Abstract
Histone lysine demethylases (KDMs) comprise a large class of enzymes that catalyze site-specific demethylation of lysine residues on histones and other proteins. They play critical roles in controlling transcription, chromatin architecture, and cellular differentiation. However, the genomic landscape and clinical significance of KDMs in breast cancer remain poorly characterized. Here, we conducted a meta-analysis of 24 KDMs in breast cancer and identified associations among recurrent copy number alterations, gene expression, breast cancer subtypes, and clinical outcome. Two KDMs, KDM2A and KDM5B, had the highest frequency of genetic amplification and overexpression. Furthermore, among the 24 KDM genes, KDM2A had the highest correlation between copy number and mRNA expression, and high mRNA levels of KDM2A were significantly associated with shorter survival of breast cancer patients. KDM2A has two isoforms: the long isoform is comprised of a JmjC domain, CXXC-zinc finger, PHD zinc finger, F-box, and the AMN1 protein domain; whereas the short isoform of KDM2A lacks the N-terminal JmjC domain but contains all other motifs. Detailed characterization of KDM2A in breast cancer revealed that the short isoform of KDM2A is more abundant than the long isoform at DNA, mRNA, and protein levels in a subset of breast cancers. Furthermore, our data indicate that the short isoform of KDM2A has oncogenic potential and functions as an oncogenic isoform in a subset of breast cancers. Taken together, our findings suggest that amplification and overexpression of the KDM2A short isoform is critical in breast cancer progression.
Keywords: breast cancer, histone lysine demethylase, KDM2A, gene amplification, copy number alteration
Introduction
Histone lysine demethylases (KDMs) regulate chromatin architecture and transcription and play critical roles in epigenetic signaling. Structurally, the KDMs can be broadly categorized into two functional enzymatic families. The first family, with two members (KDM1A/LSD1 and KDM1B/LSD2), can only remove mono- and dimethylated (me1/me2) histone lysine residues through an amine oxidative reaction [1,2]. The second family, referred to as the Jumonji C (JmjC) domain-containing demethylases, employs an oxygenase mechanism to demethylate mono-, di-, and trimethylated (me1/me2/me3) lysine residues [1,3]. In humans, 32 proteins belong to the JmjC domain-containing protein family [1]. Among them, 22 proteins have been shown to demethylate histone lysine residues in vitro and in vivo [1,4]. Based on sequence homologies and structural similarities, these 22 JmjC domain-containing demethylases can be categorized into seven functionally divergent protein subfamilies (KDM2-8) [5,6]. Notably, each subfamily of JmjC demethylases exhibits different substrate specificity toward different histone lysine residues. For example, the KDM2 subfamily specifically targets H3K36me2/me1 methylation marks, the KDM4 subfamily targets H3K9me3/me2 and H3K36me3/me2, and the KDM5 subfamily targets H3K4me3/me2 marks [1,3]. In general, H3K4 and H3K36 marks are associated with gene activation, whereas H3K9 and H3K27 marks are linked to gene repression [7]. Given the correlation between particular methyl marks and the transcriptional state of genes, it has been proposed that the activity of specific KDMs contributes to different transcriptional and biological outcomes, depending on the KDM substrate [7–9].
Breast cancer is a heterogeneous disease, consisting of five subtypes, including Luminal A, Luminal B, human epidermal growth factor receptor 2 (HER2/ERBB2)-enriched, basal-like, and normal-like breast cancers [10]. Both Luminal A and Luminal B breast cancers are estrogen receptor (ER) positive, but Luminal B cancers have poorer outcomes [11]. Basal-like breast cancer is highly aggressive, marked by high rates of relapse, visceral metastases, and poor prognoses [10,12]. Recently, dysregulation and mutations of KDMs have been found in different human tumors, including breast cancer [13–19]. The KDM4C gene, originally termed GASC1 (gene amplified in squamous cell carcinoma 1), was identified and cloned from the 9p24 amplified region of esophageal cancer cell lines [20]. We previously demonstrated that KDM4C is significantly amplified and overexpressed in aggressive basal-like breast cancers and functions as a transforming oncogene [17]. KDM5A is amplified and overexpressed in breast cancer and has been shown to be associated with a drug-resistant phenotype [21,22].
Many lines of evidence suggest that genetic alteration and dysregulation of KDMs are associated with breast cancer initiation and progression, where the effect is to activate expression of oncogenes, repress expression of tumor suppressors, alter DNA mismatch repair, disrupt chromosomal stability, or interact with key hormonal receptors which control cellular proliferation [1,23–25]. However, our knowledge regarding the specificity of KDMs in different subtypes of breast cancer remains largely incomplete. Thus, we systematically determined the genetic alteration and expression status of 24 KDMs. Moreover, we identified associations among recurrent copy number alterations, gene expression, breast cancer subtypes, and clinical outcome in breast cancer. Lastly, we focused on genomic and functional characterization of KDM2A because of its high level amplification and overexpression and, more importantly, because its overexpression was significantly associated with shorter survival of breast cancer patients.
Materials and methods
Cell culture
The cultures for the SUM series of breast cancer cell lines and the nontransformed human mammary epithelial cell line MCF10A have been described previously [26,27]. The Colo824 cell line was obtained from DSMZ (Braunschweig, Germany), SUM cell lines were obtained from Dr. Stephen P. Ethier, and all other cell lines in this study were obtained from ATCC (Manassas, VA, USA).
The Cancer Genome Atlas (TCGA) data for breast cancer
The DNA copy number, mutation, and overall survival datasets of 976 breast cancer samples used in this research were obtained from the cBio Cancer Genomics Portal [28,29]. The copy number for each KDM was generated from the copy number analysis algorithms GISTIC (Genomic Identification of Significant Targets in Cancer) and categorized as copy number level per gene: “−2” is a deep loss (possibly a homozygous deletion), “−1” is a heterozygous deletion, “0” is diploid, “1” indicates a low-level gain, and “2” is a high-level amplification. For mRNA expression data, the relative expression of an individual gene and the gene’s expression distribution in a reference population were analyzed. The reference population was either all tumors that are diploid for the gene in question, or, when available, normal adjacent tissue. The returned value indicates the number of standard deviations away from the mean of expression in the reference population (Z-score). Somatic mutation data were obtained from exome sequencing [28,29]. Breast cancer subtype information was extracted from a previous publication and the cBio Cancer Genomics Portal [12,28,29].
Semiquantitative PCR reactions
mRNA was prepared from human breast cancer cell lines and the MCF10A cell line by using an RNeasy Plus Mini Kit (QIAGEN). mRNA was mixed with qScript cDNA SuperMix (Quanta Biosciences, Gaithersburg, MD, USA) then converted into cDNA through a reverse-transcription (RT) reaction for real-time PCR reactions. Primer sets were ordered from Life Technologies (Carlsbad, CA, USA). A PUM1 primer set was used as a control. Semiquantitative RT-PCR was performed using the FastStart Universal SYBR Green Master (Roche Diagnostics Indianapolis, IN, USA). Primer sequences are provided in Supplementary Table S1.
