Abstract
High-profile studies have provided conflicting results regarding the involvement of the Omi/HtrA2 gene in Parkinson’s disease (PD) susceptibility. Therefore, we performed a large-scale analysis of the association of common Omi/HtrA2 variants in the Genetic Epidemiology of Parkinson’s disease (GEO-PD) consortium.
GEO-PD sites provided clinical and genetic data including affection status, gender, ethnicity, age at study, age at examination (all subjects); age at onset and family history of PD (patients). Genotyping was performed for the five most informative SNPs spanning the Omi/HtrA2 gene in approximately 2–3 kb intervals (rs10779958, rs2231250, rs72470544, rs1183739, rs2241028). Fixed as well as random effect models were used to provide summary risk estimates of Omi/HtrA2 variants.
The 20 GEO-PD sites provided data for 6378 cases and 8880 controls. No overall significant associations for the five Omi/HtrA2 SNPs and PD were observed using either fixed effect or random effect models. The summary odds ratios ranged between 0.98 and 1.08 and the estimates of between-study heterogeneity were not large (non-significant Q statistics for all 5 SNPs; I2 estimates 0–28%). Trends for association were seen for participants of Scandinavian descent for rs2241028 (OR 1.41, p = 0.04) and for rs1183739 for age at examination (cut-off 65 years; OR 1.17, p = 0.02), but these would not be significant after adjusting for multiple comparisons and their Bayes factors were only modest.
This largest association study performed to define the role of any gene in the pathogenesis of Parkinson’s disease revealed no overall strong association of Omi/HtrA2 variants with PD in populations worldwide.
Keywords: Omi, HtrA2, Genetics, Parkinson’s disease, PARK13
1. Introduction
The constant decline of genotyping costs and the development of robust analytical methods have contributed to the report of many susceptibility genes for complex diseases (Ioannidis et al., 2009). However, most of these genetic associations have not been replicated consistently across different populations (McCarthy et al., 2008). Indeed for Parkinson’s disease (PD), as of April 10, 2009, at least 594 studies of 402 candidate genes and 1892 polymorphisms have yielded only 16 genes with nominally significant associations (p < 0.05) upon evaluation of the published data (http://www.pdgene.org) and even these associations are often of borderline significance and almost always correspond to small genetic effects. Furthermore, at least three published genome-wide association studies of PD have queried several hundred thousand SNPs without yielding solidly reproducible associations (Elbaz et al., 2006; Evangelou et al., 2009; Fung et al., 2006; Maraganore et al., 2005; Pankratz et al., 2009). The reasons for non-replication probably include overestimation of effect size and significance by initial studies, small sample sizes, and disease and population heterogeneities (Zondervan and Cardon, 2004, 2007). Given this setting, large multi-centred pooled analyses have gained increasing interest. Large consortia can overcome the above mentioned limitations of genetic association studies and publication biases that may limit the value of meta-analyses (Chanock et al., 2007). For example, a consortium analysis has validated that a REP-1 promoter variant in the alpha-synuclein gene (SNCA) confers susceptibility to PD in populations worldwide (Maraganore et al., 2006).
A number of genetic linkage studies in high-profile journals have supported the role of mitochondria in the pathogenesis of PD. Mutations in Parkin (PARK2), DJ-1, and PTEN-induced kinase 1 (PINK1) cause autosomal recessive forms of PD, possibly via an impairment of mitochondrial function and/or dynamics (Bonifati et al., 2003; Kitada et al., 1998; Valente et al., 2004). Subsequently, point mutations in the Omi/HtrA2 gene were reported in PD (Strauss et al., 2005). These mutations cause a loss of serine pro-tease function of the human Omi/HtrA2 protein in vitro, and paralleled observations in rodents, where loss of Omi/HtrA2 function was critically related to neurodegeneration mimicking relevant clinical aspects of parkinsonism (Jones et al., 2003; Strauss et al., 2005). In this context, in vitro and in vivo studies revealed mitochondrial dysfunction in loss of function models of Omi/HtrA2 as a primary mechanism for neurodegeneration (Jones et al., 2003; Strauss et al., 2005).
Subsequently, Bogaerts and colleagues performed an Omi/HtrA2 mutation analysis in a large Belgian cohort. They identified a novel amino acid exchange that was not observed in controls, leading to potential stabilization of the inactive Omi/HtrA2 protein (Bogaerts et al., 2008). Moreover, the authors identified nine patients carrying specific heterozygous mutations in the 5′ and 3′ regulatory region, suggesting that mutations in the Omi/HtrA2 promoter affect the transcriptional activity of the gene (Bogaerts et al., 2008). However, a large sequence-based study on a North-American PD patient-control series detected the original pathogenic Omi/HtrA2 variants in healthy control subjects (Simon-Sanchez and Singleton, 2008); whether this occurrence is due to reduced penetrance as observed for other PD mutations was unclear.
Therefore, to determine the relevance of Omi/HtrA2 variation in susceptibility to PD, we conducted the largest study in terms of number of participants ever conducted in PD genetics for any gene. Our study is a large multi-centre study which includes over 15,000 subjects from 20 sites representing 14 countries and 4 continents.
2. Methods
2.1. Sampling
As of April 10, 2009 the Genetic Epidemiology of Parkinson’s Disease (GEO-PD) consortium includes investigators from 35 sites representing 22 countries, and six continents. Together, we have pledged to share clinical and genetic data and DNA bio specimens for more than 20,000 PD cases and 20,000 controls, when considering the anticipated total recruitment of all sites by the end of 2010. All GEO-PD sites were invited to participate in this study. A total of 20 teams representing 14 countries and 4 continents agreed to participate and contributed clinical and genotypic data for a total of 15,258 individuals (6378 cases and 8880 controls). Of these 2372 individuals were part of a previous study (Ross et al., 2008).
