Abstract
Frontonasal dysplasia (FND) can have severe presentations that are medically and socially debilitating. Several genes are implicated in FND conditions, including Aristaless-Like Homeobox 1 (ALX1), which is associated with FND3. Breeds of cats are selected and bred for extremes in craniofacial morphologies. In particular, a lineage of Burmese cats with severe brachycephyla is extremely popular and is termed Contemporary Burmese. Genetic studies demonstrated that the brachycephyla of the Contemporary Burmese is a simple co-dominant trait, however, the homozygous cats have a severe craniofacial defect that is incompatible with life. The craniofacial defect of the Burmese was genetically analyzed over a 20 year period, using various genetic analysis techniques. Family-based linkage analysis localized the trait to cat chromosome B4. Genome-wide association studies and other genetic analyses of SNP data refined a critical region. Sequence analysis identified a 12 bp in frame deletion in ALX1, c.496delCTCTCAGGACTG, which is 100% concordant with the craniofacial defect and not found in cats not related to the Contemporary Burmese.
Keywords: Cartilage homeo protein 1, CART1, domestic cat, facial development, frontonasal dysplasia, FND, Felis silvestris catus
Frontonasal dysplasia (FND) or median cleft syndrome is a heterogeneous group of disorders that describes an array of abnormalities affecting development of the maxilla-facial structures and the skull. The prevalence of FND is unknown and is considered a rare or “orphan” disease (ORPHA No.: ORPHA250), however affected children can have severe presentations that are life-long medically and socially debilitating. Three genes have been implicated in FND conditions. Aristaless-Like Homeobox 1 (ALX1) (OMIM:601527) is associated with FND3, which was defined in three Turkish sibs of consanguineous parents (Uz et al., 2010). ALX1 is also known as Cartilage homeoprotein-1 (CART1) (Zhao et al., 1993), which has been demonstrated to cause neural tube defects in mice (Zhao et al., 1996), presenting as acrania and meroanencephaly in mice.
Domesticated animals are often selected for craniofacial variants that become breed defining traits. Conditions that would be considered abnormalities or severe craniofacial defects in humans are desired phenotypes in cats and dogs, thus companion animals are excellent models for human facial development due to their popularity. Many dog and cat breeds are bred for brachycephaly, which is assumed to be preferred due to its neotenic effect on the animal’s face. In dogs, the definition of brachycephaly has been quantified by morphological measurements (Huber and Lups, 1968; Koch et al., 2012; Regodon et al., 1991; Schmidt et al., 2011) and two genes have been implicated for affecting head type (Haworth et al., 2001; Hunemeier et al., 2009; Schoenebeck et al., 2012). The health concerns associated with canine brachycephaly have come under strong veterinary and public scrutiny (Kruijsen and Wayop, 2011; Oechtering et al., 2010; Roberts et al., 2010), suggesting severe modifications to breeding programs to alleviate the extent of brachycephaly.
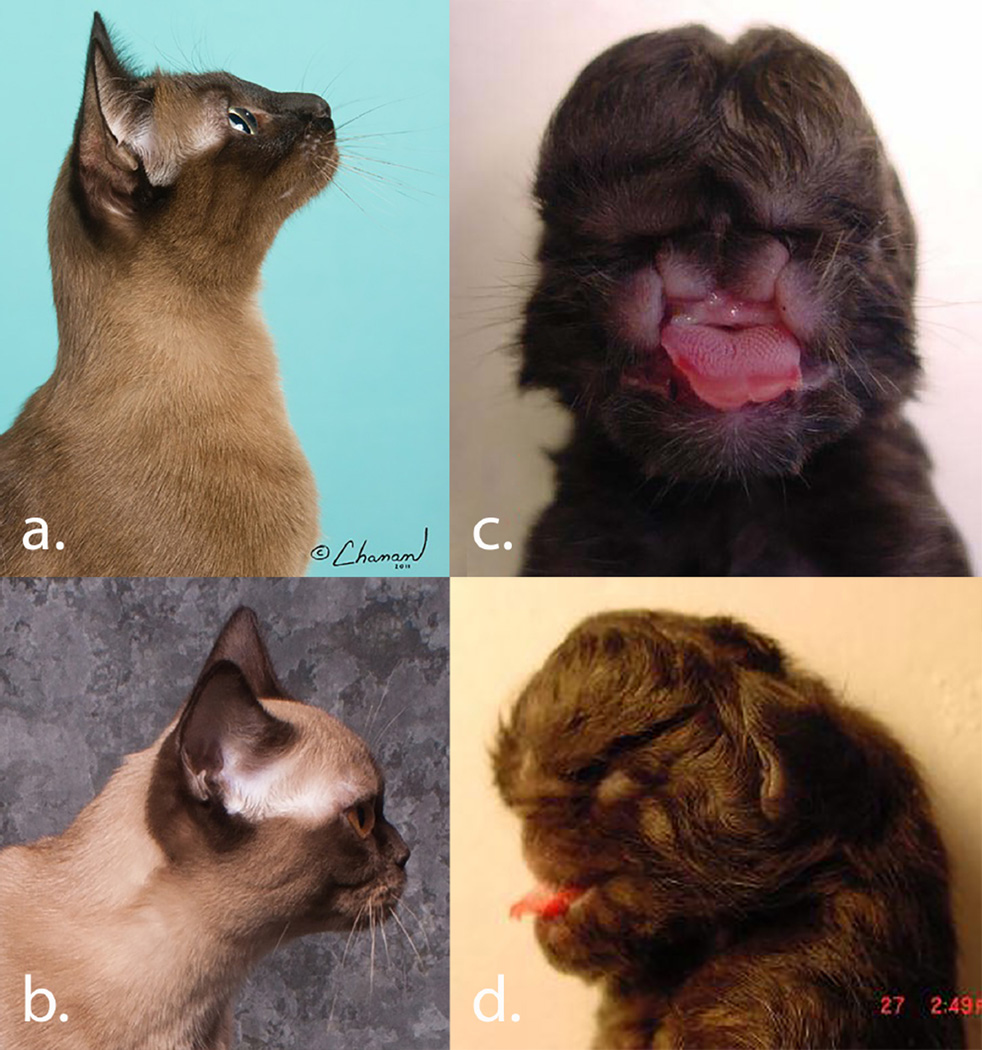
The Burmese is a cat breed with an extreme brachycephalic phenotype (Fig. 1a). In the late 1970’s, a male Burmese cat in the USA with a more brachycephalic head type became a highly popular sire and his lineage became known as the “Contemporary” Burmese (Fig. 1b). The head type was found to be heritable, however, offspring from “Contemporary” style mating produced a craniofacial defect in 25% of offspring (Noden and Evans, 1986; Sponenberg and Graf-Webster, 1986). The abnormality is characterized by agenesis of all derivatives of the medial nasal prominence; lateral duplication of most derivatives of the maxillary process; including the canine teeth and whiskers fields; telencephalic meningoencephalocele; and secondary ocular degeneration (Fig. 1c – d). The midline facial defect is autosomal recessive, however, carriers of the mutation are more brachycephalic individuals than wildtype and were positively selected in the breed, thus the trait has also been described as co-dominant. Affected kittens were generally born live and require euthanasia as the condition is incompatible with life. The heterozygous cats became the hallmark phenotype of the “Contemporary” Burmese and the predominant winners at cat shows.