Immunoblotting and antibodies
Whole-cell lysates were prepared by scraping cells from dishes into cold RIPA lysis buffer. After centrifugation at high speed, protein content was estimated by the Bradford method. A total of 20–100 μg of total cell lysate was resolved by SDS–polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane. Antibodies used in the study included anti-KDM2A (1:2000, Bethyl Laboratories A301-475A, Montgomery, TX, USA), anti-β-tubulin (1:5000, Sigma-Aldrich T8328, St. Louis, MO, USA), and anti-Vinculin (1:1000, Cell Signaling, Danvers, MA, USA).
KDM2A shRNA and siRNA knockdown
Short hairpin RNA (shRNA)-mediated KDM2A knockdown was achieved by using the Expression Arrest GIPZ lentiviral shRNAmir system (Open Biosystems, Lafayette, CO, USA). Breast cancer cells were infected with the shRNA lentivirus. Cells expressing shRNA were selected with puromycin for 7–20 days for viable cell count, protein, and RNA extraction. The infected cells were passaged for functional studies (colony formation and invasion assays). To determine distinctive roles for the KDM2A short (KDM2A-S) and long (KDM2A-L) isoforms in breast cancer, we specifically knocked down KDM2A-L and KDM2A-S with a small interfering RNA (siRNA) approach in the ZR75-1 and T47D breast cancer cell lines. siRNAs were purchased from Sigma-Aldrich (St. Louis, MO, USA). As the negative control, we used a MISSION siRNA Universal Negative Control. For transfection, cells were seeded in appropriate cell culture plates and maintained overnight under standard conditions. Plate sizes, cell densities, and siRNA quantities depended on the cell lines and the experimental setup; 10–30 nM siRNA was transfected using the MISSION siRNA transfection reagent according to the manufacturer’s protocol (Sigma-Aldrich, St. Louis, MO, USA). Five days after siRNA transfection, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays were performed. KDM2A shRNA and siRNA sequences are provided in Supplementary Table S2.
Examination of cell growth
Cell growth was assessed by using a Coulter counter or the MTT assay. For the MTT assay, cells were seeded in 24-well plates at a density of 1–2 × 104 cells per well and allowed to attach overnight. At designated time points, thiazolyl blue tetrazolium bromide (Molecular Probes, Carlsbad, CA, USA) was added to each well (final concentration 0.5 mg/ml). After removing the growth medium, dimethyl sulfoxide was added. Absorbance of the solution was read at a test wavelength of 540 nm against a reference wavelength of 570 nm. Soft agar assays were performed as previously described [17]. Cells were photographed and counted with an automated mammalian cell colony counter (Oxford Optronix GELCOUNT, Oxford, United Kingdom).
Statistical analysis
Statistical analyses were performed using R software (http://www.r-project.org) and Graphpad Prism (version 6.03). Correlations between copy numbers and mRNA levels of each KDM from 976 sequenced breast cancer specimens were analyzed using Spearman, Kendall, and Pearson correlation tests. The Spearman and Kendall tests are rank correlations—the Spearman coefficient relates the two variables while conserving the order of data points, and the Kendall coefficient measures the number of ranks that match in the data set. Although the Pearson correlation coefficient is the most widely used, it was deemed the least relevant to our study, as it measures only the strength of linear relationships and ignores all others. We used the “cor” function in R statistical software for computation, specifying in the code which type of test we wanted (Spearman, Kendall, or Pearson). The difference in mRNA expression level for each KDM between the basal-like and the other cancer subtypes was calculated using Student’s t-test. The association between the clinical outcome and individual KDM copy number and expression level was evaluated using a log-rank test. Multivariate survival analysis was conducted using the Cox regression function in R statistical software.
Results
Genetic alteration of KDMs in breast cancer
Copy number alteration (CNA) and somatic mutation are important mechanisms that activate oncogenes or inactivate tumor suppressors in human cancers [30,31]. We hypothesized that KDMs with recurrent CNA or mutation would be more likely to play important roles in breast cancer initiation and progression. The human genome encodes 24 proteins that have demonstrated abilities to demethylate histone lysine residues. Although the JmjC-containing protein JARID2 lacks histone demethylase activity, we included it in this study because JARID2 regulates recruitment of polycomb repressive complex 2 (PRC2), which is responsible for di- and trimethylation of H3K27 [32]. Except for KDM5D, which is localized on chromosome Y, we analyzed copy numbers and mutations of these 24 KDMs compiled from 976 TCGA breast cancer specimens via cBioPortal [28,29]. As shown in Table 1, we discovered distinct patterns of CNAs and mutations of KDMs in breast cancer. We found that seven KDMs exhibited high-level amplification in more than 2% of breast cancers, and two of them (KDM2A and KDM5B) exhibited high-level amplification in more than 5% of samples. No KDM genes showed homozygous deletion or somatic mutation in more than 2% of breast cancers; the highest frequency of homozygous deletion occurred in KDM6B (0.94%), and the highest frequency of mutation was found in KDM6A (1.54%) among these breast cancer samples.
Table 1.