2.2. Genotyping
A total of five SNPs were selected for genotyping: rs10779958, rs2231250, rs72470544, rs1183739 and rs2241028, listed in order from 5′ to 3′ end of the gene. It has been shown previously that tagSNPs selected from HapMap sufficiently capture the common genetic variation in different populations (de Bakker et al., 2005). Therefore, the HapMap was used to select tagSNPs with following criteria: (i) SNP with an r2 threshold of 0.8, (ii) coding SNPs or (iii) SNPs in the 5′ and 3′ UTR. Apart from that, the gene is enriched with rare variant(s) which were described previously (Strauss et al., 2005). Genotyping was done either by each site, a combination of sites, or by a commercial contract. The genotyping platforms employed include TaqMan® SNP Genotyping Assays (Applied Biosystems Inc.), MassARRAYTM Analyzer Compact (Sequenom) and Illumina GoldenGate (Illumina Inc.). Data from sites that showed missing rates >5% were excluded from further analyses. An exact test was used to test for Hardy Weinberg equilibrium (HWE) distortion in controls. Deviation from HWE was considered significant for p < 0.05. We observed deviations for different SNPs and different sites. To control for potential genotypic error, we asked sites that provided data for SNPs that were not in HWE to repeat the genotyping. Site-specific data for SNPs that still deviated significantly from HWE were removed from further analysis.
2.3. Statistical analysis
Effect estimates based on major versus minor allele contrast were computed. Results were then synthesised across different sites using both fixed effect and random effect models. Fixed effect models assume that the odds ratio is the same in all sites and that observed differences are due to chance alone. Random effect models allow that the odds ratios might be different due to heterogeneity across different sites. Random effect calculations take into account the estimated between-study heterogeneity. Heterogeneity between sites was tested using the χ2-based Q statistic and was also assessed with the I2 metric, which ranges from 0% to 100% I2 is estimated by the ratio (Q–df)/Q, where Q is χ2 based statistic and df is degree of freedom. Typically estimates of I2 < 25% are considered to reflect little or no heterogeneity, 25–50% moderate heterogeneity, 50–75% large heterogeneity and >75% very large heterogeneity. It should be acknowledged that I2 can have large uncertainty in its estimation (Ioannidis et al., 2007), especially for variants with low minor allele frequency. Thus it represents a tentative working interpretation for heterogeneity.
The main analyses considered all sites and populations regardless of ancestry. In secondary analyses, we examined separately populations of Caucasian versus Asian descent due to relevant differences in genotypic distribution; and further evaluated within Caucasians specifically in populations of Scandinavian descent. Omi/HtrA2 is located on chromosome 2p close to the PARK3 locus that was previously linked to families with Northern European descent (OMIM %602404). We also evaluated subgroup effects for PD with age at examination (cut-off 65 years).
Given that 5 SNPs were tested, nominal statistical significance was claimed for a threshold of p < 0.01 (i.e. 0.05/5). Power calculations showed that with 6000 cases and 9000 controls, our study would have at least 86% power to detect an allele-based odds ratio of 1.15 for minor allele frequencies of 10% or higher for alpha = 0.01. Power would be only 20% for a minor allele frequency of 2% and odds ratio of 1.15, but it would be 78% for the same minor allele frequency of 2% and odds ratio of 1.3. Furthermore, for associations with uncorrected p < 0.05, we also expressed the strength of the putative associations by calculating the respective Bayes factor, assuming that average genetic effects for PD susceptibility with common variants may reflect odds ratios of 1.30 and using a lump-and-smear prior, as described by Ioannidis (Ioannidis, 2008).
Meta-analyses were performed using Review Manager 4.2.7 and STATA 10.0 (Stata Corp., College Station, TX, USA). Reporting of methods and results follows the STREGA guidance (Little et al., 2009).
3. Results
3.1. Database
Twenty sites contributed 6378 cases and 8880 controls combined (Table 1). Most of the cases were of Caucasian ancestry (71.5%), but some were of Asian ancestry (28.5%). No subjects of African ancestry were included in the study, since a Nigerian site of GEO-PD could not contribute data and a South African site only recently joined the GEO-PD consortium. To maintain homogeneity in the Caucasian sample, 0.2% of those subjects who reported a mixed ancestry were removed from further analysis. The proportion of men and women ranged from 42% to 58% across different participating sites (Table 1). The median age at onset of PD in our studied population was 61 years and the median age at study in the overall sample was 67 years.
Table 1.