Figure 1.
Variation of the Burmese cat breed’s craniofacial structure. A) Traditional lines are not as extreme, but selection has continued for the past 30 years for a more extreme type that is not associated with congenital abnormalities. Some Traditional lines and contemporary lines are now difficult to distinguish phenotypically. Thus, all Burmese need to be genotyped to confirm presence or absence of the variant.
B) The Contemporary style Burmese has extreme brachycephyla and the phenotype is association with the craniofacial defect. C) Frontal view displays duplication of the maxillary processes and agenesis of the medial nasal prominence. D) Lateral view displays abnormal development of the maxillary processes and ocular degeneration. Photographs courtesy of Nancy Reeves, Isabelle Marchand and Richard Katris – Chanan Photography.
The controversy of the craniofacial defect and the recognition of other health concerns in non-USA Burmese, such as hypokalemia (Blaxter et al., 1986; Jones and Gruffydd-Jones, 1990), orofacial pain (Rusbridge et al., 2010) and diabetes (Rand et al., 1997) has led to the isolation of the USA and non-USA breeds and the USA Burmese divided into “Traditional” and “Contemporary” styles; Burmese are now one of the most genetically inbred cat populations worldwide with significantly reduced popularity due to the health concerns (Kurushima et al., 2012; Lipinski et al., 2008). Genetic studies have proven to be highly efficient in populations with high linkage disequilibrium (LD) and inbreeding, particularly companion animals. The LD of the Burmese is amongst the most extended for cat breeds (Alhaddad et al., 2013).
A long-term project that initiated with targeted linkage analysis, and, as domestic cat genomic resources improved, progressed to identity by descent mapping, homozygosity mapping and a genome-wide case – control association study (GWAS) suggests ALX1 as a major gene controlling craniofacial structure and the variant in ALX1 is associated with the Burmese brachycephaly and the craniofacial abnormality.
Materials & Methods
Burmese cat sample collection
Cadavers of affected and normal stillborn kittens were voluntarily submitted by Burmese owners from the period of twenty years (1992 – 2012). Approximately 3 ml EDTA anti-coagulated whole blood of normal parents and siblings was also collected and submitted by the owners’ veterinarians. For more recent submissions, DNA was supplied by owners on cotton swabs or cytological brushes via buccal swabbing. Pedigrees were supplied by the owners. White blood cells were isolated from the whole blood using standard techniques and DNA from white cells and tissues was isolated by phenol – chloroform extraction (Sambrook and Russel, 2001), salt precipitation (Miller et al., 1988), or using Qiagen kits (Qiagen, Valencia, CA). Genomic DNA from buccal swabs was isolated using the DNAeasy kit (Qiagen). Pedigree relationships were confirmed by parentage analyses (Lipinski et al., 2007).
Markers for Linkage Analysis
Short tandem repeat (STRs) markers for linkage analysis were selected in proximity to candidate genes, including the homeobox gene clusters (HOXA@, HOXB@, HOXC@, HOXD@) and sonic hedgehog (SHH). These genes are mapped by somatic cell hybrid studies to cat chromosomes that have conserved regions of synteny to human chromosomes 7, 11, 12, and 17, respectively (O’Brien et al., 1997b). At that time, twenty-four STRs were publically available on cat chromosomes A2q, D1, B4q, E1 in juxtaposition to the candidate genes (Menotti-Raymond et al., 2003; Menotti-Raymond et al., 1999).
Linkage Analysis
Linkage analysis was conducted using the software package LINKAGE (Lathrop and Lalouel, 1984, 1988; Lathrop et al., 1984). The kittens with the craniofacial defect were considered congenitally affected with full penetrance of the phenotype, assuming an autosomal recessive mode of inheritance. The allele frequency of 0.5 was estimated for non-genotyped founders of the pedigree since the trait is under positive selection in the Contemporary lines of the breed.
SNP array genotyping
The initial dataset for the SNP array genome-wide analysis comprised 46 cats, including affected Burmese kittens cases that were unrelated as possible and related Burmese, and cats from the closely related breed, Bombay, for controls. Approximately 600 ng of genomic DNA from tissue, blood or buccal swab was submitted to Neogene, Inc (Lincoln, NE, USA) for genotyping on the Illumina Infinium Feline 63K iSelect DNA array (Illumina, Inc., San Diego, CA). Genotyping and analysis was performed as previously described (Gandolfi et al., 2012).
Array data analyses
SNP genotyping rate and minor allele frequency was evaluated using PLINK (Purcell et al., 2007). SNPs with a MAF < 5%, genotyping rate < 90%, and individuals genotyped for < 90% of SNPs were excluded from downstream analyses. An MDS with 2 dimensions was performed using PLINK to evaluate population substructure within cases and controls. Inflation of p-values was evaluated by calculating the genomic inflation factor (λ). The P̂ for each individual was calculated using PLINK. To reduce λ, cats not tightly clustered and/or highly related with a p-hat > 0.3 were removed from downstream analyses. Moreover, selection for each case to the closest control using the values from the MDS dimensions was attempted. Linkage disequilibrium from position 106,142,990 – 114,551,706 was determined and presented as a plot produced by HAPLOVIEW (Barrett, 2009; Barrett et al., 2005). To investigate the haplotype, SNPs from the haplotype block (n = 129 SNPs) were exported and visually inspected.
Identity by descent (IBD) analysis was conducted using PLINK (Purcell et al., 2007). Segmental sharing was surveyed with the command --segment using a window of 25 SNPs (~1000 Kb). All the samples were included in the analysis using the function --all-pairs. Shared haplotypes between all sample comparisons were plotted and visually inspected.
Homozygosity analysis was conducted using PLINK (Purcell et al., 2007). A window of 25 SNPs (~1000 Kb) was surveyed for homozygosity, allowing five missing genotypes and a single heterozygous. A homozygous block was defined by five SNPs (or ~250 Kb) and the threshold of homozygosity match was selected as 0.99. The consensus homozygosity block was defined as the overlapping homozygosity block from each individual using the command (--homo-group). Minor allele frequency (MAF) was calculated for each SNP using the function --geno in PLINK, separating cases from controls. For each SNP, the MAF was plotted along the chromosomal length.
ALX1 genomic analyses
The complete CDS of ALX1 is publicly available and can be found on chromosome B4: 110145316 – 110165008 in Felis catus 6.2. ALX1 has 4 coding exons; the full CDS and the 5 UTR and 3’ UTR was analyzed on genomic DNA. Primers were tested for efficient product amplification on a DNA Engine Gradient Cycler (MJ Research, GMI, Ramsey, MN) and the final PCR magnesium concentrations, annealing temperatures, and amplicon sizes for each primer pair are shown in Supplementary Table 1. PCR and thermocycling conditions were conducted as previously described (Gandolfi et al., 2010). The PCR products were purified and directly sequenced as previously described . Sequences were verified and aligned using the software sequencer version 4.10 (Gene Codes Corp., Ann Arbor, MI).
