Abstract
Human immunoglobulin heavy chain diversity is influenced by biases in the pairing of IGHD and IGHJ genes, but these biases have not been described in detail. We have used high throughput sequencing of VDJ rearrangements to explore DJ pairing biases in twenty-nine individuals. It was possible to infer three contrasting IGHD-IGHJ haplotypes in nine of these individuals, and two of these haplotypes include deletion polymorphisms involving multiple contiguous IGHD genes. We were therefore able to explore how the underlying genetic makeup of the heavy chain locus influences the formation of particular DJ pairs. Analysis of non-productive rearrangements demonstrates that 3′ IGHD genes tend to pair preferentially with 5′ IGHJ genes, while 5′ IGHD genes pair preferentially with 3′ IGHJ genes, and the relationship between IGHD gene pairing frequencies and IGHD gene position is a near linear one for each IGHJ gene. Striking differences are seen, however, in individuals who carry deletion polymorphisms in the D locus. The absence of different blocks of IGHD genes lead to increases in the utilization frequencies of just a handful of genes, and these genes have no clear positional relationships to the deleted genes. This suggests that pairing frequencies may be influenced by additional complex positional relationships that arise perhaps from chromatin structure. In contrast to IGHD gene usage, IGHJ gene usage is unaffected by the IGHD gene deletion polymorphisms. Such an outcome would be expected if the recombinase complex associates first with an IGHJ gene, before associating with an IGHD gene partner.
Introduction
The mammalian immune system generates B cell receptors that are able to bind to almost any conceivable antigen. The genes that encode the heavy chain variable regions of human immunoglobulin molecules are formed by recombination of a variable (IGHV), a diversity (IGHD) and a joining (IGHJ) gene, each selected from ordered clusters of IGHV, IGHD and IGHJ genes (1) that map to chromosome 14 region q32.33 (2, 3). The diversity of antibodies is in part an outcome of the combinatorial diversity that results from the many permutations that are possible when these genes recombine, and this recombination process is usually said to involve essentially random rearrangement of V, D and J genes. It is clear, however, that different genes are used at widely varying yet predictable frequencies (4–7).
Many factors have been reported to influence differential gene usage, with much attention being focused upon the recombination signal sequences (RSS) that flank each V, D and J gene. DNA cleavage in the gene recombination process is catalysed by the products of recombination activation genes 1 and 2 (RAG1 and RAG2) (8, 9), and the RSS direct the RAG recombinase complex to the correct position at which to cut the DNA. Each RSS is composed of a conserved heptamer and a conserved nonamer, separated by spacers of either 12±1 or 23±1 base pairs (1). Both in vitro and in vivo studies have shown that variations in the heptamer and nonamer sequences can influence the frequency of recombination between genes (10, 11). The consequences of variation in the spacer sequences are less certain (12, 13).
The position of genes within the immunoglobulin gene locus has been reported to influence recombination frequencies. Souto-Carneiro and colleagues reported that the more 3′ D genes tend to pair preferentially with 5′ J genes, while more 5′ D genes have a tendency to pair with 3′ J genes (14). In a much larger study, Volpe and Kepler observed similar pairing trends, leading them to propose a model in which multiple DJ rearrangements can be successively generated prior to V to DJ rearrangement (15). This study utilized sequences from the Genbank sequence database, and on the assumption that there is little variation in rearrangement frequencies between individuals, these sequences were derived from thousands of different individuals.
The human genome shows substantial variation between individuals at the immunoglobulin gene loci. This has been most clearly demonstrated in studies that have compared VDJ rearrangements in different individuals using high throughput sequencing (4, 6). VDJ rearrangement is an intra-chromosomal event, and datasets of VDJ rearrangements can therefore be used to infer the genes and allelic variants that are present on each chromosome of an individual. Using this technique, contiguous deletions of a number of D genes have been inferred, and these deletion polymorphisms appear to be present at relatively high frequencies in the human population (6). A focus of analysis at the level of the chromosome is also useful for the study of rearrangement frequencies and has shown that gene usage is strongly affected by the genotype of the individual. Further evidence for genetic influence on recombination has come from twin studies, which have shown patterns of gene usage during VDJ rearrangement to be highly heritable (5, 16).
Population variation within the immunoglobulin heavy chain gene locus was recently confirmed by the sequencing of the locus from a single chromosome (17), for this second reported sequence of the complete locus was substantially different to the immunoglobulin gene locus reference sequence (18). The new sequence also highlighted gene deletions and gene duplications, as well as previously unreported allelic variants. All such variations could be of consequence for studies of recombination and repertoire development.
In order to study the consequences of individual immunogenotypic variation on recombination frequencies and biases in DJ pairing, we have studied large IGH repertoire datasets generated by deep sequencing of VDJ genes from a small number of individuals. We determined the genotype of each donor, and grouped individuals according to three dominant D gene haplotypes. Apparent positional biases in DJ pairing were seen, but there were striking differences in the rearrangement frequencies of particular D genes in individuals who carry deletion polymorphisms in the D locus. These differences suggest that rearrangement frequencies are not simply the outcome of distances between genes. They may also be a consequence of more complex positional relationships arising from chromatin structure. A proper understanding of the biases in DJ pairing may therefore require a more detailed knowledge of the chromatin remodelling and loop formation that brings distant genes into closer proximity to one another during the recombination process (19).
Materials and Methods
Specimen collection and sequence generation
Ig gene sequence data sets were generated from samples of human peripheral blood that were obtained under a Stanford University Institutional Review Board-approved protocol. Participants were 28 healthy individuals and one subject with HIV infection. Data from 27 of these individuals has previously been published (20).