Description of datasets contributed by each study site.
| Site | Country | Total sample (N) | Case | Control | Male (%) | Mean age of onset | Mean age at study | Familial PD (n) | Diagnostic criteria |
|---|---|---|---|---|---|---|---|---|---|
| Annesi | Italy | 368 | 184 | 184 | 176 (47.8) | 60.09 | 64.77 | NA | UKPDBB |
| Brice | France | 545 | 293 | 252 | 323 (59.2) | 47.53 | 57.79 | NA | UKPDBB |
| Elbaz | France | 710 | 209 | 501 | 415 (58.4) | 63.49 | 66.93 | 45 | Bower |
| Ferrarese | Italy | 182 | 87 | 95 | 108 (59.3) | 61.17 | 64.48 | 15 | Gelb |
| Lynch | Ireland | 369 | 184 | 185 | 145 (39.2) | 49.55 | 63.34 | 21 | UKPDBB |
| Opala | Poland | 223 | 101 | 122 | 145 (65.0) | 63.68 | 72.41 | 13 | UKPDBB |
| Farrer/Wszolek | US | 536 | 312 | 224 | 308 (57.4) | 63.85 | 73.67 | 102 | UKPDBB |
| Klein | Germany | 458 | 226 | 193 | 227 (49.5) | 48.51 | 61.28 | NA | UKPDBB |
| Maraganore | US | 2155 | 1076 | 1079 | 1299 (60.2) | 61.64 | 66.87 | 847 | Bower |
| Mellick | Australia | 547 | 273 | 274 | 270 (49.3) | 62.0 | 67.95 | 107 | Bower |
| Aasly | Norway | 1349 | 391 | 958 | 677 (50.1) | 58.03 | 72.65 | 97 | UKPDBB |
| Valente | Italy | 288 | 192 | 96 | 143 (49.6) | 58.30 | 67.87 | 2 | UKPDBB |
| Wirdefeldt | Sweden | 273 | 95 | 178 | 142 (52) | 65.88 | 75.20 | 69 | Gelb |
| Hadjigeorgiou | Greece | 600 | 300 | 300 | 343 (57.1) | 64.36 | 69.64 | NA | Bower |
| V. Broeckhoven | Belgium | 539 | 266 | 273 | 310 (57.5) | 59.80 | 65.14 | 38 | Pals/Engelborghs |
| Sharma/Gasser | Germany | 1700 | 678 | 1020 | 977 (57.4) | 55.33 | 54.94 | NA | UKPDBB |
| Toda | Japan | 3509 | 988 | 2521 | NA | NA | NA | NA | UKPDBB |
| Hattori | Japan | 373 | 240 | 133 | NA | 37.76 | 47.46 | NA | UKPDBB |
| Tan | Singapore | 368 | 179 | 189 | 173 (47.0) | 61.44 | 66.52 | NA | UKPDBB |
| Lin | Taiwan | 200 | 100 | 100 | 102 (51) | 59.29 | 64.59 | 6 | UKPDBB |
| Total | 15298 | 6378 | 8879 | 59.72 | 65.66 |
UKPDBB, United Kingdom Parkinson’s Disease Brain Bank; NA, not available.
The distribution of allele frequencies for each SNP and at each participating site is shown in Table 2. The genotypes were in HWE in controls for most of sites and for most of the SNPs, however, data from a total of 5 sites had to be excluded either due to HWE deviation or failure to genotype SNP rs72470544 (two sites showed HWE distortion and genotyping was not successful for remaining three sites) and 4 sites were excluded similarly for rs1183739 (two sites showed HWE distortion and genotyping was not successful for the remaining two sites).
Table 2.
Allele frequencies of the five SNP (rs10779958, rs2231250, rs1183739, rs2241028 and rs72470544) in cases and controls for each site.
| Sites | Cases (major /minor allele)
|
Controls (major /minor allele)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs10779958 | rs2231250 | rs1183739 | rs2241028 | rs72470544 | rs10779958 | rs2231250 | rs1183739 | rs2241028 | rs72470544 | |
| Annesi | 306/56 | 301/55 | 327/35 | 348/14 | NA | 293/67 | 292/68 | 318/44 | 352/14 | NA |
| Brice | 501/85 | 499/87 | NA | 551/33 | 569/15 | 434/70 | 434/70 | NA | 479/25 | 499/5 |
| V. Broekhoven | 428/94 | 423/93 | 446/78 | 491/39 | 511/15 | 448/80 | 450/86 | 453/91 | 515/31 | 533/13 |
| Elbaz | 357/57 | 357/57 | 354/62 | 399/17 | 409/7 | 839/145 | 839/145 | 855/133 | 936/62 | 966/20 |
| Ferrarese | 145/27 | 144/28 | NA | 165/7 | 171/1 | 154/34 | 153/35 | NA | 179/9 | 185/3 |
| Lynch | 316/52 | 317/53 | 280/74 | 347/25 | 350/18 | 306/64 | 304/64 | 306/52 | 340/30 | 353/15 |
| Opala | 176/26 | 176/26 | 165/27 | 192/10 | 190/8 | 216/28 | 216/28 | 188/44 | 234/12 | 231/11 |
| Farrer/Wszolek | 524/100 | 514/98 | 431/87 | 585/39 | 586/40 | 371/77 | 369/75 | 363/69 | 421/25 | 429/19 |
| Klein | 381/71 | 384/68 | 377/75 | 432/20 | 592/8 | 379/85 | 380/84 | 386/78 | 425/39 | 598/2 |
| Maraganore | 1845/307 | 1846/306 | 1814/338 | 2024/128 | NA | 1853/305 | 1853/305 | 1819/339 | 2023/135 | NA |
| Mellick | 444/94 | 442/94 | 458/82 | 503/37 | NA | 441/85 | 445/85 | 450/82 | 502/34 | NA |
| Aasly | 667/113 | 663/107 | 629/115 | 738/38 | 716/28 | 1614/302 | 1608/300 | 1495/293 | 1774/128 | 1786/110 |
| Valente | 300/62 | 300/62 | 320/42 | 344/18 | 350/12 | 124/30 | 124/30 | 135/19 | 142/12 | 147/7 |
| Larrisa | 470/114 | 472/114 | 512/74 | NA | 592/8 | 500/90 | 508/92 | 512/84 | NA | 598/2 |
| Wirdefeldt | 159/27 | 162/26 | 150/38 | 183/7 | 151/5 | 303/53 | 301/51 | 287/55 | 335/21 | 301/7 |
| Sharma/Gasser | 1114/210 | 1144/212 | 1176/182 | 1282/76 | 1315/45 | 1731/299 | 1727/295 | 1694/330 | 1912/116 | 1953/69 |
| Toda | 1684/5903 | NA | NA | 1527/5489 | NA | 292/1115 | NA | NA | 407/1487 | NA |
| Hattori | 400/618 | NA | NA | NA | NA | 80/128 | NA | NA | NA | NA |
| Tan | 289/611 | NA | NA | 319/653 | NA | 69/127 | NA | NA | 39/85 | NA |
| Lin | 165/343 | 165/343 | NA | 169/334 | NA | 29/47 | 29/47 | NA | 25/56 | NA |
NA: not applicable either due to significant HWE deviations or genotype failure that led to the exclusion of these data from further analyses.