ALX1 mutation genotyping
The cats of the multi-generational pedigree segregating for the deformity (Supplementary Figs. 1, 2), as well as Burmese and other breed cats, were genotyped to confirm segregation of the variant with the craniofacial defect and to determine allele frequency. The University of California – Davis, Veterinary Genetics Laboratory and Langford Veterinary Services has offered the Burmese craniofacial mutation genetic test for approximately three years. A PCR reaction using Alx1-Fdel with a fluorescence label and Alx1-R del (Supplementary Table 1) was performed and electrophoretically separated on an ABI DNA analyzer (Applied Biosystems). The predicted size of the wild-type allele was 198 bp and 186 for the variant allele and verified using the software STRand (Toonen and Hughes, 2001). Langford Veterinary Services, primers for pyrosequencing were designed using PyroMark Assay Design Ver 2.0 (Qiagen, UK) (Supplementary Table 1). Pyrosequencing was undertaken after PCR amplification using GoTaq Master Mix (Promega, UK) of genomic DNA isolated from mouth swabs using the Nucleospin Blood kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions (PyroGold, Qiagen) on a PyroMark Q24 (Qiagen). Pyrosequencing PCR was conducted using 95°C for 2 min, followed by 38 cycles of 95°C for 20 sec and 58°C for 40 sec.
Results
Burmese cat sample collection
The phenotype of the craniofacial defect in the affected Burmese cats is unique and distinct, with only mild variations in presentation, therefore diagnosis of affected kittens is not confounded by other congenital birth defects in cats (Fig 1c - d.). All affected cats used in the analyses were presented to the investigators and were phenotypically confirmed. Additional stillborn kitten littermates were often submitted and were used as normal siblings when determined phenotypically normal by gross examination. Cats were considered normal if a blood or buccal swab sample had been submitted. Over 488 samples from Burmese cats were ascertained, 83 were stillborn kittens with the craniofacial defect.
Linkage Analysis
A linkage analysis was conducted on two extended families consisting of 124 individuals, which included 47 affected and 62 normal offspring (Supplementary Figs. 1 and 2). Linkage was suggested by four STRs, FCA863, FCA683, FCA864 and FCA866. STR FCA864 identified significant complete linkage (Z = 4.63, Θ = 0.00) to the craniofacial phenotype suggesting the trait should be localized to cat chromosome B4 (Table 1). Fourteen additional STRs, including FCA105, FCA124, FCA298, FCA327, FCA621, RCA656, FCA785, FCA789, FCA790, FCA791, FCA792, FCA991, and FCA992, were also tested for linkage to the cranial defect data not shown). These markers did not support linkage and generally excluded 10 – 15 cM flanking the loci. No linkage was suggested with the four HOX@ clusters and SHH, although HOXC@ is on cat chromosome B4. FCA864 is located at chrB4: 91513852 – 91514204 in the cat reference assembly - Felis catus 6.2 (http://www.ncbi.nlm.nih.gov/assembly/320798).
Table 1.
Linkage of feline STRs on chromosome B4 to the Burmese craniofacial defect.
| Marker 1 | Marker 2 | θ |
Max theta, θ |
Max LOD Z |
||||
|---|---|---|---|---|---|---|---|---|
| 0.00 | 0.01 | 0.05 | 0.10 | 0.20 | ||||
| FCA683 | Defect | − ∞ | −0.82 | 2.10 | 2.87 | 2.83 | 0.140 | 3.00 |
| FCA863 | Defect | − ∞ | −10.57 | −4.66 | −2.44 | −0.75 | --------- | -------- |
| FCA864 | Defect | 4.63 | 4.56 | 4.23 | 3.81 | 2.87 | 0.000 | 4.63 |
| FCA866 | Defect | − ∞ | 0.49 | 1.56 | 1.73 | 1.41 | 0.097 | 1.73 |
| FCA683 | FCA863 | − ∞ | 1.62 | 3.22 | 3.49 | 3.02 | 0.099 | 3.49 |
| FCA683 | FCA864 | − ∞ | 1.00 | 1.51 | 1.59 | 1.41 | 0.096 | 1.59 |
| FCA683 | FCA866 | − ∞ | −0.57 | 0.55 | 0.81 | 0.75 | 0.127 | 0.84 |
| FCA863 | FCA864 | − ∞ | −−6.70 | −3.33 | −1.96 | −0.74 | --------- | -------- |
| FCA863 | FCA866 | − ∞ | −8.63 | −3.96 | −2.15 | −0.71 | --------- | -------- |
| FCA864 | FCA866 | − ∞ | −4.99 | −2.83 | −1.88 | −0.93 | --------- | -------- |
SNP data analyses
Forty-eight cats, including 23 cases (20 Burmese, three Bombay) and 23 controls (16 Burmese and seven Bombay), were submitted for SNP genotyping. Affected Burmese, Bombay and American Shorthair cats originated from United States and healthy controls from the Burmese and Bombay breeds were selected from the USA and other countries were included in the analysis. The genotyping rate was 0.995, hence all the cats were included in the downstream analysis. After evaluating the genotype qualities of 62,897 SNPs on the array, 43,087 markers passed quality control and were included in the case-control association. Approximately 19,549 SNPs were eliminated for low MAF and 314 SNPs were eliminated for poor genotyping. For haplotype analysis, ROH and IBD, only SNPs with poor genotyping rate were removed from analyses.