PMBCs were isolated as described previously (21) and genomic DNA (gDNA) and RNA isolation was performed using Allprep kits (Qiagen). PCR amplification of VDJ rearrangements from each sample were performed with six independent 100ng gDNA aliquots, generating six independent bar-coded libraries per sample. PCR was carried out with BIOMED-2 IGHV forward primers and a common IGHJ reverse primer (22) and 10-nucleotide barcode sequences for sample and replicate library identity, as previously described (21). High-throughput 454 (Roche) sequencing was performed using Titanium chemistry.
Partitioning of VDJ sequences
Sequences were sorted into individual data sets based on the presence of perfect matches to sample barcodes and to the first three bases of the J common primer. Samples were then trimmed of barcodes and V primer sequences. VDJ gene rearrangements were partitioned into V, D, and J regions and V-D (N1) and D–J (N2) junctions using the alignment program iHMMune-align (23) and the UNSWIg germline repertoire (http://www.ihmmune.unsw.edu.au/unswig.php). The UNSWIg repertoire does not include IGHD1-14 and IGHD6-25 as there is no compelling evidence that the genes are capable of rearrangement (24). The IGHD repertoire includes three IGHD2-2 alleles, but these only differ at the ends of the sequences, and exonuclease removals of gene ends during the recombination process frequently removes the critical nucleotides that make it possible to distinguish between them. No attempt was therefore made to identify IGHD2-2 alleles. Sequences containing D alignments of less than 8 nucleotides in length were removed from the analysis, as were non-IGH artefacts, duplicate sequences and chimeric sequences. Out of frame J genes were used to define non-productive sequences. As the sequences were amplified from peripheral blood B cells, some sequences may have resulted from clonal expansions in response to antigen. PCR and 454 sequencing errors can also give rise to sets of sequences that appear to have arisen through the process of somatic point mutation. To reduce possible biases arising from the inclusion of multiple sequences from such clonal lineages, all apparent lineages were subjected to filtering. Clones were defined as groups of related sequences that shared V and J genes, and that had identical CDR3 sequences following the exclusion of mutations within the IGHD. One representative sequence from each clone was then included in the final dataset. Where sequences within a clone set carried variable numbers of mutations, the least mutated sequence was selected as the representative sequence.
Genotyping and haplotyping
Determination of genotypes and inference of haplotypes were carried out as described previously (6). Some D genes were not included in the genotype analysis, and sequences that appeared to include these genes were removed from the analysis. Three members of the IGHD1 gene family (IGHD1-1, IGHD1-7, and IGHD1-20) were excluded from genotype analysis as these genes are short and highly similar and therefore cannot be identified with certainty. IGHD5-5 and IGHD5-18 were excluded as they share identical coding regions. This makes it impossible to be certain of the germline source of this coding region within a VDJ rearrangement. IGHD4-4 and IGHD4-11 also share identical coding regions and were excluded for the same reason. IGHD7-27 is so short that the loss of more than a few nucleotides by exonuclease removals makes it essentially undetectable. The number of detected VDJ rearrangements that include IGHD7-27 provides no means by which the number of undetectable IGHD7-27-containing rearrangements can be estimated. IGHD7-27 was therefore also excluded from genotype analysis. Although IGHD2-2 alleles were not identified in the partitioning of VDJ rearrangements, the IGHD2-2 gene was included in the genotype analysis.
Of the twenty-nine individuals studied, nine were heterozygous at either the IGHJ4 locus or the IGHJ6 locus, and sufficient sequences were available from these nine individuals to use likelihood ratios to determine haplotypes (6). Haplotypes containing deletions of six and five contiguous D genes were inferred in several individuals. As IGHD1-7, IGHD4-4, IGHD5-5 and IGHD6-25 were excluded from the genotype analysis, their absence from certain haplotypes was presumed from the absence of neighbouring genes. Rearrangement frequencies were calculated for each gene, and this frequency was based upon the total number of rearrangements involving one of the included D genes.
Statistical analysis
Comparison of D and J gene usage between productive and non-productive datasets was performed using paired t tests. Multiple t tests were then performed to examine differences in individual genes. Comparison of D and J gene usage between different D genotypes was also carried out using paired t tests, with multiple t tests used to examine differences in individual genes. The desired false discovery rate was set to 1% for all multiple t tests. All t tests were performed using GraphPad Prism version 6.05 for Windows.
Contingency tables were used to test whether pairing frequencies were dependent or independent of the overall utilization frequencies of IGHD and IGHJ. Adjusted residuals were used to measure the degree of departure from independence of each pair. Values greater than 1.96 show there was a statistically significant (p<0.05) over-representation of pairs of particular D and J genes, while values less than −1.96 show there was a statistically significant (p<0.05) under-representation of pairs of particular D and J genes (25).