Frequency of KDM genetic alterations and expression levels in breast cancer
| DNA Alterations | mRNA Expression Levels | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||
| Gene | Location | Amp | Gain | Diploid | Hetloss | Homdel | Mutation | Z Score>=1 | 1>Z Score>−1 | Z Score<=−1 |
|
|
|
|||||||||
| KDM5B | 1q32.1 | 13.20 | 62.68 | 22.56 | 1.56 | 0.00 | 1.23 | 45.57 | 48.87 | 5.57 |
| KDM2A | 11q13.2 | 5.20 | 22.35 | 58.11 | 14.35 | 0.00 | 1.02 | 26.80 | 57.22 | 15.98 |
| KDM8 | 16p12.1 | 4.26 | 47.30 | 40.75 | 7.69 | 0.00 | 0.00 | 27.32 | 58.87 | 13.81 |
| KDM5A | 12p11 | 4.05 | 22.14 | 60.60 | 12.99 | 0.21 | 0.61 | 24.23 | 58.56 | 17.22 |
| KDM4C | 9p24.1 | 2.29 | 14.97 | 53.12 | 29.00 | 0.62 | 0.82 | 18.35 | 55.15 | 26.49 |
| KDM1B | 6p22.3 | 2.18 | 24.32 | 59.98 | 13.31 | 0.21 | 0.10 | 21.44 | 65.46 | 13.09 |
| JARID2 | 6p22.3 | 2.08 | 24.53 | 59.98 | 13.10 | 0.31 | 0.31 | 18.56 | 68.76 | 12.68 |
| KDM4A | 1p34.1 | 1.77 | 13.51 | 57.07 | 27.65 | 0.00 | 0.82 | 16.49 | 59.59 | 23.92 |
| JMJD1C | 10q21.3 | 1.46 | 12.79 | 64.03 | 21.52 | 0.21 | 1.02 | 13.61 | 69.79 | 16.60 |
| KDM7A | 7q34 | 1.46 | 26.20 | 58.63 | 13.62 | 0.10 | 0.41 | 17.32 | 72.27 | 10.41 |
| KDM4D | 11q21 | 1.25 | 11.54 | 47.30 | 39.19 | 0.73 | 0.20 | 11.86 | 65.67 | 22.47 |
| MINA | 3q11.2 | 1.25 | 20.79 | 68.61 | 9.25 | 0.10 | 0.10 | 20.21 | 67.84 | 11.96 |
| KDM4B | 19p13.3 | 1.25 | 14.45 | 58.63 | 25.47 | 0.21 | 0.41 | 13.71 | 60.31 | 25.98 |
| KDM5C | Xp11.22 | 1.14 | 15.90 | 66.53 | 16.01 | 0.42 | 0.82 | 17.94 | 67.94 | 14.12 |
| KDM4E | 11q21 | 1.14 | 11.64 | 47.30 | 39.19 | 0.73 | 0.00 | 7.42 | 92.58 | 0.00 |
| PHF8 | Xp11.22 | 1.14 | 16.53 | 66.74 | 15.38 | 0.21 | 0.92 | 12.99 | 76.91 | 10.10 |
| KDM6A | Xp11.2 | 0.83 | 14.86 | 66.22 | 17.57 | 0.52 | 1.54 | 16.49 | 66.91 | 16.60 |
| KDM2B | 12q24.31 | 0.52 | 16.94 | 62.68 | 19.75 | 0.10 | 0.51 | 13.30 | 78.45 | 8.25 |
| PHF2 | 9q22.31 | 0.42 | 12.37 | 59.15 | 27.65 | 0.42 | 0.31 | 14.02 | 59.59 | 26.39 |
| KDM3B | 5q31 | 0.31 | 20.27 | 57.90 | 21.41 | 0.10 | 0.92 | 15.26 | 62.58 | 22.16 |
| KDM3A | 2p11.2 | 0.21 | 15.18 | 69.44 | 15.07 | 0.10 | 0.61 | 14.02 | 74.43 | 11.55 |
| KDM6B | 17p13.1 | 0.10 | 5.30 | 33.58 | 60.08 | 0.94 | 0.20 | 8.04 | 67.32 | 24.64 |
| NO66 | 14q24.3 | 0.10 | 12.16 | 56.03 | 31.39 | 0.31 | 0.31 | 13.71 | 64.43 | 21.86 |
| KDM1A | 1p36.12 | 0.00 | 5.82 | 53.53 | 40.23 | 0.42 | 0.31 | 11.65 | 74.02 | 14.33 |
Amp=high-level amplification; Gain=low-level gain; Hetloss=heterozygous deletion; Homdel= homozygous deletion. Genes were ranked based on the frequency of high-level amplification.
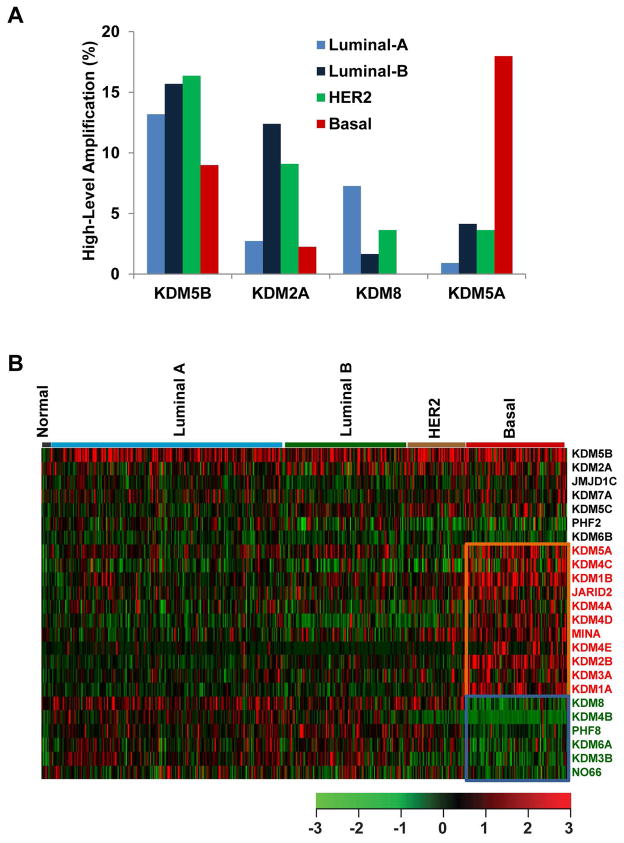
In TCGA breast cancer samples, the average CNA rate was 0.297 (range 4.85×10−7 to 0.997) based on the segmented copy number scores of the tumor samples and the paired-normal control (data updated on December 9, 2014). Notably, we found that the basal-like subtype has the highest CNA rate (average 0.472), followed by Luminal B (average 0.401); in contrast, the normal-like subtype has the lowest CNA rate (average 0.068) in breast cancer. To determine whether the genetic alteration of each KDM is specific to a breast cancer subtype, we performed an independent analysis of copy number across subtypes. Among the 976 breast cancer samples, 504 had available subtype data, including 8 normal-like, 225 Luminal-A, 121 Luminal-B, 56 HER2+, and 94 basal-like breast cancers [12]. Due to the small sample size of the normal-like subtype (n=8), those samples were excluded from this analysis. As shown in Supplementary Table S3, basal-like breast cancer had the highest frequencies of KDM gene amplification and deletion, whereas Luminal A had the lowest frequencies in every category of genetic alteration. Among the seven most frequently amplified KDMs, the frequencies of KDM5A (17.98%), KDM4C (12.36%), KDM1B (10.11%), and JARID2 (10.11%) amplification were dramatically higher in basal-like breast cancer compared with the other three subtypes (Figure 1A and Supplementary Table S3). In contrast, KDM5B (16.36%) exhibited the highest frequency of amplification in the HER2+ subtype, KDM2A (12.4%) in the Luminal B subtype, and KDM8 (7.26%) in Luminal A breast cancer samples (Figure 1A and Supplementary Table S3).
Figure 1.
Copy number and expression levels of KDMs in breast cancer. (A) Frequencies of high-level amplification of KDM2A, KDM5A, KDM5B, and KDM8 in Luminal A, Luminal B, HER2+ and basal-like breast cancers, based on TCGA database. (B) Heatmap of KDM expression profiles in different types of breast cancer. Genes with significantly higher expression (p<0.001) in basal-like tumors are highlighted in red, and genes with lower expression (p<0.001) in basal-like tumors are indicated with green.
Expression profiling of KDMs in breast cancer
We next examined the relative mRNA expression levels for each KDM based on RNA-sequencing data in TCGA breast cancer samples [28,29]. Based on the mRNA expression Z-scores [RNA-Seq V2 RSEM (RNA-Seq by Expectation-Maximization)] of each KDM, mRNA expression level was divided into three categories: Z-score ≥ 1 (overexpression), Z-score between 1 and -1, and Z-score ≤ -1 (low expression). Table 1 shows the frequency of the three expression categories for each KDM in 976 TCGA breast cancer samples. We found that six KDMs (KDM5B, KDM8, KDM2A, KDM5A, KDM1B, and MINA) were overexpressed in more than 20% of breast cancers. Among 24 KDMs, KDM5B and KDM2A exhibited the highest frequency of amplification (>5%) and overexpression (>25%) among all breast cancer samples.