Association Studies
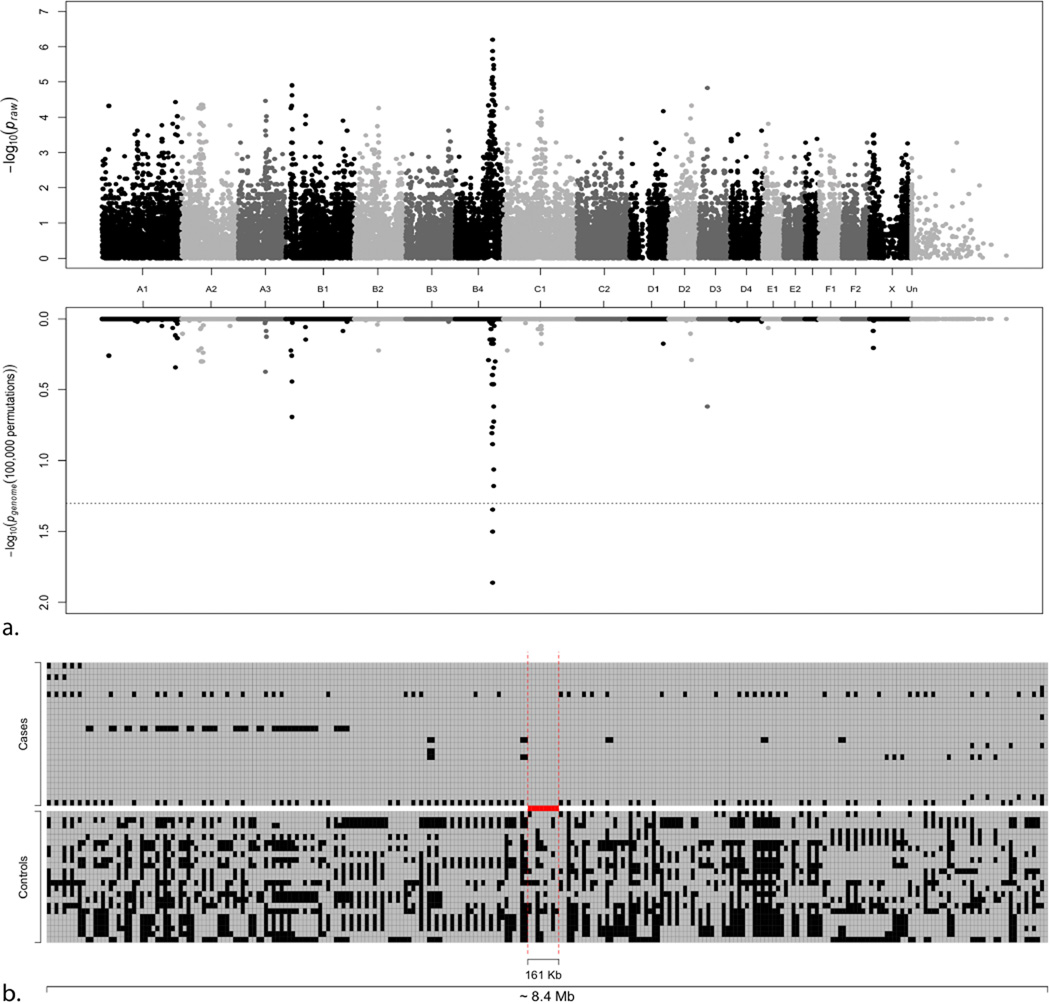
Multi-dimensional scaling (MDS) revealed stratification of the cats used in the analysis (Supplementary Fig. 3). Two main clusters, one containing Burmese cases and controls and some Bombay, a second tight cluster containing only Burmese controls, and some isolated Bombay were observed. The MDS removed one case and 11 controls leaving 22 cases and 12 controls for the analysis, reducing the genomic inflation from 2.95 – 2.13. Seven cases showed a P̂ > 0.3, and were removed, with a decrease in inflation to 1.84. Finally, 15 cases (ten Burmese, five Bombay) and 12 controls (11 Burmese, one Bombay) were included in the association analysis and a significant association was identified with several SNPs on chromosome B4 (Fig. 2). After permutation testing, only three SNPs on chromosome B4 remained genome – wide significant. SNPs B4.128525117 (position 111,895,171) and B4.128576912 (position 111,938,566) had the most significant association with the trait (Table 2). Using the solid spine of LD analysis in Haploview (Barrett, 2009; Barrett et al., 2005), a haplotype with 92% frequency in the cases from position 106,871,872 – 111,795,395 (~ 5 Mb) was identified. In the controls, smaller blocks are detected as shown in Fig. 2, Supplementary Fig. 4. The haplotype was inspected from position 106,142,990 – 114,551,706 and four affected Burmese were key in refining the area containing the gene associated with the phenotype. Two Burmese cats refined the area of association from position 108,534,662 – 112,980,578, while three Bombay showed heterozygous SNPs in the haplotype and the remaining two Bombay were heterozygous for SNPs across the entire ~ 4.5 Mb region, leaving only short block for visual examination, including a 161 Kb block that contains ALX1 (Fig. 2, Supplementary Fig. 4).
Figure 2. Manhattan plot of the Burmese head deformity GWAS and SNPs genotypes within chromosome B4 haplotype.
a. The plot represents the Praw (top) and Pgenome (bottom) values of each SNP included in the case-control association study. The association study compared the affected Burmese and Bombay cats. A significant association with chromosome B4 was detected. b. The area from SNP B4.121572441 (position 106,142,990) to SNP B4.114551707 (position 114,551,707) spans ~ 8.4 Mb. The two red vertical dashed lines represent the region of the single haplotype containing ALX1, from SNP B4.126353636 (position 110,094,604) to SNP B4.126530474 (position 110,255,914) spanning 161 Kb. Each SNP is represented by two squares where markers are on the x-axis and individuals on the y-axis. Gray boxes represent the major allele in the cases and black squares represent the minor.
Table 2.
Genome-wide significantly associated SNPS after 100,000 permutation testing.
| SNP ID | Chromosome | Position | Pvalue | Pgenome value |
|---|---|---|---|---|
| B4.128525117 | B4 | 111895171 | 6.38e−7 | 0.014 |
| B4.128576912 | B4 | 111938566 | 1.33e−6 | 0.031 |
| B4.128654054 | B4 | 112016581 | 2.23e−6 | 0.045 |
When comparing each case to all the other cases included in the IBD analysis, a region on chromosome B4 is shared across the majority of the cats (Supplementary Fig. 5). Four cats did not have the complete common ancestral allele. Other regions, such as chromosome D1 showed an extended shared allele across all the Burmese to Burmese comparisons (Supplementary Fig. 6).
ROH analysis was conducted on all the available cases (n = 23) and controls (n = 23) separately. Excluding the ROHs detected on the X chromosome shared in 23 cases as well as in the controls (data not shown), a ROH was detected in 23 cases on chromosome B4 (Supplementary Figs. 5 & 7). The ROH spanned 162 SNPs (position 106,754,478 – 112,937,278) and covers ~ 6.2 Mb. No ROH was identified for the control group in this location (Supplementary Figs. 5 & 7). Other shorter ROHs were identified on several other chromosomes (Supplementary Table 2). Several other reductions in MAF are detected, but none exclusive to cases compared to controls (Supplementary Fig. 7).
ALX1 genomic analysis and variant genotyping
The entire ALX1 CDS sequence was analyzed in ten cats, including five affected Burmese and five controls (domestic shorthair, one Persian, and three Burmese). ALX1 has one isoform and the length of the coding region of the transcript is 981 bp in human and cats, translating into 326 amino acids. The average CDS homology between human and cat is 93.8% and the protein identity is 97.5%. A 12 bp deletion (c.496delCTCTCAGGACTG) was identified in the coding region of ALX1 (XM_011288799.1). The variant is predicted in silico to be responsible for the lack of four amino acids in the homeobox domain of the protein (Supplementary Fig. 8) was further investigated. No other variants were identified during the sequencing effort.
All the unaffected cats in the pedigree were confirmed to be homozygous wild-type or carrier of the 12 bp deletion while all the affected cats were homozygous for the identified variant (Supplementary Figs. 2 and 3). Genotyping of over 3,000 Burmese type cats suggests the allele frequency is ~6% in the Burmese population. However, this estimation is biased as breeders know the at-risk cats. The variant was also genotyped in ~2400 cats from other breeds with brachycephaly, such as Persian, Exotic Shorthair, Scottish Fold, Selkirk Rex and British shorthair, as well as random bred cats. None of the tested cats from other breeds or populations showed the deletion (Table 3).
Table 3.
Frequency of the ALX1 variant in different cat breeds*.
| Group | Tested | Carrier | Wildtype |
|---|---|---|---|
| Pedigree | 230 | 119 | 28 |
| Asian | 2 | 0 | 2 |
| Australian Mist | 1 | 0 | 1 |
| Bombay | 133 | 43 | 90 |
| Burmese | 3250 | 151 | 3099 |
| Burmilla | 5 | 0 | 5 |
| Tonkinese | 6 | 0 | 6 |
| Burmese Breeds | 3264 | 194 | 3070 |
| Other Breeds | 2456 | 0 | 2456 |
Cats homozygous for the variant are affected and stillborn. From the pedigree, 83 affected kittens were all homozygous for the ALX1 variant. The total number of pedigree cats tested was increased after the linkage analyses. Testing of other cat breeds was performed at the University of California, Veterinary Genetics Laboratory, California, USA and the Langford Veterinary Services, Bristol, UK.