Results
VDJ amplicons from 29 individuals were investigated using high throughput sequencing, and after filtering, a total of 943,978 VDJ rearrangements remained. Of these, 192,290 were non-productive based on out of frame IGHJ, and 751,685 were productive. Individual sample sizes ranged from 19,279 to 110,344 sequences, with an average of 32,551. Non-productive sequences in the samples ranged from 3,737 to 26,614 sequences, with an average of 6,631. The data sets are available at dbGap (http://www.ncbi.nlm.nih.gov/gap) under accession numbers phs000760.v1.p1 and phs000666.v1.p1, or by request from the authors. Nine individuals were heterozygous at an IGHJ locus, allowing the sets of genes on each of their chromosomes to be determined as haplotypes. From these nine samples, 140,706 sequences were obtained from two individuals who were inferred to carry a single copy of a deletion polymorphism involving six contiguous D genes spanning IGHD3-3 to IGHD2-8 inclusive. Analysis of two other individuals showed striking differences between rearrangements of their two chromosomes at one or two genes. On one chromosome, just a handful of VDJ rearrangements suggested the possible presence of D genes that are part of a deletion polymorphism spanning the genes IGHD3-22 to IGHD1-26 inclusive. The implied utilization frequencies, if the genes were truly present in the genomes of these individuals, were so low that an alternative possibility was addressed. The identification of D genes within VDJ rearrangements is notoriously difficult, so the data sets were manually reviewed to determine whether or not the critical rearrangements could have been mis-aligned against the IGHD germline gene repertoire. We concluded that all the alignments to the genes in question were probably in error. The alignments were all short and included mismatches to the germline. We therefore inferred that these two individuals, as well as a third individual, carry a single copy of a deletion polymorphism involving the five contiguous genes. 69,837 sequences were derived from the three individuals. A further 109,486 sequences came from four individuals with no apparent D gene deletions. The DJ pairing frequencies identified in the HIV-infected subject in this study were not appreciably different from those of the healthy subjects. Data from this individual was included in the analysis because they are heterozygous for the contiguous 6 D gene deletion and because of the large number of rearrangements (111,387) in their data set.
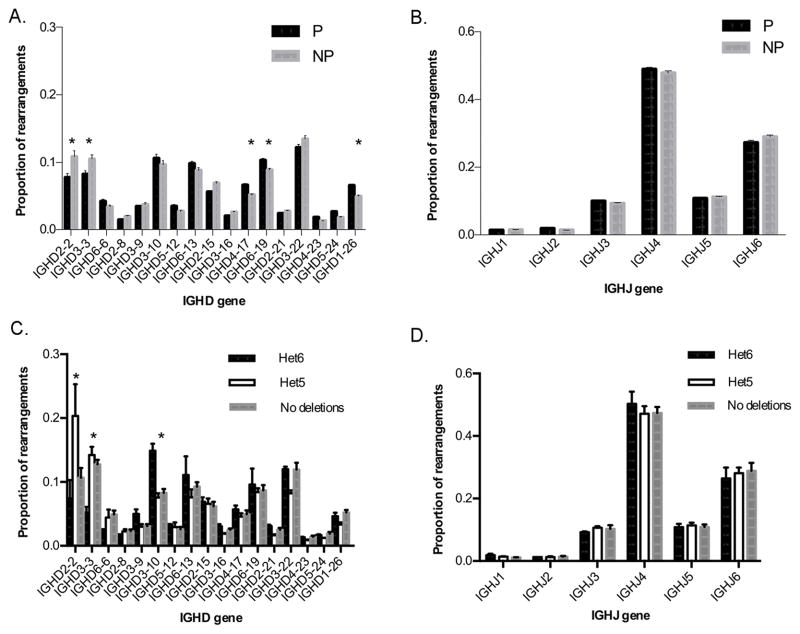
IGHD gene usage was analysed in pooled data from the twenty-nine individuals, and clear differences were seen between productive and non-productive sequences (Figure 1A). Although an overall difference between the two datasets was not detected using a paired t test, there were differences for 5 of the 19 genes examined – IGHD1-26, IGHD2-2, IGHD3-3, IGHD4-17 and IGHD6-19 (multiple t test, p<0.01). An overall difference between productive and non-productive sequences was not detected for J genes (Figure 1B) and no differences were detected for individual genes. Frequencies of gene usage were consistent with previous reports (4, 5). Because of the differences between the productive and non-productive datasets, subsequent analysis of D and J gene usage focused on non-productive sequence data. Sequence data from different individuals was pooled for each of the three IGHD genotypes – those with a single copy of the inferred six D gene deletion (Het6), those with a single copy of the inferred five D gene deletion (Het5) and those with no apparent D gene deletions (No deletions). Significant differences in usage of IGHD3-10, IGHD3-3 and IGHD2-2 were detected between individuals with the differing genotypes (Figure 1C) (multiple t test, p<0.01 in each case). No significant differences were detected for individual J genes (Figure 1D).
FIGURE 1. Comparison of the frequency of (A) IGHD gene usage in productive and non-productive sequences (B) IGHJ gene usage in productive and non-productive sequences (C) IGHD gene usage in individuals with different IGHD genotypes (D) IGHJ gene usage in individuals with different IGHD genotypes.
Het6 refers to IGHD gene usage in pooled rearrarangements from individuals who carried a single copy of a deletion polymorphism involving the six contiguous IGHD genes. Het5 refers to IGHD gene usage in pooled rearrarangements from individuals who carried a single copy of a deletion polymorphism involving the five contiguous IGHD genes. Rearrangements using each IGHD gene are shown as the proportion of the total number of rearrangements for which the IGHD gene could be identified. All genes are ordered as they appear in the genome, from 5′ (left) to 3′ (right). *p < 0.01. Error bars are SEM.
To explore the relationship between gene usage frequency and gene pairing among the non-productive rearrangements, we used contingency tables to test whether the pairing frequencies were dependent or independent of the overall utilization frequencies of the D and J genes (Table 1). A clear trend was observed for the more 5′ D genes to pair with the more distant 3′ J genes, and for the more 3′ D genes to pair with the nearer 5′ J genes.
Table 1.