Because mRNA overexpression can better translate the effect of elevated copy number to cancer initiation and progression, correlation between gene expression and copy number has been used widely as an indicator to rank candidate driver oncogenes in human cancer. Therefore, we next analyzed the correlation between copy number and mRNA level of 24 KDMs from 976 sequenced breast cancer specimens. To weigh the benefits of different statistical analyses, we compared three different correlation tests—Spearman, Kendall, and Pearson. As shown in Supplementary Table S4, all 24 KDM genes showed positive correlation between copy number and mRNA expression, with five genes (KDM2A, KDM4C, PHF2, KDM3B, and NO66) having a Spearman correlation coefficient (r) greater than 0.5 (Supplementary Table S4 and Figure S1). Among the 24 KDMs, KDM2A had the highest correlation according to both Spearman (r=0.617, p=2.20 × 10−16) and Kendall (r=0.506, p=2.20 × 10−16) analyses.
Previously, we determined that amplification and overexpression of KDM4C is more prevalent in aggressive, basal-like breast cancer [17]. To determine whether mRNA expression is associated with a specific subtype of breast cancer, we compared expression levels of each KDM in the 504 TCGA breast cancer samples with subtype information. The significance of difference for each KDM between the basal-like versus other subtypes was calculated using Student’s t-test. We found that the expression levels of 11 KDMs (KDM5A, KDM4C, KDM1B, JARID2, KDM4A, KDM4D, MINA, KDM4E, KDM2B, KDM3A, and KDM1A) were significantly higher (p<0.001), and expression levels of 6 KDMs (KDM8, KDM4B, PHF8, KDM6A, KDM3B, and NO66) were significantly lower (p<0.001) in the basal-like subtype compared with non-basal subtypes (Figure 1B and Supplementary Table S5). Two commonly amplified genes, KDM2A and KDM5B, showed the highest overexpression in both basal and non-basal breast cancers (Figure 1B). These data demonstrate that different subtypes of breast cancer have different expression patterns for each KDM gene and that KDM5B and KDM2A are the most commonly amplified/overexpressed KDM genes in breast cancer.
Association of KDMs expression with breast cancer patient survival
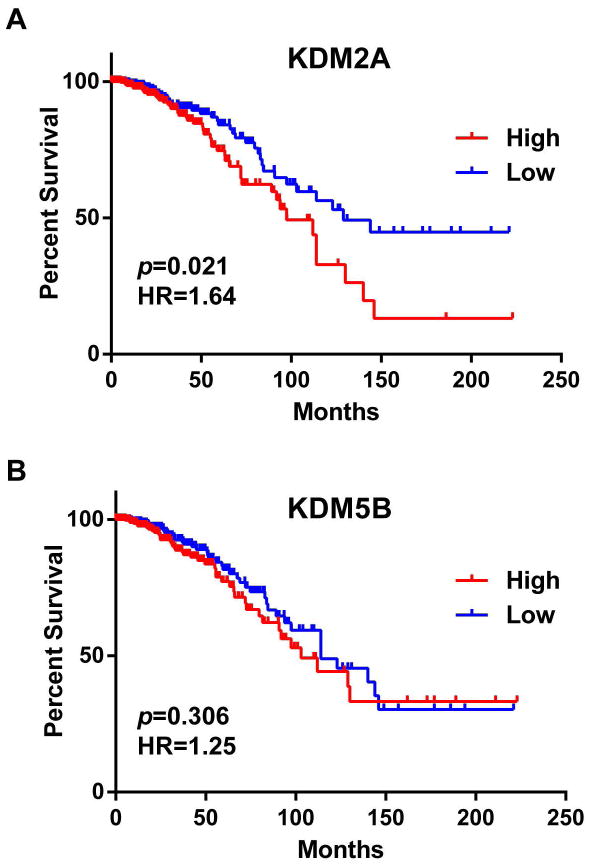
Next, we examined the relationship between KDM mRNA expression and overall patient survival in 781 of 976 breast cancer samples for which survival data were available [28,29]. Accordingly, samples were divided, based on the mRNA expression Z-scores of each KDM, into low (n=391) and high (n=390) expression groups. Supplementary Table S6 summarizes the results of a log-rank statistical analysis of overall survival correlated with expression levels of 24 KDMs in breast cancer. High mRNA levels of KDM2A, KDM3A, KDM6A, and MINA were significantly associated (p<0.05) with shorter survival in breast cancer patients (Figure 2A and Supplementary Table S6). Conversely, low mRNA levels of KDM4B were significantly associated (p<0.05) with shorter survival. Notably, KDM5B, which has the highest genetic amplification and overexpression rate for KDMs in breast cancer, was not significantly correlated with patient survival in terms of mRNA expression levels (Figure 2B). For KDM2A, the group with higher mRNA expression had a hazard ratio (HR), a ratio of the probability of death, of 1.64 (95% confidence interval, 1.085 to 2.578) compared to the group with lower expression in breast cancer.
Figure 2.
Kaplan-Meier plots of overall survival associated with mRNA expression levels of (A) KDM2A and (B) KDM5B in breast cancer.
We next examined the relationship between KDM mRNA expression and overall patient survival in 346 Luminal breast cancer samples with available survival data. We found that high mRNA levels of MINA and KDM4E were significantly associated (p<0.05) with shorter survival in Luminal breast cancer patients. Conversely, low mRNA levels of KDM4B and NO66 were significantly associated (p<0.05) with shorter survival in Luminal breast cancer patients (Supplementary Table S7). Supplementary Table S8 summarizes the results of a log-rank statistical analysis of overall survival correlated with expression levels of 24 KDMs in basal breast cancer patients (n=94), which revealed that no KDM was positively associated with shorter survival. Furthermore, we performed a multivariate analysis (Cox model, n=468) to investigate whether the mRNA expression level of each KDM was predictive of poor prognosis when compared with standard prognostic markers, including age at diagnosis, ER status, progesterone-receptor (PR) status, HER2 status, tumor size, lymph node status, metastasis status, and molecular subtype (basal vs. non-basal). We found that high mRNA levels of JARID2 (p=0.0009, HR=2.435) and JMJD1C (p=0.0264, HR=2.219) were significantly associated with shorter survival in breast cancer patients (Supplementary Table S9).
Amplification and overexpression of KDM2A isoforms in a subset of breast cancers
In our analysis of the genetic alterations of KDMs in breast cancer, we were surprised by the importance of KDM2A, because it exhibited higher frequency of amplification and overexpression in breast cancer, and its overexpression was associated with shorter survival. Thus, we further examined the genetic alterations and transforming roles of KDM2A in breast cancer.