Discussion
Frontonasal dysplasias are a heterogeneous group of disorders (Sedano et al., 1970; Twigg et al., 2009). Cases can be sporadic, however, several familial cases have been reported (Fryburg et al., 1993; Moreno Fuenmayor, 1980; Nevin et al., 1999; Toriello et al., 1985; Twigg et al., 2009; Wu et al., 2007), with two or more of the following clinical signs: true ocular hypertelorism, broadening of the nasal root, median facial cleft affecting the nose and/or upper lip and palate, unilateral or bilateral clefting of the alae nasi, lack of formation of the nasal tip, anterior (rostral) cranium bifidum occultum and a V-shaped or widow’s peak frontal hairline (Sedano and Gorlin, 1988). The Burmese craniofacial defect has the same constellation of dysmorphologies as in humans and is a biomedical model for FND (Fig 1 and 3). The Burmese craniofacial abnormality was originally described as either maxillonasal hypoplasia (Zook, 1983) or incomplete diprosopus (Sekeles, 1981) and a mechanism of transformation of the medial nasal part of the frontonasal process was suggested. The dysmorphology was declared a telencephalic meningohydroencephalocele. Defects in ALX1 are the cause of frontonasal dyspasia type 3 (FND3; OMIM: 613456) (Uz et al., 2010).
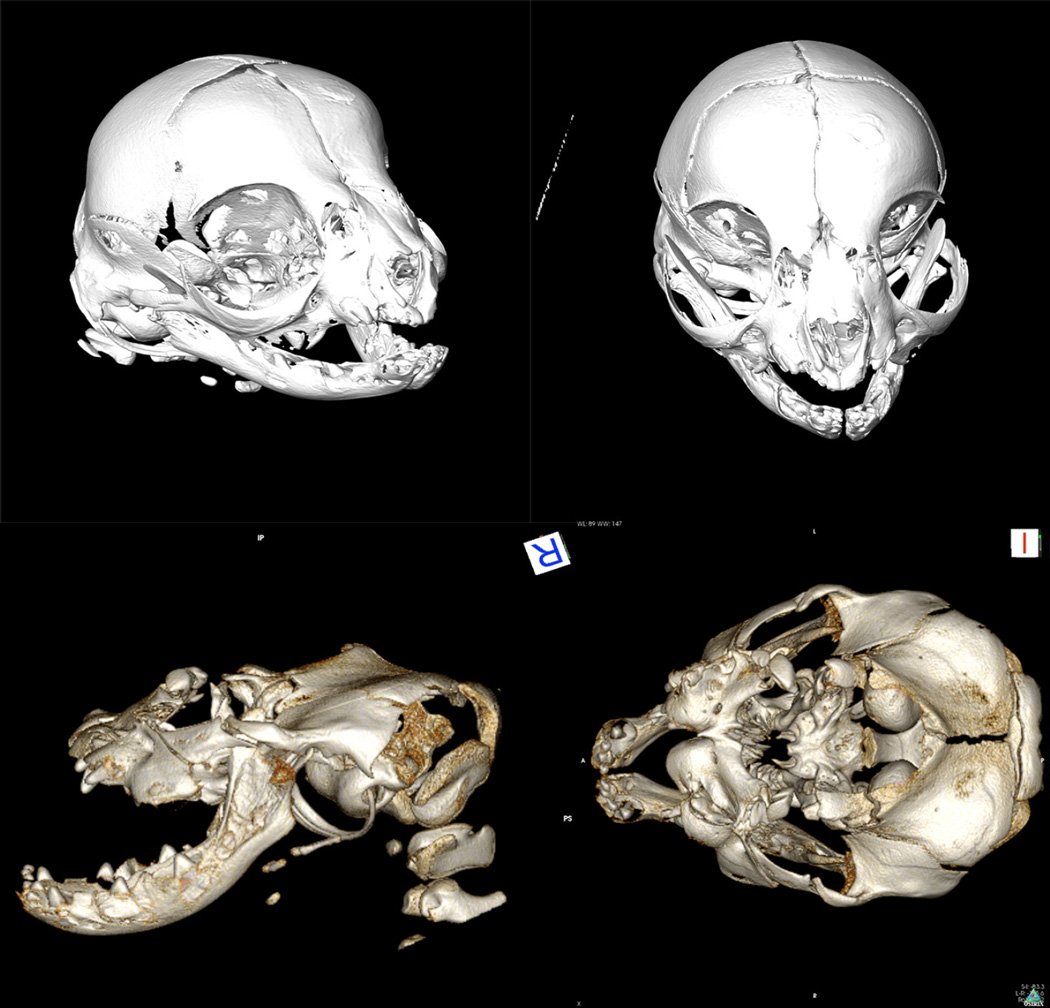
Figure 3.
Three dimensional CT reconstructions of Burmese cat crania. Top) Frontal views of normal stillborn littermate of a Burmese with the craniofacial defect. Bottom) Lateral and dorsal-ventral view of Burmese kitten with a craniofacial defect. Normal kittens may carry the ALX1 variant. Affected kittens are homozygous for the variant and have the hallmark features of FND.
The genetic analyses of the craniofacial abnormality in the Burmese cats were initiated prior to the development of valuable genetic resources for the cat. Progress was incremental as the positions of the HOX@ in the cat were identified by somatic cell hybrid analyses (O’Brien et al., 1997a), STRs developed, linkage and radiation hybrid maps constructed (Menotti-Raymond et al., 1999; Murphy et al., 1999) and synteny between the cat and human genomes established (O’Brien et al., 1997b). Before the understanding of the function of ALX1, candidate genes on cat chromosome B4, such as SHH, were sequenced and eliminated (data not shown). The development of the cat BAC library, and later the first cat genome assembly also allowed the identification of regional STRs and repeated linkage studies continued to eliminate candidate genes on cat chromosome B4 (data not shown). The cat samples submitted by the Burmese breeders produced a larger, more extended pedigree containing over 300 genotyped cats that included the Burmese, Bombay and American Shorthair breeds (data not shown). Eventually, several different genetic methods identified the same chromosomal region for the craniofacial defect in the Burmese cats and the newly recognized function of ALX1 suggested a strong candidate gene (Dee et al., 2013).