3.2. Overall results
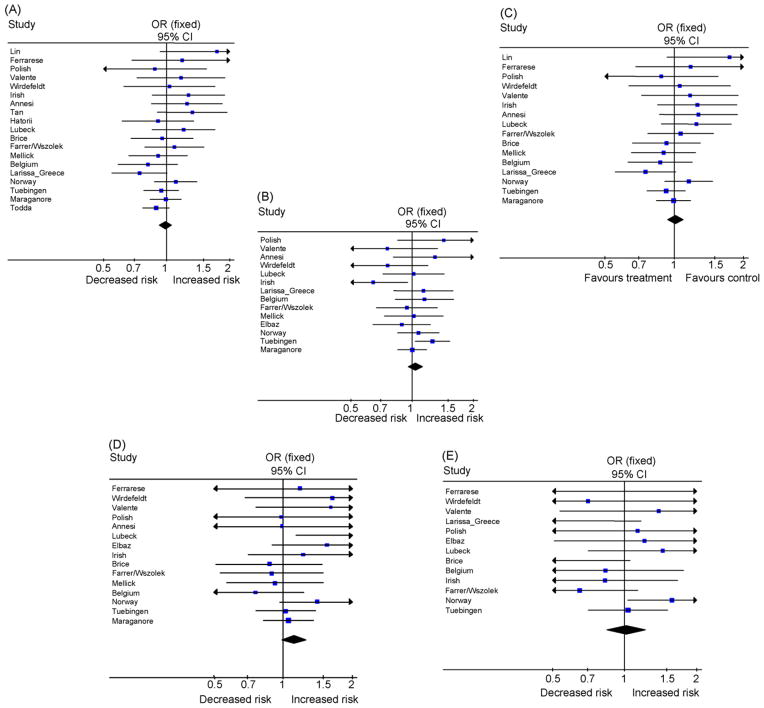
The overall results did not reveal nominally significant associations for any of the SNPs with PD, either for random effect or fixed effect models (Table 3). The odds ratios ranged from 0.98 to 1.02 (Fig. 1) and no p-value was less than 0.07, even without adjustment for 5 comparisons. We observed no substantial heterogeneity for three SNPs: rs2241028, rs2231250 and rs10779958 (I2 estimates ranged from 0% to 8%), while for rs1183739 and rs72470544 the I2 estimates were larger (23% and 28%, respectively), but not significant. The Q test was non-statistically significant for all 5 SNPs.
Table 3.
Summary effect estimates and confidence interval for Omi/HtrA2 gene.
| SNP | Overall | Het. P (I2) | Scandinavian | Age at examination* | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||||
| Sites | RE OR (95% CI) | FE OR (95% CI) | RE OR (95% CI) | FE OR (95% CI) | Het. P (I2) | RE OR (95% CI) | FE OR (95% CI) | Het. P (I2) | |||
| rs72470544 | 13 | 0.98(0.77–1.26) | 1.02(0.84–1.22) | 0.17(28%) | 1.27(0.63–2.56) | 1.45(0.97–2.15) | 0.20(39%) | 0.96(0.72–1.27) | 0.96(0.72–1.27) | 0.35(9%) | |
| rs10779958 | 20 | 0.99 (0.93–1.06) | 0.99(0.93–1.06) | 0.41(3%) | 1.02(0.82–1.27) | 1.03(0.85–1.25) | 0.33(9%) | 1.03(0.91–1.16) | 1.03(0.92–1.17) | 0.56(0%) | |
| rs2231250 | 17 | 1.01(0.94–1.09) | 1.02(0.94–1.09) | 0.49(0%) | 1.03(0.79–1.34) | 1.07(0.87–1.30) | 0.27(24%) | 1.02(0.91–1.16) | 1.03(0.91–1.16) | 0.56(0%) | |
| rs1183739 | 14 | 1.03(0.93–1.13) | 1.04(0.96–1.12) | 0.20(23%) | 1.01(0.83–1.23) | 1.01(0.83–1.23) | 0.39(0%) | 1.17(1.03–1.33) | 1.17(1.03–1.33) | 0.76(0%) | |
| rs2241028 | 18 | 1.07(0.99–1.17) | 1.08(0.99–1.17) | 0.58(0%) | 1.41(1.02–1.94) | 1.41(1.02–1.94) | 0.90(0%) | 1.05(0.76–1.44) | 1.01(0.81–1.26) | 0.18(36%) | |
FE, fixed effects; RE, random effects; CI, confidence interval; Het., heterogeneity (Q statistic); (*) age at examination <65 years; bold: p < 0.05.