Breast cancer cell lines retain many of the molecular characteristics of the tumors from which they were derived. Thus, they provide excellent models for studying cancer biology and for preclinical assessment of novel therapeutic strategies [33]. To further investigate the genetic alterations and expression levels of KDMs and to identify cell line models to research the therapeutic implications of KDMs, particularly KDM2A, we utilized 18 breast cancer cell lines: nine basal-like lines, three HER2+ lines, and six Luminal lines. We performed quantitative RT-PCR (qRT-PCR) assays to measure the mRNA expression level of the 24 KDMs in these breast cancer cell lines. MCF10A, an immortalized but nontumorigenic breast epithelial cell line, was used as the control. Supplementary Table S10 shows the relative expression of 24 KDM genes in 18 breast cancer cell lines compared to expression in MCF10A cells. We found that 5 (KDM2A, KDM4C, KDM5B, KDM8, and JARID2) of 7 KDM genes with highest frequencies (>2%) of amplification exhibited more than two-fold increase in mRNA expression levels in at least four breast cancer cell lines.
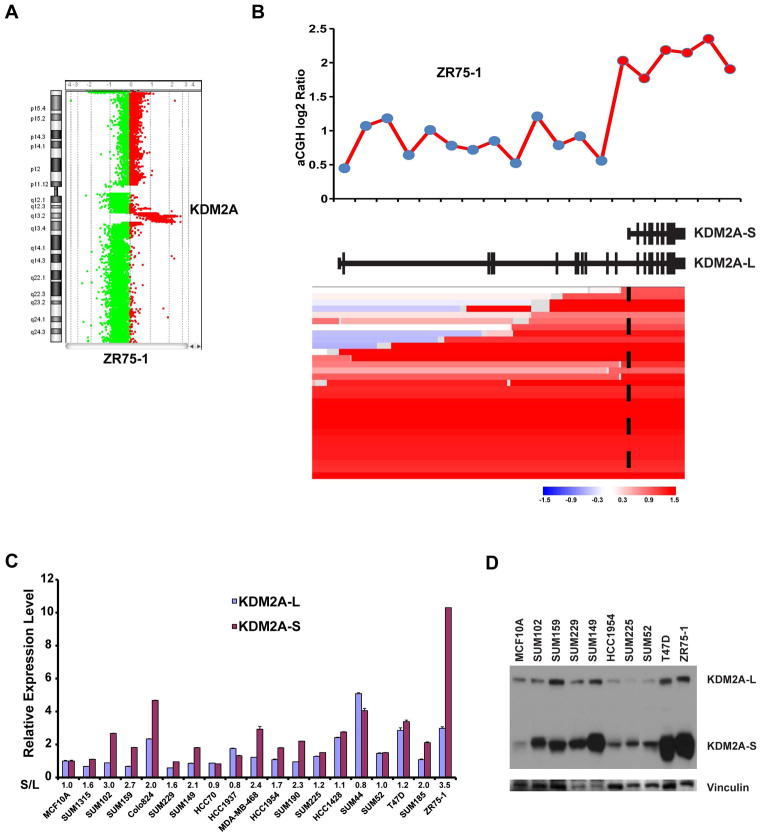
The KDM2A gene is localized to 11q13.2, a chromosomal region that is commonly altered in breast cancer, particularly in the Luminal B subtype [34]. The KDM2A gene has two alternatively spliced variants—a long isoform (21 exons) and a short isoform (10 exons)—that are derived from alternative splicing of exon 12. The KDM2A long isoform encodes an 1162-amino-acid protein containing a JmjC domain, a CXXC zinc finger (zf-CXXC) domain, a plant homeodomain (PHD) zinc finger, an F-box-like domain, and an antagonist of mitotic exit network protein 1 (AMN1) domain. The short isoform encodes a 723-amino-acid protein that lacks the N-terminal JmjC enzymatic domain (Supplementary Figure S2). Inspection of comparative genomic hybridization (CGH) array data in the ZR75-1 Luminal B cell line revealed high-level amplification of the KDM2A short isoform but only low-level gain for exons 1–12 of the KDM2A long isoform (Figures 3A and B) [35]. Based on CGH data of breast cancer cell lines, derived from the UCSC Cancer Genomics Browser, we identified two additional cell lines (ZR75B and 600MBE) that also demonstrated high-level amplification of the KDM2A short isoform (Supplementary Figure S3). This prompted us to analyze 1064 TCGA breast cancer samples from the UCSC Cancer Genomics Browser with segmented copy number information [36]. We found 16 samples with KDM2A amplification that affected only the short isoform region, without amplification or low-level gain for exons 1–12 of the KDM2A long isoform (Figure 3B). Supplementary Figure S4 shows the breast cancer subtypes, mRNA expression Z-scores and estimated exon-level transcription (RPKM: reads per kilobase of transcript per million reads mapped) of these 16 samples.
Figure 3.
Amplification and overexpression of KDM2A isoforms in breast cancer. (A) Genomic view of chromosome 11 analyzed using the Agilent oligonucleotide array (Agilent Technologies, Santa Clara, CA) in ZR75-1 cells. (B) Top: Genome plot of the KDM2A region in the array-CGH data of ZR75-1 breast cancer cells. The X-axis identifies genomic position, and Y-axis is the log2 ratio for each Agilent probe. Bottom: Heatmap of segmented copy number data of TCGA breast cancer samples, which contains the amplification of the KDM2A short isoform. The genomic structure of both the long and short isoforms of KDM2A was based on the NCBI database (middle). (C) mRNA expression levels of the short and long isoforms of KDM2A, measured by qRT-PCR, in a panel of 18 breast cancer cell lines. mRNA expression levels in MCF10A cells, an immortalized but nontumorigenic breast epithelial cell line, were arbitrarily set as 1. Relative expression levels are shown as fold changes compared with that in MCF10A cells. The relative ratio (S/L = short/long) is shown at the bottom. Data are expressed as mean ± SD. (D) Protein level of long and short isoforms of KDM2A was analyzed by western blot in nine breast cancer cell lines and the MCF10A line. Vinculin was used as the loading control. L=long isoform; S=short isoform.
We then examined the expression of the KDM2A short and long isoforms at the mRNA level in 18 breast cancer cell lines relative to MCF10A cells. We found that 7 of 18 breast cancer cell lines showed relatively high abundance of the KDM2A short isoform (ratio [short/long] > 2) (Figure 3C). Notably, ZR75-1 cells expressed a higher level of KDM2A-S (10.3-fold higher) and KDM2A-L (2.98-fold higher) than MCF10A cells at the mRNA level (Figure 3C). Next, we performed western blotting to detect KDM2A protein levels in a panel of breast cancer cell lines by using an antibody (A301-475A, Bethyl Laboratories, Montgomery, TX, USA) that is generated using an epitope that maps to a common region in both the long and short KDM2A isoforms. As shown in Figure 3D, we found that both isoforms of KDM2A are overexpressed at the protein level, but the short isoform is significantly dominant in several breast cancer cell lines, including the KDM2A-amplified ZR75-1, T47D, and SUM149 lines. Thus, our results indicate that, at DNA, mRNA, and protein levels, the short isoform of KDM2A is more abundant than the long isoform in a subset of breast cancers.