Initially, extended pedigrees of Burmese and Bombay cats segregating for the craniofacial defect supported linkage analyses with STRs, suggesting the craniofacial defect was linked to markers that had been mapped to cat chromosome B4. Recent, intense and rapid selection for the brachycephalic muzzle in the Burmese breed suggested that the region with the causative locus may have high linkage disequilibrium (LD). LD analyses across several cat breeds showed the Burmese had the highest LD amongst cats (~200 Kb) (Alhaddad et al., 2013), implying the Burmese would be an efficient breed for analyses on the 63K cat DNA array. The first successful genome-wide association study used non-USA Burmese cats to localize the variant causing hypokalemia with 25 cases and 35 controls and a genomic inflation of 1.8 (Gandolfi et al., 2012). As a disease trait, hypokalemia was not under positive selection, thus, the craniofacial defect, also autosomal recessive, would likely require fewer cats since the trait was under intense positive selection and had heterozygous advantage. After correction for sub-structure and relatedness, a GWAS with a genomic inflation of 1.84 using 15 cases and 12 controls also suggested localization of the craniofacial defect to cat chromosome B4. Both haplotype and identity by descent analysis revealed a 5 – 10 Mb region on chromosome B4 in which four recombinant cats reduced the critical region to 161 Kb, which included the candidate gene ALX1. The same region on chromosome B4 was confirmed by a reduction in MAF in the cases versus the controls. Other regions also showed a remarkable reduction in MAF, but the decrease was not unique to the cases; the presence of these regions was expected, since Burmese and the closely related Bombay have other unique phenotypic features under selection. The region that contains the temperature sensitive coloration pattern in TYR (Lyons et al., 2005) showed a reduction spanning almost 40 Mb, indicating a historic and positive selective pressure for the trait.
The candidate gene, homeobox transcription factor ALX1, is within the short homozygous block spanning 161 Kb. Sequencing of the gene identified a 12 bp deletion, 496delCTCTCAGGACTG. ALX1 contains two domains: the homeobox domain at position 133 – 191 and the OAR domain at positions 302 – 321 of the protein. The homeobox family transcription factor domain is defined by a highly conserved 60 amino acid sequence that encodes for a helix-turn-helix DNA binding domain (Gehring et al 1994). In vertebrates, ALX1 regulates the development and survival of mesenchyme-derived elements of the face and neck and complete gene loss of ALX1 prevents the fusion of frontonasal, nasomedial, nasolateral, and maxillary elements (Uz et al., 2010; Zhao et al., 1994). Uz et al. (2010) identified a 3.7 Mb chromosomal deletion of a region containing ALX1 that is associated with a frontonasal dysplasia. While normal development of structures originating from the frontal and nasomedial prominences are observed, the presence of bilateral cleft is suggests lack of fusion of nasomedial and nasolateral prominences. In humans, the lack of fusion of the apices of the palatal shelves suggests that embryonic development might be disrupted before the seventh week of gestation (Uz et al., 2010) or earlier from studies in mice (Zhao et al., 1996) due to the similar phenotype is hypothesized that ALX1 has a similar role in early embryogenesis in the feline model. ALX1 is tuned by several primary mesenchyme cell signals, and controls ingression genes, several skeletogenic differentiation genes and secondary mesenchyme cells specification genes (Ettensohn et al., 2003). The 4 amino acid deletion in the mutated feline protein from position 68 – 71, within the homeobox binding domain alters the activity of the element, disrupting the normal development of affected Burmese craniofacial mesenchyme. This 12 bp deletion causes a desired brachcephalic presentation in the heterozygous state and the severely dysmorphic congenital abnormality when homozygous (Fig 1 and 3).
ALX1 is expressed during embryogenesis in mesenchyme of craniofacial primordia (Zhao et al., 1993). In vivo studies of ALX1 have demonstrated the aristaless domain of ALX1 functions to restrain activity of this transcription factor mainly or completely through its effect on DNA binding (Brouwer et al., 2003). The aristaless domain (OAR) is essential for correct morphogenesis of the cranium and other regions of the body. The deletion of 4 amino acids of the homeobox domain in ALX1 in the cat demonstrates disruption of the cranium morphogenesis, but only in the homozygous state. Heterozygous cats do have a brachycephalic appearance, thus all variants in ALX1 may not be lethal.
The Burmese has its origins in cats of Thailand, historically known as the Supilak or the Copper Cat of Siam (Clutterbuck, 2004). The Burmese was accepted for stud book registration by the Cat Fancy Association (CFA) in 1936. The foundation of this breed in the United States originated the importation of a single female, “Wong Mau”, from the capital of Rangoon. Wong Mau was phenotypically distinct from the Siamese cats of her homeland in that she had a distinctively more cobby body frame with a walnut-brown coat color, exhibiting darker brown points. The Burmese has a breed defining coloration mutation at the Color (C) locus, all cats being homozygous, cbcb, for a temperature sensitive mutation in Tyrosinase (TYR) that causes the sable coloration (Lyons et al., 2005). Genetic studies support Burmese origins from South –East Asia (Lipinski et al., 2008), as well as other closely related breeds such as Bombay, Singapura, Siamese and Korats. The Bombay, Singapura, Asian and Burmilla cat breeds are derived from the Burmese and need to be screened for the craniofacial defect, as well as hypokalemia.
Animal models offer a useful tool to understand the effects of single gene defects. Moreover, breed phenotypes under strong positive selective pressure facilitates the localization of the loci that harbor gene(s) controlling aesthetic features (Gandolfi 2013, Gandolfi 2013). The Persians are one of the oldest cat breeds, presented as the Angora in early cat shows of the late 1800’s and early 1900’s (1871a; 1871b). Besides Burmese, the craniofacial structure of the breed was also drastically modified after World War II, replacing the moderate facial structure with the drastically brachcephalic structure of the Peked-faced Persian during the 1960’s (Morris, 1999). Persians have influenced many breeds and are the major craniofacial type contributor to the Exotic Shorthair, Himalayan, Scottish Fold, Selkirk Rex and even the modern British Shorthair. Combined, these breeds represent over 60% of the registered cats in the Cat Fanciers’ Association in the USA (CFA, 2004). The Burmese ALX1 variant was not identified in these brachycephalic breeds. The drastic and rapid change in the Persian family of cat breeds suggests a second gene affecting the craniofacial structure in these breeds. Traditional Burmese have also become more brachycephalic over the past 3 decades and their phenotype cannot be clearly distinguished from Burmese heterozygous for the ALX1 variant. Thus, all Burmese need to be genotyped to confirm presence or absence of the variant.
Supplementary Material
Highlights.
Cat breeds are models for mammalian frontonasal development.
A 12 bp in frame deletion in ALX1, c.496 delCTCTCAGGACTG is 100% concordant with the craniofacial defect in cats.
The ALX1 variant in cats has a heterozygous advantage in Burmese cat breeding.
The cat model for frontonasal dysplasia could facilitate therapeutics directed to early developmental stages.