Fig. 1.
A panel showing the forest plot for each SNP. The summary effect estimate is indicated by the diamond (A) Forest plot for SNPs rs10779958, (B) rs1183739, (C) rs2231250, (D) rs2241028 and (E) rs72470544.
3.3. Subgroup analyses
Subgroup analyses on Caucasian versus Asian ancestry samples did not reveal any subgroup differences that would be beyond chance (data not shown). We observed a non-significant trend for association with SNP rs2241028 in our Caucasian samples (p = 0.07 uncorrected for multiple comparisons). When restricting the analysis to the Scandinavian sites, including Sweden and Norway, the odds ratio was 1.44 (95% CI, 1.02–2.02, p = 0.04 uncorrected for multiple comparisons). Furthermore, we identified a subgroup of 238 subjects (143 cases and 95 controls) of only Swedish or Norwegian descent from our North-American site (Mayo Clinic /Maraganore). Incorporating these data also into the analysis of Scandinavian descent population lead to marginal change in odds ratio from 1.44 to 1.41 (95% CI, 1.02–1.94, p = 0.04 uncorrected for multiple comparisons). Another SNP (rs72470544) also had a trend for association by fixed effects only in Scandinavian populations, but this was not formally significant (odds ratio 1.45, p = 0.07 uncorrected). These effects were not different beyond chance compared with the effects in non-Scandinavian ancestry participants.
Finally, we observed a nominally significant association of SNP rs1183739 with age at examination (cut-off 65 years) (odds ratio 1.17 95% CI 1.03–1.33; p = 0.02), but this was also not significant when corrected for multiple comparisons and the difference against non-young age at examination (odds ratio 0.97 95% CI 0.87–1.07; p = 0.52) was not beyond chance.
For both the rs2241028 and rs1183739 subgroup effects in Scandinavian descent populations and young onset PD, the estimated Bayes factors were 0.4, suggesting that the nominally significant results increased 2.5-fold (1/0.4) the odds that the association was true compared with before running the study.
4. Discussion
Our study revealed no evidence for an overall association of common variants in the Omi/HtrA2 gene with PD across populations worldwide. Our generally “negative” findings employing a candidate gene approach are also consistent with the findings of three published genome-wide association studies of PD, none of which highlighted Omi/HtrA2 as a susceptibility gene in PD (Fung et al., 2006; Maraganore et al., 2005; Pankratz et al., 2009).
Our subgroup analyses should be interpreted with caution, however, we cannot fully exclude the possibility that Omi/HtrA2 may confer susceptibility to PD in persons of Scandinavian descent. As shown in a large study on genetic variation among Europeans the geographic distribution of a given sample needs to be considered during the evaluation of genetic association studies (Novembre et al., 2008). In this context the observed trend for marker rs2241028 as a susceptibility factor for PD in Caucasians was more pronounced in the Scandinavian population and was also seen when we included participants of Scandinavian descent from the USA. The same tentative level of support is offered by the data for the borderline association of rs1183739 in the subgroup of PD patients with an earlier age at examination. The results on both of these subgroup effects increase the odds of these associations being true only by about 2.5-fold compared with before running the study, therefore they are still very likely to reflect false-positive results in subgroup analyses, but they should not be entirely dismissed and it would be interesting to have some additional studies in Scandinavian populations and young-age PD respectively.
Our genetic study shows, that as for other established PD genes, i.e. Parkin (PARK2) and PINK1 (PARK6), also for Omi/HtrA2 no clear genetic association of sequence variations with PD was observed among different populations. By contrast in vitro and in vivo studies strongly implicate loss of Omi/HtrA2 protein in disrupted mitochondrial homeostasis and subsequent cell death; these studies have included a knockout mouse model that recapitulated a range of relevant clinical aspects of PD (Jones et al., 2003; Strauss et al., 2005). Moreover, recent pathoanatomical studies indicate that Omi/HtrA2 represents a consistent pathological marker for neurodegeneration in different alpha-synucleinopathies (Kawamoto et al., 2008). Loss of Omi/HtrA2 function may contribute to a broader spectrum of neurodegeneration, as decreased levels of Omi/HtrA2 were demonstrated in brains of Huntington’s disease patients (Inagaki et al., 2008). Therefore, while our genetic association studies provide no consistent support for an association of Omi/HtrA2 and PD, functional studies suggest that further study of this gene in the context of neurodegenerative disorders is justified. We cannot exclude the possibility that other neurodegenerative diseases besides PD may be influenced by Omi/HtrA2 variations. Moreover, we should acknowledge that here we have examined SNPs that have allele frequencies of 2% or higher. We cannot exclude the possibility that rare mutations with frequency of 1% or less may associate with PD.
Acknowledging these caveats, our study is the largest in sample size conducted on PD genetics for any gene to date and the results are strong enough to suggest that these variants are unlikely to be a clinically important determinant of PD risk. For the most uncommon variants among those examined (those with minor allele frequency in the range of 2%), we cannot exclude subtle associations, but power was adequate to exclude odds ratios of 1.30 or higher even for these relatively uncommon variants. Our approach demonstrates the utility of large-scale consortia in clarifying controversial associations that have been hotly debated in the literature.
Acknowledgments
Australia: G.T. Sutherland (Eskitis Institute for Cell and Molecular Therapies, School of Biomolecular & Physical Sciences, Griffith University, Nathan, QLD), G.A. Siebert (Eskitis Institute for Cell and Molecular Therapies, School of Biomolecular & Physical Sciences, Griffith University, Nathan, QLD).