Knockdown of KDM2A inhibits cell proliferation and transforming phenotypes in breast cancer
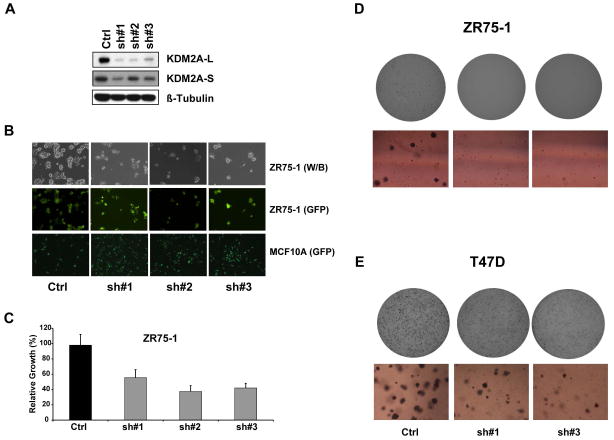
To assess the contribution of endogenous KDM2A overexpression on the transformation of human breast cancer, we examined the effects of knocking down KDM2A in ZR75-1 cells, which exhibit high-level amplification of KDM2A, particularly the short isoform. To perform shRNA knockdown experiments, we obtained three pGIPZ-KDM2A shRNA expression constructs. As a negative control, we used nonsilencing shRNA lentivirus at the same titer as KDM2A shRNA. qRT-PCR and western blot assays revealed that all three shRNAs decreased the abundance of both the long and short isoforms of KDM2A at mRNA and protein levels (Figure 4A). As shown in Figures 4B and 4C, KDM2A knockdown slowed ZR75-1 cell growth to around 40–70% of the growth of the non-silenced control. KDM2A knockdown also inhibited growth of T47D breast cancer cells (Supplementary Figure S5). In contrast, knocking down KDM2A had no impact on cell growth in SUM52 breast cancer cells without KDM2A amplification (Supplementary Figure S6). Interestingly, when the same batch of KDM2A shRNAs and the nonsilencing shRNA were used to transduce MCF10A control cells, we observed that knockdown of KDM2A expression increased the growth of MCF10A cells (Figure 4B).
Figure 4.
Effect of KDM2A knockdown on growth and transforming phenotypes of breast cancer cells. (A) Knockdown of KDM2A in ZR75-1 cells with three different shRNAs was confirmed by western blot. Beta-tubulin was used as the loading control. (B) Top row shows black and white images and middle row shows TurboGFP images of ZR75-1 cells after viral infection with control shRNA and KDM2A shRNA#1, #2, and #3. Bottom row shows TurboGFP images of MCF10A cells with or without KDM2A knockdown. In the pGIPZ shRNA vector, TurboGFP and shRNA are part of a bicistronic transcript that allows the visual marking of shRNA-expressing cells. (C) Bar graph shows relative cell growth after knocking down KDM2A in ZR75-1 breast cancer cells (p<0.05). Data are expressed as mean ± SD. Effects of KDM2A knockdown on anchorage-independent growth of (D) ZR75-1 and (E) T47D breast cancer cells. Top, representative images of one whole plate with the Oxford Optronix counter. Bottom, optical sections of representative colonies (also see Supplementary Figure S7A and B).
Anchorage-independent growth of cancer cells in vitro is a key characteristic of the tumor phenotype, particularly with respect to metastatic potential. Therefore, we sought to examine the role of KDM2A knockdown in anchorage-independent colony formation. We found that KDM2A knockdown also significantly inhibited the colony-forming ability of ZR75-1 and T47D cells in soft agar, and notably, colonies barely formed after the KDM2A knockdown of ZR75-1 cells (Figure 4D, 4E, and Supplementary Figure S7A, B). We also found that KDM2A knockdown significantly inhibited the invasive capacity of ZR75-1 cells (Supplementary Figure S7C).
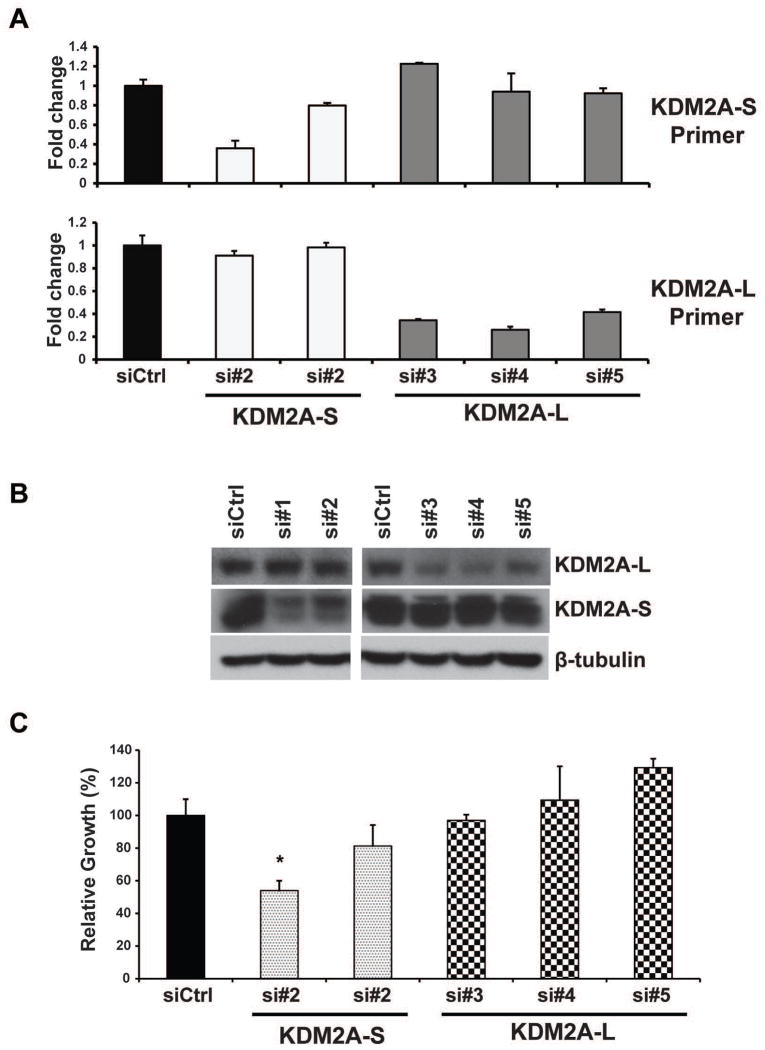
To further determine the distinctive roles of the KDM2A-S and KDM2A-L isoforms in breast cancer, we specifically knocked down KDM2A-L and KDM2A-S with a siRNA approach in ZR75-1 and T47D breast cancer cells. The specific knockdown of KDM2A-S (siRNA#1 and #2) and KDM2A-L (siRNA#3, #4 and #5) in ZR75-1 cells was confirmed by qRT-PCR and western blot assays (Figure 5A and B). We found that knockdown of the KDM2A-S isoform, but not the KDM2A-L isoform, inhibited ZR75-1 breast cancer cell growth. Knockdown of the KDM2A-S isoform with siRNA #1 slowed ZR75-1 cell growth to around 50% of the growth of the siRNA control (Figure 5C). We observed similar results in the T47D breast cancer cell line (Supplementary Figure S8). In contrast, knockdown of the KDM2A-S isoform had only a minor effect on MCF10A cell growth (Supplementary Figure S9). We also found that knockdown of the KDM2A-S isoform, but not the KDM2A-L isoform, inhibited the invasive capacity of ZR75-1 cells (Supplementary Figure S10). In summary, knockdown of KDM2A, particularly its short isoform, reduced cellular proliferation and blocked the colony-forming and invasive abilities of breast cancer cells in vitro.