Acknowledgements
This work was supported by funding by the National Center for Research Resources R24 RR016094 and is currently supported by the Office of Research Infrastructure Programs/OD R24OD010928 (LAL), NIH-NIDCR (R03-DE014965-01) and the Winn Feline Foundation (NAB – RFA 1994; 1997, 2001, W10 – 014, W11-041), Cat Health Network (D12FE-506, D12FE-508, D12FE-559), the Center for Companion Animal Health (2009-05-F, 2008-36-F) and the Veterinary Genetics Laboratory, School of Veterinary Medicine, University of California - Davis. We greatly appreciate the efforts of the cat breeders (nearly 80 different cat breeders and owners of affected and non-affected lines) for their supplemental funding and donation of samples over the past 20 years, especially Karen Thomas, DVM, JoAnn Arnett, Erica Graf-Webster, Art Graafmanns, Hilary Helmrich, Nancy Reeves, Roger and Rochelle Hornstein for long lasting and continued appreciation and support. We appreciate the technical assistance of Leslie Bach, Amy Young, Mark Ruhe, and Jennifer Grahn.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- The Cat-Show. Penny Illustrated Paper, The Naturalist. 1871a Jul 22;511:22. [Google Scholar]
- Crystal Palace - Summer concert today Cat Show on July 13. Penny Illustrated Paper, Amusement. 1871b Jul 08;510:11. [Google Scholar]
- Alhaddad H, Khan R, Grahn RA, Gandolfi B, Mullikin JC, Cole SA, Gruffydd-Jones TJ, Haggstrom J, Lohi H, Longeri M, Lyons LA. Extent of linkage disequilibrium in the domestic cat, Felis silvestris catus, and its breeds. PLoS One. 2013;8:e53537. doi: 10.1371/journal.pone.0053537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blaxter A, Lievesley P, Gruffydd-Jones T, Wotton P. Periodic muscle weakness in Burmese kittens. Vet Rec. 1986;118:619–620. doi: 10.1136/vr.118.22.619. [DOI] [PubMed] [Google Scholar]
- Brouwer A, ten Berge D, Wiegerinck R, Meijlink F. The OAR/aristaless domain of the homeodomain protein Cart1 has an attenuating role in vivo . Mech Dev. 2003;120:241–252. doi: 10.1016/s0925-4773(02)00416-1. [DOI] [PubMed] [Google Scholar]
- CFA. Cat Fanciers’ Association Registration Totals by Color and Breed - 2003, and 1/1/58 to 12/31/03. Cat Fanciers’ Almanac. 2004;20:72–86. [Google Scholar]
- Clutterbuck MR. Siamese Cats: Legends and Reality. Bangkok, Thailand: White Lotus Press; 2004. [Google Scholar]
- Dee CT, Szymoniuk CR, Mills PE, Takahashi T. Defective neural crest migration revealed by a Zebrafish model of Alx1-related frontonasal dysplasia. Hum Mol Genet. 2013;22:239–251. doi: 10.1093/hmg/dds423. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Alx1, a member of the Cart1/Alx3/Alx4 subfamily of Paired-class homeodomain proteins, is an essential component of the gene network controlling skeletogenic fate specification in the sea urchin embryo. Development. 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- Fryburg JS, Persing JA, Lin KY. Frontonasal dysplasia in two successive generations. Am J Med Genet. 1993;46:712–714. doi: 10.1002/ajmg.1320460623. [DOI] [PubMed] [Google Scholar]
- Gandolfi B, Gruffydd-Jones TJ, Malik R, Cortes A, Jones BR, Helps CR, Prinzenberg EM, Erhardt G, Lyons LA. First WNK4-hypokalemia animal model identified by genome-wide association in Burmese cats. PloS one. 2012;7:e53173. doi: 10.1371/journal.pone.0053173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandolfi B, Outerbridge CA, Beresford LG, Myers JA, Pimentel M, Alhaddad H, Grahn JC, Grahn RA, Lyons LA. The naked truth: sphynx and Devon rex cat breed mutations in KRT71 . Mamm Genome. 2010;21:509–515. doi: 10.1007/s00335-010-9290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haworth KE, Islam I, Breen M, Putt W, Makrinou E, Binns M, Hopkinson D, Edwards Y. Canine TCOF1; cloning, chromosome assignment and genetic analysis in dogs with different head types. Mamm Genome. 2001;12:622–629. doi: 10.1007/s00335-001-3011-0. [DOI] [PubMed] [Google Scholar]
- Huber W, Lups P. [Biometric and mechanism of development of the symptoms of brachycephaly in the dog] Archiv der Julius Klaus-Stiftung fur Vererbungsforschung, Sozialanthropologie und Rassenhygiene. 1968;43-44:57–65. [PubMed] [Google Scholar]
- Hunemeier T, Salzano FM, Bortolini MC. TCOF1 T/Ser variant and brachycephaly in dogs. Anim Genet. 2009;40:357–358. doi: 10.1111/j.1365-2052.2008.01838.x. [DOI] [PubMed] [Google Scholar]
- Jones BR, Gruffydd-Jones TJ. Hypokalemia in the cat. Cornell Vet. 1990;80:13–16. [PubMed] [Google Scholar]
- Koch D, Wiestner T, Balli A, Montavon P, Michel E, Scharf G, Arnold S. Proposal for a new radiological index to determine skull conformation in the dog. Schweizer Archiv fur Tierheilkunde. 2012;154:217–220. doi: 10.1024/0036-7281/a000331. [DOI] [PubMed] [Google Scholar]
- Kruijsen N, Wayop I. [Does the bulldog still have a future?] Tijdschrift voor diergeneeskunde. 2011;136:268–269. [PubMed] [Google Scholar]
- Kurushima JD, Lipinski MJ, Gandolfi B, Froenicke L, Grahn JC, Grahn RA, Lyons LA. Variation of cats under domestication: genetic assignment of domestic cats to breeds and worldwide random-bred populations. Anim. Genet. 2012 doi: 10.1111/age.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM. Easy calculations of lod scores and genetic risks on small computers. American Journal Human Genetics. 1984;36:460–465. [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM. Efficient computations in multilocus linkage analysis. Am. J. Hum. Genet. 1988;42:498–505. [PMC free article] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J. Strategies for multilocus linkage analysis in humans. Proc Nat Acad Sci USA. 1984;81:3443–3446. doi: 10.1073/pnas.81.11.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski M, Amigues Y, Blasi M, Broad T, Cherbonnel C, Cho G, Corley S, Daftari P, Delattre D, Dileanis S. An international parentage and identification panel for the domestic cat (Felis catus) Animal genetics. 2007;38:371–377. doi: 10.1111/j.1365-2052.2007.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski MJ, Froenicke L, Baysac KC, Billings NC, Leutenegger CM, Levy AM, Longeri M, Niini T, Ozpinar H, Slater MR, Pedersen NC, Lyons LA. The ascent of cat breeds: genetic evaluations of breeds and worldwide random-bred populations. Genomics. 2008;91:12–21. doi: 10.1016/j.ygeno.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons LA, Imes DL, Rah HC, Grahn RA. Tyrosinase mutations associated with Siamese and Burmese patterns in the domestic cat (Felis catus) Animal Genetics. 2005;36:119–126. doi: 10.1111/j.1365-2052.2005.01253.x. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Agarwala R, Schaffer AA, Stephens R, O’Brien SJ, Murphy WJ. Radiation hybrid mapping of 304 novel microsatellites in the domestic cat genome. Cytogenet Genome Res. 2003;102:272–276. doi: 10.1159/000075762. [DOI] [PubMed] [Google Scholar]