Adjusted residuals from chi-squared analysis of non-productive sequence data from 29 individuals1
| IGHJ1 | IGHJ2 | IGHJ3 | IGHJ4 | IGHJ5 | IGHJ6 | |
|---|---|---|---|---|---|---|
| IGHD2-2 | −5.955 | −6.350 | −14.360 | −45.811 | 17.891 | 50.521 |
| IGHD3-3 | −9.958 | −7.175 | −15.192 | −34.661 | 5.625 | 48.573 |
| IGHD6-6 | −2.984 | 2.507 | −5.856 | −4.968 | 2.087 | 7.912 |
| IGHD2-8 | −2.418 | −0.540 | −6.436 | −4.002 | 3.332 | 6.975 |
| IGHD3-9 | −5.193 | −2.075 | 1.646 | −5.470 | −1.804 | 8.197 |
| IGHD3-10 | −8.198 | −5.455 | −5.667 | −11.378 | 7.945 | 14.249 |
| IGHD5-12 | −6.190 | −1.568 | −5.436 | 10.234 | −8.502 | 0.140 |
| IGHD6-13 | 6.723 | 0.360 | −11.343 | 2.785 | 8.630 | −3.809 |
| IGHD2-15 | 3.601 | −4.382 | −5.648 | −11.307 | 15.368 | 5.520 |
| IGHD3-16 | −4.655 | −1.712 | 2.920 | 15.306 | −5.199 | −13.453 |
| IGHD4-17 | −0.616 | 11.112 | 0.485 | 5.182 | −4.348 | −5.699 |
| IGHD6-19 | 7.155 | 2.102 | −1.859 | 26.519 | −8.275 | −24.839 |
| IGHD2-21 | 12.438 | 9.271 | 9.899 | 4.469 | −4.795 | −13.621 |
| IGHD3-22 | 10.787 | 4.534 | 32.214 | 32.964 | −16.193 | −49.667 |
| IGHD4-23 | 1.821 | 6.453 | 10.120 | 6.955 | −5.514 | −12.418 |
| IGHD5-24 | −0.852 | 3.037 | 7.012 | 18.699 | −10.423 | −18.403 |
| IGHD1-26 | 1.253 | 0.127 | 14.378 | 19.411 | −11.739 | −22.766 |
Values greater than 1.96 (dark grey) and less than −1.96 (light grey) represent significant departures from the expected values at a 95% confidence level.
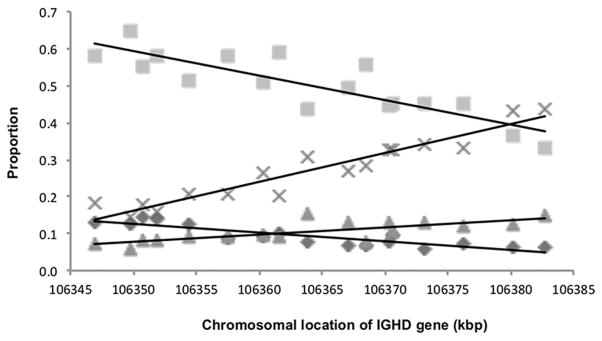
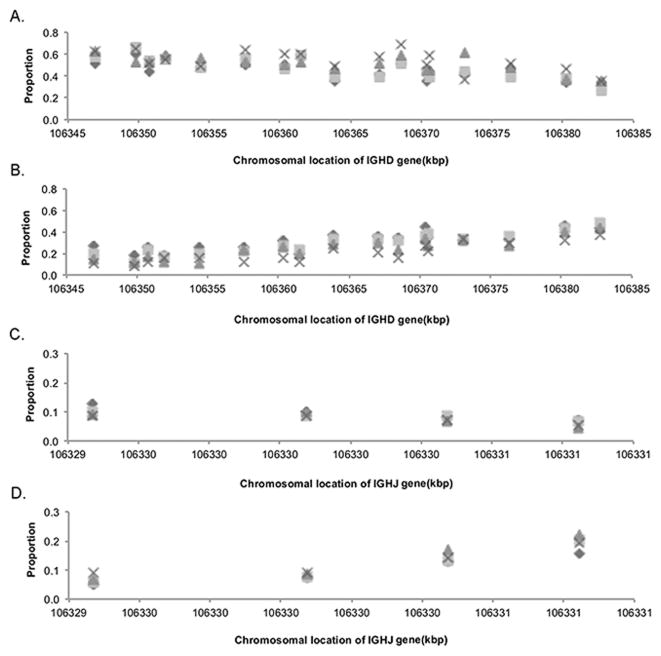
In order to uncover any underlying relationships that could have been obscured by the variable rearrangement frequencies of individual D genes, we examined the proportion of rearrangements of each of the D genes that were associated with the four most commonly utilized J genes (Figure 2). This analysis shows, for example, the proportion of all the IGHD2-2-containing alignments that also utilized IGHJ4. The patterns shown suggest a linear relationship between chromosomal position and utilization frequencies, and interestingly, the pairing frequencies of IGHD3-9 and IGHD3-10, which are located close together in the genome, were almost identical. More surprisingly, the direction of the relationship between chromosomal location and utilization frequencies varied between IGHJ5 and IGHJ6 on the one hand, and IGHJ3 and IGHJ4 on the other. This suggests that the relationship is not simply a measure of the linear distance between the joining genes. The departures from the regression lines seen in Figure 2 also suggest the possibility that there are complex, reproducible but non-linear relationships between chromosomal location and gene utilization frequency. To explore this possibility, we investigated whether the patterns were consistent between individuals, focussing on four of the most frequently used genes: IGHJ4, IGHJ6, IGHD3-10 and IGHD3-22. Figure 3 shows the results of analysis from four individuals who each carried the full complement of D genes on each chromosome. Each of the two J genes is plotted against all the genotyped D genes (Figure 3A and B), and each of the two D genes are plotted against the four most abundant J genes (Figure 3C and D). There is a remarkable consistency in the rises and falls between adjacent genes for the four individuals. The trends for IGHJ4 and IGJHJ6 again go in opposite directions to one another, with neither relationship being strictly linear. Similarly, while IGHD3-22 was seen in association with increasing proportions of the more 3′ J genes, IGHD3-10 was seen in association with increasing proportions of the more 5′ J genes.