Figure 5.
Effect of KDM2A isoforms on ZR75-1 breast cancer cell growth. (A) qRT-PCR and (B) immunoblot detection of KDM2A-S and -L isoforms in ZR75-1 cells at day 5 after siRNA transfection. si=siRNA. (C) MTT assays show that knockdown of the KDM2A-S isoform, but not the KDM2A-L isoform, inhibits ZR75-1 breast cancer cell growth (*p<0.05).
To understand the role of KDM2A in breast cancer, we performed RNA sequencing (RNA-Seq) in ZR75-1 breast cancer cells treated with KDM2A shRNA#1 and #3 that knocked down both isoforms of KDM2A (Figure 4A). We found that 538 genes were upregulated and 357 genes were downregulated in breast cancer cells after knockdown of KDM2A (log2 ≥ 0.45 or ≤0.45) (Supplementary Table S11). We found that several cancer-promoting factors, including BRAF, fatty acid binding protein 5 (FABP5), and Aquaporin 3 (AQP3), were all moderately downregulated by KDM2A knockdown. We validated our RNA-Seq data for BRAF, FABP5, and AQP3 with qRT-PCR assays in ZR75-1 cells after KDM2A shRNA knockdown (Supplementary Figure S11). Future investigations are required to determine whether BRAF, FABP5, and AQP3 are direct targets of KDM2A, and the subsequent role of each candidate in KDM2A-amplified breast cancers.
Discussion
In this study, we performed comprehensive genomic and transcriptomic analysis of 24 human KDMs in a panel of 18 breast cancer cell lines and 976 primary breast cancer specimens. We demonstrated that KDM5B and KDM2A are the most commonly amplified/overexpressed KDM genes in breast cancers. KDM2A had the highest correlation between copy number and mRNA expression among the 24 KDM genes studied, and higher expression of KDM2A, but not KDM5B, was significantly associated with shorter survival of breast cancer patients. Detailed characterization of KDM2A revealed that the short isoform of KDM2A at DNA, mRNA, and protein levels is more abundant than the long isoform in a subset of breast cancers. Furthermore, knockdown of the KDM2A short isoform inhibits cell proliferation and transforming phenotypes in breast cancer.
Accumulating evidence indicates that breast cancer exhibits subtype-associated mutations and CNAs, and that these alterations might contribute to the development of more aggressive forms of the disease and higher recurrence rates [12,17,37]. For example, we previously demonstrated that KDM4C is prevalently amplified and overexpressed in the basal-like subtype, one of the most deadly forms of breast cancer [17,19]. Stable KDM4C overexpression in MCF10A cells induces transformative phenotypes, whereas KDM4C knockdown in breast cancer cells inhibits proliferation in vitro and in vivo [17,19]. Using a large-scale cancer genomics data set, we found that four KDM genes (KDM1B, KDM4C, KDM5A, and JARID2) were highly amplified (>10%) and overexpressed in basal-like breast cancer. KDM1B, also called LSD2, regulates gene transcription by modulating intragenic H3K4me2 methylation [38]. Recent studies demonstrated that KDM1B plays an important role in regulating DNA methylation and gene silencing in breast cancer and that combination therapies targeting KDM1B and DNA methyltransferases exhibit great synergy in growth inhibition of breast cancer cells, including the MDA-MB-231 basal-like cell line [39]. KDM5A is an interacting partner of the retinoblastoma protein, a key regulator of cell cycle control and differentiation [40]. In human cancer, upregulation of KDM5A was found in different tumor types and implicated in promoting drug tolerance and maintenance of tumor-initiating cells [21,22,41,42]. JARID2 is required for the genomic recruitment of the PRC2 complex, which has a critical role in stem-cell identity, differentiation, and cancer [43–45]. Given the fact that KDM1B, KDM4C, KDM5A, and JARID2 are highly amplified and overexpressed in basal-like breast cancer, they might play important roles in this aggressive subtype.
Among the 24 KDMs, KDM5B was the most commonly amplified and overexpressed gene in TCGA breast cancer samples. The frequency of KDM5B amplification was higher in HER2+ (16.36%) and Luminal B (15.7%) and lower in basal-like (8.99%) breast cancers. KDM5B was initially identified as a gene upregulated by HER2 in breast cancer cells [46]. Inhibiting KDM5B in breast cancer cells has been shown to reduce proliferation and to inhibit anchorage-independent growth in soft agar. In addition, knockdown of KDM5B reduced mammary tumor growth in vivo [47,48]. KDM5B is an H3K4 demethylase that functions as a transcriptional repressor. In breast cancer, it was demonstrated that KDM5B repressed several tumor-suppressor genes [47]. Recently, KDM5B was identified as a Luminal lineage-driving oncogene in breast cancer [49]. Gene expression analysis demonstrated that there are two subgroups of HER2+ breast cancer: Luminal HER2+ tumors that cluster with Luminal B and basal HER2+ tumors that cluster with the basal-like tumors [12,50]. Here, we found that KDM5B was more commonly amplified in Luminal cancers and a subset of HER2+ breast cancers. Thus, we speculate that KDM5B plays a role in promoting Luminal lineage phenotypes in a set of HER2+ breast cancers.
Previous studies demonstrated that KDM4B is highly expressed in ER-positive, luminal subtypes of breast cancer [51–54]. KDM4B binds to the ER, which together demethylate repressive H3K9me3 marks and recruit members of the SWI/SNF-B and MLL2 chromatin remodeling complexes to induce gene expression in an estrogen-dependent manner [51]. Downregulation of KDM4B in ER-positive MCF7 or T47D cells reduced cell proliferation and tumor formation in nude mice, whereas no changes were reported in ER-negative MDA-MB-231 cells upon KDM4B knockdown [51–53]. Furthermore, KDM4B is a transcriptional target of hypoxia-inducible factor HIF [52]. Immunohistochemistry on tissue microarrays revealed that KDM4B and the hypoxia marker CA9 together stratify a subclass of breast cancer patients and predict a worse outcome of these breast cancers, even though KDM4B alone was not significantly associated with overall survival [52].
