity.41 It is also interesting to note that these lipid signals resemble those discussed in the earlier section on evolution. Sphingolipids are ubiquitous molecules in both bacteria and eukaryotes so it is likely not a coincidence that they would serve as excellent interkingdom signaling molecules.
Another very intriguing example of the ability of the human gut microbiome to influence human development comes from a study on the corrective effects of B. fragilis in a maternal immune activation mouse (MIA) – a model that recapitulates several key features of autism spectrum disorders (ASD). This study revealed that B. fragilis colonization of the gut could modulate the levels of several key metabolites known to be altered in the ASD mouse model.42 It would be interesting to see if B. fragilis small molecule metabolites could also generate the same metabolomic regulation – and such a study, or a similar study, would likely require a natural products chemist as part of the interdisciplinary team.
5. Future prospects
Since every animal – not to mention every plant and fungus – has its own microbiome, the number of possible interactions in these multilaterial systems is effectively unlimited. Study of these interactions will undoubtedly reveal dynamic chemical conversations – such as the production of metabolites in response to inducer molecules from another organism. These inducer molecules alter secondary metabolite expression to reveal previously “cryptic” molecules – expanding opportunities for novel structure discovery and enhancing our understanding of how ecological cues can regulate expression of secondary metabolites. A particularly promising set of interactions exists in the human microbiome and as the first round of DNA sequencing draws to a close, the task of annotating the incredibly complex but important set of chemical interactions that literally make our life on Earth possible now begins.
Acknowledgments
This article reflects the profound influence that Jerry Meinwald and Tom Eisner, two pioneers in chemical ecology, had on JC during two decades they were colleagues at Cornell University. The article has benefited from numerous conversations with Nicole King and members of her laboratory at the University of California, Berkeley and Cameron Currie and members of his laboratory at the University of Wisconsin. The insights and results of former and current members of the Clardy laboratory at Harvard Medical School are also acknowledged.
References
- 1.Newman DJ, Cragg GM. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cragg GM, Newman DJ. Biochim Biophys Acta. 2013;1830:3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter GT. Nat Prod Rep. 2014;31:711–717. doi: 10.1039/c3np70085b. [DOI] [PubMed] [Google Scholar]
- 4.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Charlop-Powers Z, Owen JG, Reddy BVB, Ternei MA, Brady SF. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1318021111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouslimani A, Sanchez LM, Garg N, Dorrestein PC. Nat Prod Rep. 2014;31:718–729. doi: 10.1039/c4np00044g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies J. J Antibiot. 2013;66:361–364. doi: 10.1038/ja.2013.61. [DOI] [PubMed] [Google Scholar]
- 8.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rokas A. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- 10.Richter DJ, King N. Annu Rev Genet. 2012;47:130919210319006. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 11.Fairclough SR, Dayel MJ, King N. Curr Biol. 2010;20:R875–6. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, King N. elife. 2012;1 doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beemelmanns C, Woznica A, Alegado RA, Cantley AM, King N, Clardy J. J Am Chem Soc. 2014;136:10210–10213. doi: 10.1021/ja5046692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corcelli A, Lattanzio VMT, Mascolo G, Babudri F, Oren A, Kates M. Appl Environ Microbiol. 2004;70:6678–6685. doi: 10.1128/AEM.70.11.6678-6685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takikawa H, Nozawa D, Kayo A, Muto SE, Mori K. J Chem Soc, Perkin Trans. 1999;1:2467–2477. [Google Scholar]
- 16.Hadfield MG. Annu Rev Marine Sci. 2011;3:453–470. doi: 10.1146/annurev-marine-120709-142753. [DOI] [PubMed] [Google Scholar]
- 17.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. Science. 2014 doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jabrane A, Sabri A, Compère P, Jacques P, Vandenberghe I, Van Beeumen J, Thonart P. Appl Environ Microbiol. 2002;68:5704–5710. doi: 10.1128/AEM.68.11.5704-5710.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst MRH, Glare TR, Jackson TA. J Bacteriol. 2004;186:5116–5128. doi: 10.1128/JB.186.15.5116-5128.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strauch E, Kaspar H, Schaudinn C, Dersch P, Madela K, Gewinner C, Hertwig S, Wecke J, Appel B. Appl Environ Microbiol. 2001;67:5634–5642. doi: 10.1128/AEM.67.12.5634-5642.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bode HB. Angew Chem Int Ed Engl. 2009;48:6394–6396. doi: 10.1002/anie.200902152. [DOI] [PubMed] [Google Scholar]
- 22.Kroiss J, Kaltenpoth M, Schneider B, Schwinger MG, Hertweck C, Maddula RK, Strohm E, Svatos A. Nat Chem Biol. 2010;6:261–263. doi: 10.1038/nchembio.331. [DOI] [PubMed] [Google Scholar]
- 23.Seipke RF, Kaltenpoth M, Hutchings MI. FEMS Microbiology Reviews. 2012;36:862–876. doi: 10.1111/j.1574-6976.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 24.Currie CR. Annu Rev Microbiol. 2001;55:357–380. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- 25.Oh DC, Poulsen M, Currie CR, Clardy J. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterfield NR, Ciche T, Clarke D. Annu Rev Microbiol. 2009;63:557–574. doi: 10.1146/annurev.micro.091208.073507. [DOI] [PubMed] [Google Scholar]
- 27.Goodrich-Blair H, Clarke DJ. Mol Microbiol. 2007;64:260–268. doi: 10.1111/j.1365-2958.2007.05671.x. [DOI] [PubMed] [Google Scholar]
- 28.Schild HA, Fuchs SW, Bode HB, Grünewald B. Appl Environ Microbiol. 2014;80:2484–2492. doi: 10.1128/AEM.04049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brachmann AO, Joyce SA, Jenke-Kodama H, Schwär G, Clarke DJ, Bode HB. Chembiochem. 2007;8:1721–1728. doi: 10.1002/cbic.200700300. [DOI] [PubMed] [Google Scholar]
- 30.Crawford JM, Portmann C, Zhang X, Roeffaers MBJ, Clardy J. Proc Natl Acad Sci USA. 2012;109:10821–10826. doi: 10.1073/pnas.1201160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crawford JM, Kontnik R, Clardy J. Curr Biol. 2010;20:69–74. doi: 10.1016/j.cub.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kontnik R, Crawford JM, Clardy J. ACS Chem Biol. 2010;5:659–665. doi: 10.1021/cb100117k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, Fischbach MA. Cell. 2014;158:1402–1414. doi: 10.1016/j.cell.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannun YA, Obeid LM. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 35.Olsen I, Jantzen E. Anaerobe. 2001;7:103–112. [Google Scholar]
- 36.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MREI, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 37.Brennan CA, Hunt JR, Kremer N, Krasity BC, Apicella MA, McFall-Ngai MJ, Ruby EG. elife. 2014;3:e01579. doi: 10.7554/eLife.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 39.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL. Cell. 2014;156:123–133. doi: 10.1016/j.cell.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, Fischbach MA. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