FIGURE 2. Proportion of rearrangements of each IGHD gene that pair with the four most frequently utilized IGHJ genes, using pooled data from the non-productive sequence datasets of 29 individuals.
The proportion of rearrangements of each IGHD gene is plotted against chromosomal location, and the proportions are calculated with respect to the number of rearrangements in which the IGHD gene could be identified. As the genes are found in the genome in reverse orientation, the most 3′ gene (IGHD1-26) is shown at the extreme left and the most 5′ gene (IGHD2-2) is shown at the extreme right.
◆IGHJ3
 IGHJ4 ▲IGHJ5 X IGHJ6
IGHJ4 ▲IGHJ5 X IGHJ6
FIGURE 3. Consistency of rearrangement frequencies across four individuals who had no apparent IGHD gene deletions.
Proportion shown are the proportions of all non-productive IGHJ4-containing (A) and IGHJ6-comtaining (B) rearrangements that include each IGHD gene; and the proportion of all non-productive IGHD3-10-containing (C) and IGHD3-22-containing (D) that include each of the four most highly utilized IGHJ genes (IGHJ3 – IGHJ6). Each individual is represented by a different symbol. As the IGHD and IGHJ genes are found in the genome in reverse orientation, the most 3′ gene (IGHD1-26) is shown at the extreme left and the most 5′ gene (IGHD2-2) is shown at the extreme right. The most 3′ IGHJ gene (IGHJ6) is shown at the extreme left and the most 5′ gene (IGHJ3) is shown at the extreme right.
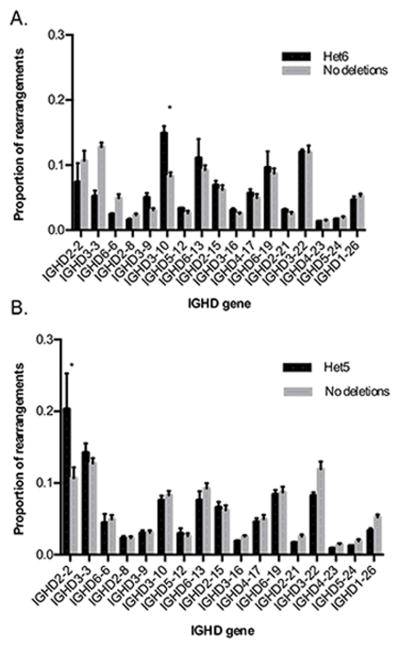
Analysis of DJ pairing in individuals whose genotype included D gene deletion polymorphisms showed very different patterns of D gene usage (Figure 4). Although an absence of certain genes on one chromosome could be expected to result in corresponding increases in the utilization frequencies of all other genes present on the same chromosome, the patterns seen were not of this kind. A substantial and statistically significant increase in the use of IGHD3-10 was observed in individuals who carried a single copy of the deletion polymorphism involving the six genes IGHD3-3 to IGHD2-8 (multiple t test, p<0.00001). A similar increase in the utilization of IGHD2-2 was seen in individuals who carried a single chromosomal deletion polymorphism involving the five genes IGHD3-22 to IGHD1-26 (multiple t test, p<1.0×10−9). Individuals with shared genotypes had no differences in J utilization frequencies (data not shown).
FIGURE 4. IGHD gene utilization frequencies in individuals with different IGHD genotypes.
Proportions of rearrangements using each IGHD gene were compared between four individuals who carried complete sets of IGHD genes on both their chromosomes, with (A) pooled non-productive sequence data for two individuals who carried a single copy of a deletion polymorphism involving the six contiguous IGHD genes, IGHD3-3 to IGHD2-8 (Het6); and (B) pooled non-productive sequence data for three individuals who carried a single copy of a deletion polymorphism involving the five contiguous IGHD genes, IGHD3-22 to IGHD1-26 (Het5). Proportions of IGHD genes are a proportion of the total number of rearrangements in which the IGHD gene could be identified. IGHD genes are ordered as they appear in the genome, from 5′ (left) to 3′ (right). *p < 0.00001. **p < 1.0×10−9.
The consequence of pairing biases on the primary repertoire of individuals with different IGHD genotypes is considerable. The most abundant pairing of D and J genes in individuals who carry a single copy of the six gene deletion polymorphism was the pairing of IGHD3-10 with IGHJ4. The usage frequency of this pairing was more than twice that of either of the other immunogenotypes. In individuals with a single copy of the five gene deletion polymorphism, the most abundant pairing (IGHD2-2 with IGHJ6) accounted for 7.8% of all pairings, but was present in 2.1% of sequences in individuals with the heterozygous six gene deletion polymorphism and 4.6% in individuals with the complete complement of D genes. In individuals with the complete complement of D genes, the most abundant pairing (IGHD3-22 with IGHJ4) accounted for 6.7% of all pairings, and was present in 7.1% of sequences in individuals with the heterozygous six gene deletion polymorphism and 5.0% in individuals with the heterozygous five gene deletion polymorphism.