Belgium: Christine Van Broeckhoven PhD DSc (Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; and Laboratory of Neurogenetics, Institute Born-Bunge; and University of Antwerp, Antwerpen), Jessie Theuns, PhD (Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; and Laboratory of Neurogenetics, Institute Born-Bunge, and University of Antwerp, Antwerpen), David Crosiers MD (Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; and Laboratory of Neurobiology, Institute Born-Bunge; and University of Antwerp; and Department of Neurology, University Hospital of Antwerp, Antwerpen), Barbara Pickut, MD (Memory Clinic and Division of Neurology, ZNA Middelheim, Antwerpen), Philippe Pals, MD, PhD (Laboratory of Neurobiology, Institute Born-Bunge; and Department of Neurology, University Hospital Antwerp; Antwerpen), Sebastiaan Engelborghs, MD, PhD (Laboratory of Neurochemistry and Behavior, Institute Born-Bunge; and University of Antwerp; and Memory Clinic and Division of Neurology, ZNA Middelheim, Antwerpen), Karen Nuytemans (Neurodegenerative Brain Diseases Group, Department of Molecular Genetics, VIB; and Laboratory of Neurogenetics, Institute Born-Bunge; and University of Antwerp, Antwerpen), Peter P. De Deyn, MD, PhD (Laboratory of Neurochemistry and Behavior, Institute Born-Bunge; and University of Antwerp; and Memory Clinic and Division of Neurology, ZNA Middelheim, Antwerpen), Patrick Cras, MD, PhD (Laboratory of Neurobiology, Institute Born-Bunge; and University of Antwerp; and Department of Neurology, University Hospital of Antwerp, Antwerpen).
Canada: Ekaterina Rogaeve, MD (Centre for Research in Neurodegenerative Diseases, Department of Medicine, Division of Neurology, University of Toronto, ON, Canada).
France: From the French Parkinson’s Disease Genetics Study Group: Y. Agid, A-M. Bonnet, M. Borg, A. Brice, E. Broussolle, P. Damier, A. Destée, A. Dürr, F. Durif, S. Lesage, E. Lohmann, P. Pollak, O. Rascol, F. Tison, C. Tranchant, F. Viallet, M. Vidailhet. Also, Dr Marie-Christine Chartier- Harlin (Inserm U837), Christophe Tzourio (Inserm U708, Paris), Philippe Amouyel (Inserm U744, Lille), Marie-Anne Loriot (Inserm UMRS775, Paris).
Germany: Rejko Krüger (Department of Neurology, University Hospital Tuebingen), Manu Sharma (Department of Neurology, University Hospital Tuebingen), Thomas Gasser, Prof (Medical Genetics, Unversity of Tübingen) Olaf Riess, PhD (Department of Neurology, University Hospital Tuebingen) Daniela Berg MD (Department of Neurology, University Hospital Tuebingen), Claudia Schulte, MSc (Department of Neurology, University Hospital Tuebingen) and Christine Klein, MD (Section of Clinical and Molecular Neurogenetics at the Department of Neurology, University of Lübeck), Ana Djarmati, PhD (Department of Neurology, University of Lübeck), Johann Hagenah, MD (Department of Neurology, University of Lübeck), Katja Lohmann, PhD (Section of Clinical and Molecular Neurogenetics at the Department of Neurology, University of Lübeck).
Greece: Efthimios Dardiotis, MD (Department of Neurology, Faculty of Medicine, University of Thessaly and Institute of Biomedical Research & Technology, CERETETH, Larissa), Vaia Tsimourtou (Department of Neurology, Faculty of Medicine, University of Thessaly, Larissa), Styliani Ralli (Department of Neurology, Faculty of Medicine, University of Thessaly, Larissa), Persa Kountra, MD (Department of Neurology, Faculty of Medicine, University of Thessaly, Larissa), Konstantinos Aggelakis (Institute of Biomedical Research & Technology, CERETETH, Larissa).
Japan: Nobutaka Hattori, MD, PhD (Department of Neurology, Juntendo University School of Medicine, Tokyo), Hiroyuki Tomiyama, MD, PhD (Department of Neurology, Juntendo University School of Medicine, Tokyo), Manabu Funayama, PhD (Department of Neurology, Juntendo University School of Medicine, Tokyo), Hiroyo Yoshino, BS (Research Institute for Diseases of Old Age, Juntendo University School of Medicine, Tokyo) Yuanzhe Li, MD, PhD (Department of Neurology, Juntendo University School of Medicine, Tokyo), Yoko Imamichi (Department of Neurology, Juntendo University School of Medicine, Tokyo), Tatsushi Toda, MD (Division of Neurology/Molecular Brain Science, Kobe University Graduate School of Medicine, Kobe, Japan), Wataru Satake (Division of Neurology/Molecular Brain Science, Kobe University Graduate School of Medicine, Kobe, Japan).
Ireland: Prof Tim Lynch FRCPI, FRCP (The Dublin Neurological Institute at the Mater Misericordiae University Hospital, Clinical Investigator at the Conway Institute, University College Dublin, Ireland) and Dr J. Mark Gibson, MD (Department of Neurology, Royal Victoria Hospital, Belfast, Ireland).