- Menotti-Raymond M, David VA, Lyons LA, Schaffer AA, Tomlin JF, Hutton MK, O’Brien SJ. A genetic linkage map of microsatellites in the domestic cat (Felis catus) Genomics. 1999;57:9–23. doi: 10.1006/geno.1999.5743. [DOI] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nuc. Acid Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno Fuenmayor H. The spectrum of frontonasal dysplasia in an inbred pedigree. Clinical Genetics. 1980;17:137–142. doi: 10.1111/j.1399-0004.1980.tb00122.x. [DOI] [PubMed] [Google Scholar]
- Morris D. Cat Breeds of the World. New York: Penguin Books; 1999. [Google Scholar]
- Murphy WJ, Menotti-Raymond M, Lyons LA, Thompson MA, O’Brien SJ. Development of a feline whole genome radiation hybrid panel and comparative mapping of human chromosome 12 and 22 loci. Genomics. 1999;57:1–8. doi: 10.1006/geno.1998.5695. [DOI] [PubMed] [Google Scholar]
- Nevin NC, Leonard AG, Jones B. Frontonasal dysostosis in two successive generations. Am J Med Genet. 1999;87:251–253. doi: 10.1002/(sici)1096-8628(19991126)87:3<251::aid-ajmg10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Noden DM, Evans HE. Inherited homeotic midfacial malformations in Burmese cats. J Craniofac Genet Dev Biol Suppl. 1986;2:249–266. [PubMed] [Google Scholar]
- O’Brien SJ, Cevario SJ, Martenson JS, Thompson MA, Nash WG, Chang E, Graves JA, Spencer JA, Cho KW, Tsujimoto H, Lyons LA. Comparative gene mapping in the domestic cat (Felis catus) J Hered. 1997a;88:408–414. doi: 10.1093/oxfordjournals.jhered.a023127. [DOI] [PubMed] [Google Scholar]
- O’Brien SJ, Wienberg J, Lyons LA. Comparative genomics: lessons from cats. Trends Genet. 1997b;13:393–399. doi: 10.1016/s0168-9525(97)01297-3. [DOI] [PubMed] [Google Scholar]
- Oechtering GU, Schluter C, Lippert JP. [Brachycephaly in dog and cat: a “human induced” obstruction of the upper airways] Pneumologie. 2010;64:450–452. doi: 10.1055/s-0030-1255513. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand JS, Bobbermien LM, Hendrikz JK, Copland M. Over representation of Burmese cats with diabetes mellitus. Australian veterinary journal. 1997;75:402–405. doi: 10.1111/j.1751-0813.1997.tb14340.x. [DOI] [PubMed] [Google Scholar]
- Regodon S, Robina A, Franco A, Vivo JM, Lignereux Y. [Radiological and statistical determination of the morphological type of the canine skull: dolichocephaly, mesocephaly and brachycephaly] Anatomia, histologia, embryologia. 1991;20:129–138. doi: 10.1111/j.1439-0264.1991.tb00752.x. [DOI] [PubMed] [Google Scholar]
- Roberts T, McGreevy P, Valenzuela M. Human induced rotation and reorganization of the brain of domestic dogs. PloS one. 2010;5:e11946. doi: 10.1371/journal.pone.0011946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusbridge C, Heath S, Gunn-Moore DA, Knowler SP, Johnston N, McFadyen AK. Feline orofacial pain syndrome (FOPS): a retrospective study of 113 cases. Journal of feline medicine and surgery. 2010;12:498–508. doi: 10.1016/j.jfms.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Preparation and analysis of eukaryotic genomic DNA., Molecular Cloning. 3rd ed. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 2001. pp. 6.1–6.30. [Google Scholar]
- Schmidt MJ, Neumann AC, Amort KH, Failing K, Kramer M. Cephalometric measurements and determination of general skull type of Cavalier King Charles Spaniels. Veterinary radiology & ultrasound : the official journal of the American College of Veterinary Radiology and the International Veterinary Radiology Association. 2011;52:436–440. doi: 10.1111/j.1740-8261.2011.01825.x. [DOI] [PubMed] [Google Scholar]
- Schoenebeck JJ, Hutchinson SA, Byers A, Beale HC, Carrington B, Faden DL, Rimbault M, Decker B, Kidd JM, Sood R, Boyko AR, Fondon JW, 3rd, Wayne RK, Bustamante CD, Ciruna B, Ostrander EA. Variation of BMP3 contributes to dog breed skull diversity. PLoS genetics. 2012;8:e1002849. doi: 10.1371/journal.pgen.1002849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedano HO, Cohen MM, Jr, Jirasek J, Gorlin RJ. Frontonasal dysplasia. J Pediatr. 1970;76:906–913. doi: 10.1016/s0022-3476(70)80374-2. [DOI] [PubMed] [Google Scholar]
- Sedano HO, Gorlin RJ. Frontonasal malformation as a field defect and in syndromic associations. Oral Surg Oral Med Oral Pathol. 1988;65:704–710. doi: 10.1016/0030-4220(88)90014-x. [DOI] [PubMed] [Google Scholar]
- Sekeles E. Craniofacial and skeletal malformaitons in a cat. Feline Prac. 1981:11. [Google Scholar]
- Sponenberg DP, Graf-Webster E. Hereditary meningoencephalocele in Burmese cats. J Hered. 1986;77:60. doi: 10.1093/oxfordjournals.jhered.a110173. [DOI] [PubMed] [Google Scholar]
- Toonen RJ, Hughes S. Increased throughput for fragment analysis on an ABI PRISM 377 automated sequencer using a membrane comb and STRand software. Biotechniques. 2001;31:1320–1324. [PubMed] [Google Scholar]
- Toriello HV, Higgins JV, Walen A, Waterman DF. Familial occurrence of a developmental defect of the medial nasal processes. American Journal Medical Genetics. 1985;21:131–135. doi: 10.1002/ajmg.1320210119. [DOI] [PubMed] [Google Scholar]
- Twigg SR, Versnel SL, Nurnberg G, Lees MM, Bhat M, Hammond P, Hennekam RC, Hoogeboom AJ, Hurst JA, Johnson D, Robinson AA, Scambler PJ, Gerrelli D, Nurnberg P, Mathijssen IM, Wilkie AO. Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am J Hum Genet. 2009;84:698–705. doi: 10.1016/j.ajhg.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uz E, Alanay Y, Aktas D, Vargel I, Gucer S, Tuncbilek G, von Eggeling F, Yilmaz E, Deren O, Posorski N, Ozdag H, Liehr T, Balci S, Alikasifoglu M, Wollnik B, Akarsu NA. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: expanding the spectrum of autosomal-recessive ALX-related frontonasal dysplasia. Am J Hum Genet. 2010;86:789–796. doi: 10.1016/j.ajhg.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu E, Vargevik K, Slavotinek AM. Subtypes of frontonasal dysplasia are useful in determining clinical prognosis. Am J Med Genet A. 2007;143A:3069–3078. doi: 10.1002/ajmg.a.31963. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Eberspaecher H, Seldin MF, de Crombrugghe B. The gene for the homeodomain-containing protein Cart-1 is expressed in cells that have a chondrogenic potential during embryonic development. Mech Dev. 1994;48:245–254. doi: 10.1016/0925-4773(94)90063-9. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Zhou X, Eberspaecher H, Solursh M, de Crombrugghe B. Cartilage homeoprotein 1, a homeoprotein selectively expressed in chondrocytes. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:8633–8637. doi: 10.1073/pnas.90.18.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Behringer RR, de Crombrugghe B. Prenatal folic acid treatment suppresses acrania and meroanencephaly in mice mutant for the Cart1 homeobox gene. Nat Genet. 1996;13:275–283. doi: 10.1038/ng0796-275. [DOI] [PubMed] [Google Scholar]
- Zook BC. Encephalocele and other congenital craniofacial anomalies in Burmese cats. Vet Med/Small Anim Clin. 1983;78:695–701. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.