Discussion
The aim of this study was to investigate DJ pairing biases in individuals with different IGHD locus genotypes using large IGH repertoire datasets generated using high throughput sequencing. The size of the datasets enabled us to focus on non-productive sequence data, allowing us to exclude the possibility that observed biases were an outcome of selection acting upon transcribed genes. Significant differences in gene usage between productive and non-productive sequences were seen for some but not all D genes, confirming recent reports (26). These differences between IGHD genes may in part be explained by their varying utilization frequencies. The low utilization frequencies of some IGHD genes could be preventing differences from reaching statistical significance. IGHD2-2 and IGHD3-3 are seen in a high proportion of rearrangements, and appear to be selected against in the productive repertoire. The stronger selection against these genes could be indicative of their greater self-reactivity. Selection against these genes would also result from the fact that all three IGHD2-2 alleles contain two stop codons in reading frame one and several hydrophobic residues in reading frame three. The most commonly used allele of IGHD3-3, IGHD3-3*01, also encodes several hydrophobic residues when translated in reading frame three. However, stop codons and hydrophobic amino acids are also observed in the corresponding reading frames of IGHD3-22, IGHD4-17 and IGHD4-23, amongst other genes, and no selection was apparent against these genes in the productive repertoire. A better understanding of selection will require the direct investigation of the repertoire before and after selection.
The DJ pairing biases that were detected were much more complex than those that have previously been suggested. The analysis of the non-productive repertoires from individuals in this study aligns with previous reports that there is a tendency for more 5′ D genes to pair with more 3′ J genes, and for more 3′ D genes to pair with closer 5′ J genes (14, 15, 21). The datasets revealed more intricate patterns that were shared by individuals with shared IGHD genotypes, but which differed between individuals who carried different IGHD genotypes.
Investigation of the effects of genotypic variation on DJ pairing clearly shows that when blocks of D genes are absent from the IGHD locus, the increase in usage of other D genes is not spread evenly amongst the remaining genes. Sequences amplified from individuals who carry a single copy of a deletion polymorphism involving six contiguous D genes from IGHD3-3 to IGHD2-8 utilized the IGHD3-10 gene at a significantly higher frequency than that seen in individuals with a full complement of genes. We do not believe that it is simply the ‘proximity’ of this gene to the genomic position that is normally occupied by the deletion polymorphism that gave rise to this increase, for the deletion polymorphism involving five contiguous genes (IGHD3-22 to IGHD1-26) tells a different story. Individuals who carry a single copy of this deletion polymorphism showed a highly significant increase in the use of IGHD2-2. IGHD2-2 is a 5′ IGHD gene that is far removed from the normal location of the five deleted IGHD genes.
Interestingly, the changed utilization frequencies of D genes that were detected in individuals who carried D deletion polymorphisms were not accompanied by detectable changes in the use of any J genes. In this study, the four more frequently expressed J genes could be analysed, while eighteen different D genes could be analysed. It is easier to demonstrate that biases are statistically significant when the number of categories is low. The observation of significant differences in D gene utilization frequencies, and the failure to detect significant differences in the utilization of J genes was therefore surprising, and it may point to the order of events that lead up to DJ recombination.
A model has been proposed in which the assembly of a synaptic complex is initiated by the binding of the recombinase to a 12-bp spacer RSS. This is followed by the capture of a 23-bp spacer RSS (27). In the case of DJ recombination, this implies that the recombinase first binds to the 3′ D RSS, before capturing the J RSS. However, a recent study in mice suggests that the recombinase is first recruited to the J region (28). The results of the present study provide indirect evidence that the human recombinase is also first recruited to the J region. If the recombinase binds first to the 3′ D RSS, the disruptions to the utilization frequencies of D genes would be likely to be accompanied by disruptions to the utilization frequencies of the J genes. This was not observed and therefore is consistent with initial binding of the recombinase to the J RSS.
It has been suggested that persistent RAG protein expression leads to successive rounds of DJ recombination, prior to V-DJ recombination (29). It has also been suggested that the tendency for more 5′ D genes to be seen with more 3′ J genes (IGHJ4 to IGHJ6) could be an outcome of this process (15). The results of the present study, using a very large dataset of non-productive VDJ rearrangements, suggest that successive rounds of rearrangement could only account for some of the biases in the rearrangements of IGHJ5 and IGHJ6, and cannot account for the overall patterns of IGHJ4 rearrangements. Our results are consistent with the existence of more complex positional biases, though variability in RSS could also be contributing to the variability in the utilization of different genes.
There is no doubt that variations in both the heptamer and nonamer sequences of RSS can influence recombination frequencies. This has been shown both in vivo (11) and in vitro (10). Opinions differ regarding the effect of spacer sequences on the frequency of recombination. Wei and colleagues concluded that the sequence of the spacer had little effect on the frequency of gene recombination (13), whilst others have reported that differential rates of rearrangement are associated with different spacer sequences (12). The crystal structure of the mouse RAG1 nonamer binding domain (NBD) bound to DNA highlights, for the mouse at least, the importance of contacts with the spacer sequence in RAG-mediated activity (30). The entire crystal structure of the mouse RAG1-RAG2 complex has now been reported (31) and appears to corroborate the view that some sequence-specific recognition of the recombinase by the NBD is important (30). Differences in RSS, including unreported variations in the RSS, however, are at most just part of the explanation for variable recombination frequencies, for there are many examples of genes with identical RSS that have quite different recombination frequencies (18).