Italy: Enza Maria Valente, MD, PhD (Mendel Institute, Casa Sollievo della Sofferenza Hospital, Rome), Alessandro Ferraris, MD (Mendel Institute, Casa Sollievo della Sofferenza Hospital, Rome), Bruno Dallapiccola (Mendel Institute, Casa Sollievo della Sofferenza Hospital, Rome), Anna Rita Bentivoglio, MD, PhD (Institute of Neurology, Catholic University, Rome), Tamara Ialongo, MD, PhD (Institute of Neurology, Catholic University, Rome), Chiara Riva, PhD (Department of Neuroscience, University of Milano-Bicocca, Monza), Barbara Corradi, PhD (Department of Paediatrics, University of Milano-Bicocca, Monza), Patrizia Tarantino, PhD (Institute of Neurological Sciences, National Research Council) and Ferdinanda Annesi, PhD (Institute of Neurological Sciences, National Research Council).
Norway: J Aasly MD (Department of Neurology, University of Trondheim, Norway).
Poland: Grzegorz Opala, MD, PhD (Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice), Barbara Jasinska-Myga, MD, PhD (Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice), Gabriela Klodowska-Duda, MD, PhD (Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice), Magdalena Boczarska-Jedynak, MD, PhD (Department of Neurology, Aging, Degenerative and Cerebrovascular Disorders, Medical University of Silesia, Katowice).
Singapore: Dr. Eng King Tan, MD, PhD (National Medical and Biomedical Research Councils, and the Duke-NUS Graduate Medical School, Singapore Millenium Foundation).
Sweden: Andrea Carmine Belin, PhD (Department of Neuroscience, Karolinska Institutet, Stockholm), Lars Olson, Professor (Department of Neuroscience, Karolinska Institutet, Stockholm), Dagmar Galter, PhD (Department of Neuroscience, Karolinska Institutet, Stockholm), Marie Westerlund, PhD (Department of Neuroscience, Karolinska Institutet, Stockholm), Olof Sydow, PhD (Department of Clinical Neuroscience, Karolinska University Hospital, Stockholm), Nancy Pedersen, PhD, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Nancy Pedersen, PhD, Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm. Also Christer Nilsson, MD, PhD (Department of Geriatric Psychiatry, Lund University), Andreas Puschmann, MD (Department of Neurology, Lund University Hospital, Department of Geriatric Psychiatry, Lund University).
Taiwan: JJ Lin, MD, Department of Neurology, Cushang Show-Chwan Hospital, Taiwan.
USA: Demetrius M. Maraganore, MD (Department of Neurology, Mayo Clinic, Rochester, MN) J, Eric Ahlskog PhD, MD (Department of Neurology, Mayo Clinic, Rochester, MN, USA), Mariza de Andrade, PhD (Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA), Timothy G. Lesnick, MS (Department of Health Sciences Research, Mayo Clinic, Rochester, MN, USA) and Walter A. Rocca, MD, MPH (Departments of Neurology and Health Sciences Research, Mayo Clinic, Rochester, MN, USA). Also, Harvey Checkoway, PhD (Department of Environmental and Occupational Health Sciences, University of Washington, Seattle, WA), M Farrer PhD, OA Ross PhD (Division of Neurogenetics, Mayo Clinic, Jacksonville, USA).
Footnotes
Disclosure statement: All authors have reported no actual or potential conflict of interest and have certified that all data contained in this manuscript have not been previously published, have not been submitted elsewhere and will not be submitted elsewhere while under the consideration of Neurobiology of Aging. Appropriate approval and procedures were used concerning human subjects. All authors have reviewed the contents of the manuscript being submitted, approve of its content and validate the accuracy of the data. Should a significant conflict of interest be present, the Editors reserve the right to reject the article on that basis. Funding to investigators was provided by the German Federal Ministry for Education and Research (BMBF, NGFNplus; 01GS08134) to T.G.O.R and R.K. and the German Research Council (DFG, KR2119/3-1) to R.K.; “High-Tech Research Center” Project, Grant-in-Aid for Scientific Research (to N.H., 17390256, and to H.T., 21591098) from the Japanese Ministry of Education, Culture, Sports, Science and Technology; Italian Ministry of Health, Ricerca Finalizzata ex. art. 56, grant nr. ART56-OGR-704795 to E.M.V., the Volk-swagen Foundation and the Hermann and Lilly Schilling Foundation to C.K. and a Rapid Response Innovation Award from the Michael J. Fox Foundation to O.A.R. ; The Morris K. Udall Center, NIH/NINDS P50NS40256, and by P01AG17216, R01NS057567, R01AG15866, R01NS042859, R01NS039422, CIHR121849, and PARF to Z.W.; the Special Research Fund of the University of Antwerp, the Fund for Scientific Research Flanders (FWO-V), the Institute for Science and Technology – Flanders (IWT-V), the Interuniversity Attraction Poles program P6/43 of the Belgian Science Policy Office, the Medical Research Foundation Antwerp and Neurosearch Antwerp and a Methusalem Excellence Grant of the Flemish Government, Flanders, Belgium to the Antwerp site.
References
- Bogaerts V, Nuytemans K, Reumers J, Pals P, Engelborghs S, Pickut B, Corsmit E, Peeters K, Schymkowitz J, De Deyn PP, Cras P, Rousseau F, Theuns J, Van Broeckhoven C. Genetic variability in the mitochondrial serine protease HTRA2 contributes to risk for Parkinson disease. Hum Mutat. 2008;29(6):832–840. doi: 10.1002/humu.20713. [DOI] [PubMed] [Google Scholar]
- Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447(7145):655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37(11):1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Elbaz A, Nelson LM, Payami H, Ioannidis JP, Fiske BK, Annesi G, Carmine Belin A, Factor SA, Ferrarese C, Hadjigeorgiou GM, Higgins DS, Kawakami H, Kruger R, Marder KS, Mayeux RP, Mellick GD, Nutt JG, Ritz B, Samii A, Tanner CM, Van Broeckhoven C, Van Den Eeden SK, Wirdefeldt K, Zabetian CP, Dehem M, Montimurro JS, Southwick A, Myers RM, Trikalinos TA. Lack of replication of thirteen single-nucleotide polymorphisms implicated in Parkinson’s disease: a large-scale international study. Lancet Neurol. 2006;5(11):917–923. doi: 10.1016/S1474-4422(06)70579-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelou E, Maraganore DM, Annesi G, Brighina L, Brice A, Elbaz A, Ferrarese C, Hadjigeorgiou GM, Krueger R, Lambert JC, Lesage S, Markopoulou K, Mellick GD, Meeus B, Pedersen NL, Quattrone A, Van Broeckhoven C, Sharma M, Silburn PA, Tan EK, Wirdefeldt K, Ioannidis JP. Non-replication of association for six polymorphisms from meta-analysis of genome-wide association studies of Parkinson’s disease: Large-scale collaborative study. Am J Med Genet B: Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung HC, Scholz S, Matarin M, Simon-Sanchez J, Hernandez D, Britton A, Gibbs JR, Langefeld C, Stiegert ML, Schymick J, Okun MS, Mandel RJ, Fernandez HH, Foote KD, Rodriguez RL, Peckham E, De Vrieze FW, Gwinn-Hardy K, Hardy JA, Singleton A. Genome-wide genotyping in Parkinson’s disease and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol. 2006;5(11):911–916. doi: 10.1016/S1474-4422(06)70578-6. [DOI] [PubMed] [Google Scholar]
- Inagaki R, Tagawa K, Qi ML, Enokido Y, Ito H, Tamura T, Shimizu S, Oyanagi K, Arai N, Kanazawa I, Wanker EE, Okazawa H. Omi/HtrA2 is relevant to the selective vulnerability of striatal neurons in Huntington’s disease. Eur J Neurosci. 2008;28(1):30–40. doi: 10.1111/j.1460-9568.2008.06323.x. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Calibration of credibility of agnostic genome-wide associations. Am J Med Genet B: Neuropsychiatr Genet. 2008;147B(6):964–972. doi: 10.1002/ajmg.b.30721. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ (Clin Res ed) 2007;335(7626):914–916. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat Rev. 2009;10(5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, Davies E, Hajnoczky G, Saunders TL, Van Keuren ML, Fernandes-Alnemri T, Meisler MH, Alnemri ES. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425(6959):721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- Kawamoto Y, Kobayashi Y, Suzuki Y, Inoue H, Tomimoto H, Akiguchi I, Budka H, Martins LM, Downward J, Takahashi R. Accumulation of HtrA2/Omi in neuronal and glial inclusions in brains with alpha-synucleinopathies. J Neuropathol Exp Neurol. 2008;67(10):984–993. doi: 10.1097/NEN.0b013e31818809f4. [DOI] [PubMed] [Google Scholar]
- Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, van Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N. Strengthening the reporting of genetic association studies (STREGA)—an extension of the STROBE statement. Eur J Clin Investig. 2009;39(4):247–266. doi: 10.1111/j.1365-2362.2009.02125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Kruger R, Rocca WA, Schneider NK, Lesnick TG, Lincoln SJ, Hulihan MM, Aasly JO, Ashizawa T, Chartier-Harlin MC, Checkoway H, Ferrarese C, Hadjigeorgiou G, Hattori N, Kawakami H, Lambert JC, Lynch T, Mellick GD, Papapetropoulos S, Parsian A, Quattrone A, Riess O, Tan EK, Van Broeckhoven C. Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA. 2006;296(6):661–670. doi: 10.1001/jama.296.6.661. [DOI] [PubMed] [Google Scholar]
- Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, Stephens M, Bustamante CD. Genes mirror geography within Europe. Nature. 2008;456(7218):98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N, Wilk JB, Latourelle JC, DeStefano AL, Halter C, Pugh EW, Doheny KF, Gusella JF, Nichols WC, Foroud T, Myers RH. Genomewide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross OA, Soto AI, Vilarino-Guell C, Heckman MG, Diehl NN, Hulihan MM, Aasly JO, Sando S, Gibson JM, Lynch T, Krygowska-Wajs A, Opala G, Barcikowska M, Czyzewski K, Uitti RJ, Wszolek ZK, Farrer MJ. Genetic variation of Omi/HtrA2 and Parkinson’s disease. Parkinsonism Relat Disorders. 2008;14(7):539–543. doi: 10.1016/j.parkreldis.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon-Sanchez J, Singleton AB. Sequencing analysis of OMI/HTRA2 shows previously reported pathogenic mutations in neurologically normal controls. Hum Mol Genet. 2008;17(13):1988–1993. doi: 10.1093/hmg/ddn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss KM, Martins LM, Plun-Favreau H, Marx FP, Kautzmann S, Berg D, Gasser T, Wszolek Z, Muller T, Bornemann A, Wolburg H, Downward J, Riess O, Schulz JB, Kruger R. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14(15):2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, Romito L, Albanese A, Dallapiccola B, Bentivoglio AR. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56(3):336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR. The complex interplay among factors that influence allelic association. Nat Rev. 2004;5(2):89–100. doi: 10.1038/nrg1270. [DOI] [PubMed] [Google Scholar]
- Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2(10):2492–2501. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]