The contrasting variation that was seen in the pairing of IGHJ4 and IGHJ6 with different D genes, and the variation in gene utilization frequencies seen in individuals with differing D genotypes gives credence to the view that DJ recombination is subject to strong positional biases. However, the relative positions of the genes along the chromosome are insufficient to explain the observed biases. Enhancer activity has been proposed to facilitate looping of the IGHV region DNA in mice, bringing distal genes into closer proximity to the other genes (19). Our data, in particular the undulating patterns observed for the proportions of each D gene rearranging with a particular J gene, suggest the possibility that multiple loops of D genes are active in the human. Such loops could provide an explanation for the increase in use of IGHD2-2 that accompanies the deletion of a distant block of IGHD genes.
Ontogeny is associated with a progressive increase in the addition of N nucleotides in DJ junctions in both the human (14) and the mouse (32). The general lack of N addition during murine fetal and neonatal development ensures that IGHD genes are the dominant influence on the repertoire of fetal and neonatal CDR3 sequences. This, together with strong pairing biases, could ensure that critical murine specificities are generated at high frequency (33). Critical specificities may also be preferentially generated in early human development, and this could explain the observation that the proportion of 5′D – 3′J pairings increases progressively from human fetal to neonatal to adult rearrangements (14). These increases could possibly be achieved by chromatin remodelling leading to changes in accessibility of certain genes. The loss of critical D genes, as well as changes in DJ pairing frequencies that are associated with differing IGHD locus immunogenotypes could therefore be particularly important during fetal and neonatal development. Certainly murine IGHD gene deletions have been shown to increase susceptibility to infection (34). Alterations in IGHD gene sequences can also increase the likelihood of the production of self-reactive antibodies (35).
Biases in DJ pairing and differences in the pairing biases that result from genotypic variation are firmly established by the results of this study, and these biases join a growing list of processes that we now know can shape the antibody repertoire of an individual. An inevitable outcome of immunogenotypic differences between individuals must therefore be that particular V(D)J rearrangements and particular heavy and light chain pairs must be represented at different frequencies in the repertoires of different individuals. The view that the formation of any particular antibody specificity is simply the result of stochastic processes is thus challenged again. In light of the accumulating evidence that the human genome shapes the emergence of the repertoire of each individual, the possibility that the resulting immunocompetence of individuals may be different must be given credence. If more large datasets of VDJ rearrangements from different individuals at varying stages of development are analysed with reference to the underlying genotypes of the individuals, a new view of immunocompetence may emerge.
Acknowledgments
This research was supported in part by the Duke University Center for AIDS Research (CFAR), an NIH funded program (5P30 AI064518), NIAID-5U19AI090019, NIH CHAVI U19AI067854, and a grant from the National Health and Medical Research Council.
The authors thank Dr. Georgia Tomaras for sharing of materials and helpful discussions.
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM, Shander M, Martinis J, Cicurel L, D’Ancona GG, Dolby TW, Koprowski H. Chromosomal location of the genes for human immunoglobulin heavy chains. Proc Natl Acad Sci USA. 1979;76:3416–3419. doi: 10.1073/pnas.76.7.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsuda F, Shin EK, Nagaoka H, Matsumura R, Haino M, Fukita Y, Taka-ishi S, Imai T, Riley JH, Anand R, et al. Structure and physical map of 64 variable segments in the 3’0.8-megabase region of the human immunoglobulin heavy-chain locus. Nat Genet. 1993;3:88–94. doi: 10.1038/ng0193-88. [DOI] [PubMed] [Google Scholar]
- 4.Boyd SD, Gaeta BA, Jackson KJ, Fire AZ, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Collins AM. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184:6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glanville J, Kuo TC, von Budingen HC, Guey L, Berka J, Sundar PD, Huerta G, Mehta GR, Oksenberg JR, Hauser SL, Cox DR, Rajpal A, Pons J. Naive antibody gene-segment frequencies are heritable and unaltered by chronic lymphocyte ablation. Proc Natl Acad Sci USA. 2011;108:20066–20071. doi: 10.1073/pnas.1107498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kidd MJ, Chen Z, Wang Y, Jackson KJ, Zhang L, Boyd SD, Fire AZ, Tanaka MM, Gaeta BA, Collins AM. The inference of phased haplotypes for the immunoglobulin H chain V region gene loci by analysis of VDJ gene rearrangements. J Immunol. 2012;188:1333–1340. doi: 10.4049/jimmunol.1102097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Briney BS, Willis JR, McKinney BA, Crowe JE., Jr High-throughput antibody sequencing reveals genetic evidence of global regulation of the naive and memory repertoires that extends across individuals. Genes Immun. 2012;13:469–473. doi: 10.1038/gene.2012.20. [DOI] [PubMed] [Google Scholar]
- 8.Schatz DG, Oettinger MA, Baltimore D. The V(D)J Recombination Activating Gene, Rag-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 9.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 10.Cowell LG, Davila M, Ramsden D, Kelsoe G. Computational tools for understanding sequence variability in recombination signals. Immunol Rev. 2004;200:57–69. doi: 10.1111/j.0105-2896.2004.00171.x. [DOI] [PubMed] [Google Scholar]
- 11.Feeney AJ, Atkinson MJ, Cowan MJ, Escuro G, Lugo G. A defective Vkappa A2 allele in Navajos which may play a role in increased susceptibility to haemophilus influenzae type b disease. J Clin Invest. 1996;97:2277–2282. doi: 10.1172/JCI118669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nadel B, Tang A, Escuro G, Lugo G, Feeney AJ. Sequence of the spacer in the recombination signal sequence affects V(D)J rearrangement frequency and correlates with nonrandom Vkappa usage in vivo. J Exp Med. 1998;187:1495–1503. doi: 10.1084/jem.187.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Z, Lieber MR. Lymphoid V(D)J recombination. Functional analysis of the spacer sequence within the recombination signal. J Biol Chem. 1993;268:3180–3183. [PubMed] [Google Scholar]
- 14.Souto-Carneiro MM, Sims GP, Girschik H, Lee J, Lipsky PE. Developmental changes in the human heavy chain CDR3. J Immunol. 2005;175:7425–7436. doi: 10.4049/jimmunol.175.11.7425. [DOI] [PubMed] [Google Scholar]
- 15.Volpe JM, Kepler TB. Large-scale analysis of human heavy chain V(D)J recombination patterns. Immunome Res. 2008;4:3. doi: 10.1186/1745-7580-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watson CT, Breden F. The immunoglobulin heavy chain locus: genetic variation, missing data, and implications for human disease. Genes Immun. 2012;13:363–373. doi: 10.1038/gene.2012.12. [DOI] [PubMed] [Google Scholar]
- 17.Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, Willsey AJ, Joy JB, Scott JK, Graves TA, Wilson RK, Holt RA, Eichler EE, Breden F. Complete Haplotype Sequence of the Human Immunoglobulin Heavy-Chain Variable, Diversity, and Joining Genes and Characterization of Allelic and Copy-Number Variation. Amer J Hum Genet. 2013;92:530–546. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C, Liu Y, Cavanagh MM, Le Saux S, Qi Q, Roskin KM, Looney TJ, Lee JY, Dixit V, Dekker CL, Swan GE, Goronzy JJ, Boyd SD. B-cell repertoire responses to varicella-zoster vaccination in human identical twins. Proc Natl Acad Sci USA. 2015;112:500–505. doi: 10.1073/pnas.1415875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo CY, Gerasimova T, Hao HP, Ivanova I, Chakraborty T, Selimyan R, Oltz EM, Sen RJ. Two Forms of Loops Generate the Chromatin Conformation of the Immunoglobulin Heavy-Chain Gene Locus. Cell. 2011;147:332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C, Liu Y, Xu LT, Jackson KJ, Roskin KM, Pham TD, Laserson J, Marshall EL, Seo K, Lee JY, Furman D, Koller D, Dekker CL, Davis MM, Fire AZ, Boyd SD. Effects of aging, cytomegalovirus infection, and EBV infection on human B cell repertoires. J Immunol. 2014;192:603–611. doi: 10.4049/jimmunol.1301384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, Fire AZ. Measurement and Clinical Monitoring of Human Lymphocyte Clonality by Massively Parallel V-D-J Pyrosequencing. Sci Transl Med. 2009;1 doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 23.Gaeta BA, Malming HR, Jackson KJ, Bain ME, Wilson P, Collins AM. iHMMune-align: hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23:1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 24.Lee CE, Gaeta B, Malming HR, Bain ME, Sewell WA, Collins AM. Reconsidering the human immunoglobulin heavy-chain locus: 1. An evaluation of the expressed human IGHD gene repertoire. Immunogenetics. 2006;57:917–925. doi: 10.1007/s00251-005-0062-5. [DOI] [PubMed] [Google Scholar]
- 25.Everitt BS. The Analysis of Contingency Tables. Chapman and Hall; 1977. [Google Scholar]
- 26.Hansen TO, Lange AB, Barington T. Sterile DJH rearrangements reveal that distance between gene segments on the human Ig H chain locus influences their ability to rearrange. J Immunol. 2015;194:973–982. doi: 10.4049/jimmunol.1401443. [DOI] [PubMed] [Google Scholar]
- 27.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: Evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 28.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 30.Yin FF, Bailey S, Innis CA, Ciubotaru M, Kamtekar S, Steitz TA, Schatz DG. Structure of the RAG1 nonamer binding domain with DNA reveals a dimer that mediates DNA synapsis. Nat Struct Mol Biol. 2009;16:499–508. doi: 10.1038/nsmb.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim MS, Lapkouski M, Yang W, Gellert M. Crystal structure of the V(D)J recombinase RAG1–RAG2. Nature. 2015;518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feeney AJ. Lack of N regions in fetal and neonatal mouse immunoglobulin V-D-J junctional sequences. J Exp Med. 1990;172:1377–1390. doi: 10.1084/jem.172.5.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feeney AJ. Predominance of the prototypic T15 anti-phosphorylcholine junctional sequence in neonatal pre-B cells. J Immunol. 1991;147:4343–4350. [PubMed] [Google Scholar]
- 34.Vale AM, Kapoor P, Skibinski GA, Elgavish A, Mahmoud TI, Zemlin C, Zemlin M, Burrows PD, Nobrega A, Kearney JF, Briles DE, Schroeder HW., Jr The link between antibodies to OxLDL and natural protection against pneumococci depends on D(H) gene conservation. J Exp Med. 2013;210:875–890. doi: 10.1084/jem.20121861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva-Sanchez A, Liu CR, Vale AM, Khass M, Kapoor P, Elgavish A, Ivanov, Ippolito GC, Schelonka RL, Schoeb TR, Burrows PD, Schroeder HW., Jr Violation of an evolutionarily conserved immunoglobulin diversity gene sequence preference promotes production of dsDNA-specific IgG antibodies. PLoS One. 2015;10:e0118171. doi: 10.1371/journal.pone.0118171. [DOI] [PMC free article] [PubMed] [Google Scholar]