A finding of particular interest from our current study is that amplification and overexpression of the KDM2A short isoform occurs in a subset of breast cancers. KDM2A has two isoforms: the long isoform is comprised of a JmjC domain, zf-CXXC, PHD zinc finger, F-box, and the AMN1 domain; whereas the short isoform of KDM2A lacks the N-terminal JmjC enzymatic domain but contains all other motifs. The JmjC domain is the catalytic core of demethylation, whereas the zf-CXXC interacts with target genes and recognizes unmethylated DNA concentrated in CpG islands, which often overlap with a large fraction (up to 70%) of promoter regions and transcription start sites [55,56]. Previous studies using different model cells revealed that KDM2A has both oncogenic and tumor-suppressive properties. Recent elegant studies demonstrated the tumor-suppressive roles of KDM2A demethylase activity. Cheng et al. demonstrated that KDM2A long isoform depletion increased the ability of fibrosarcoma and transformed mouse embryonic fibroblast cells to grow in soft agar, and structure-guided substitutions of residues in the KDM2A catalytic pocket nullified the KDM2A-mediated functions that are important for suppressing cancer cell phenotypes [57]. Furthermore, KDM2A represses NF-κB via demethylation of p65. Overexpression of KDM2A-L inhibited proliferation and colony formation of HT-29 colon cancer cells [58,59]. Silencing of KDM2A using siRNAs increased invasion and migration by suppressing a subset of matrix metalloproteinases in breast cancer cells [60]. In contrast, it was shown that KDM2A has tumorigenic capabilities in lung and gastric cancer cells [61,62]. The existence of two KDM2A isoforms and the ratio of their expression might be the reason for contrasting observations regarding the role of KDM2A in different model cells. Indeed, we have demonstrated that silencing of KDM2A inhibited the growth of ZR75-1 and T47D breast cancer cells but enhanced the growth of nontumorigenic MCF10A mammary epithelial cells. Furthermore, specific knockdown of KDM2A-S inhibited ZR75-1 breast cancer cell growth. Collectively, our data suggest that the short isoform of KDM2A has oncogenic potential and functions as an oncogenic isoform in a subset of breast cancers.
The long isoform of KDM2A is known as an H3K36me2/me1 demethylase that generally represses gene expression [5]. In this study, we identified several KDM2A target gene candidates, including FABP5 and BRAF, which were downregulated by KDM2A knockdown in ZR75-1 breast cancer cells. FABP5 is involved in fatty acid uptake, transport, and metabolism, and it is highly upregulated in breast cancers [63]. Genetic ablation of FABP5 was shown to suppress HER2-induced mammary tumorigenesis [63]. BRAF, which is a commonly mutated gene that functions as a driver oncogene in melanoma, was also identified as a potential target of KDM2A. Mutation of BRAF is rare in breast cancer (approximately 0.5%, based on the TCGA database) [64,65]. However, BRAF was found to be upregulated in breast cancer, particularly in breast cancer metastases to the brain [66]. We surveyed KDM2A chromatin immunoprecipitation sequencing (ChIP-Seq) data generated in mouse embryonic stem cells [34]. We found that KDM2A directly bound to the braf and fabp5 genomic loci (Supplementary Figure S12). We also found that the short isoform of KDM2A is more abundant at mRNA and protein levels as compared to the long isoform, even without KDM2A short isoform amplification, in several breast cancer cell lines. Alternative splicing of KDM2A in breast cancer cells can be regulated at different steps of spliceosome assembly by different splicing factors and by many different mechanisms that rely on cis-acting elements [67,68]. Future investigations using various cellular and animal experimental models are required to more precisely address the mechanism of how KDM2A short and long isoforms are involved in breast tumorigenesis.
In conclusion, our findings have added significant information to the genomic and transcriptomic profiles of KDMs in different breast cancer subtypes. We revealed that breast cancer subtypes show distinct copy number alteration patters and differential expression for each KDM gene. KDM2A and KDM5B had the highest frequency of genetic amplification in breast cancer, and elevated mRNA levels of KDM2A were significantly associated with shorter survival of breast cancer patients. The short isoform of KDM2A, at DNA, mRNA, and protein levels, is more abundant than the long isoform in a subset of breast cancers. Knockdown of the KDM2A short isoform inhibited cell proliferation and transforming phenotypes in breast cancer. Mechanistically, KDM2A might regulate several oncogenic factors. Taken together, our findings suggest that amplification and overexpression of the KDM2A short isoform is critical in the progression of breast cancer.
Supplementary Material
Acknowledgments
We thank Sarah Kimball for technical contributions. We also thank Dr. Stephen P. Ethier for providing the SUM breast cancer cell lines and for his continuous encouragement.
Grant Support: This work was partially supported by grants from the NIH/NCI R21CA175244 and the Office of Research on Women’s Health (OWRH), the Mary Kay Foundation Cancer Research Grant Program, and the Karmanos Cancer Institute-SRIG to Dr. Z-Q.Y.; by funding from the Cancer Biology Graduate Program, Wayne State University School of Medicine, to A.H.; and by funding from the China Scholarship Council Program fellowship to H.L. The Genomics Core is supported, in part, by NIH Center grant P30 CA022453 to the Karmanos Cancer Institute at Wayne State University.
Abbreviations
- KDM
histone lysine demethylase
- me1/me2/me3
mono-, di-, and trimethylated
- JmjC
Jumonji C
- HER2/ERBB2
human epidermal growth factor receptor 2
- TCGA
The Cancer Genome Atlas
- KDM2A-S
KDM2A short isoform
- KDM2A-L
KDM2A long isoform
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- RNA-Seq
RNA-sequencing
- CNA
copy number alteration
- PRC2
polycomb repressive complex 2
- zf-CXXC
CXXC zinc finger domain
- PHD
plant homeodomain
- AMN1
antagonist of mitotic exit network protein 1 domain
- BRAF
B-Raf proto-oncogene
- FABP5
fatty acid binding protein 5
- AQP3
aquaporin 3
- qRT-PCR
quantitative reverse-transcriptase PCR
- ChIP-Seq
chromatin immunoprecipitation sequencing
References
- 1.Labbe RM, Holowatyj A, Yang ZQ. Histone lysine demethylase (KDM) subfamily 4: structures, functions and therapeutic potential. American journal of translational research. 2014;6(1):1–15. [PMC free article] [PubMed] [Google Scholar]
- 2.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 3.Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nature reviews Molecular cell biology. 2012;13(5):297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- 4.Johansson C, Tumber A, Che KH, et al. The roles of Jumonji-type oxygenases in human disease. Epigenomics. 2014;6(1):89–120. doi: 10.2217/epi.13.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7(9):715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 6.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 7.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nature reviews Molecular cell biology. 2005;6(11):838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 8.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25(1):15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 9.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA : the journal of the American Medical Association. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 11.Creighton CJ. The molecular profile of luminal B breast cancer. Biologics : targets & therapy. 2012;6:289–297. doi: 10.2147/BTT.S29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinatzer U, Gollinger M, Mullauer L, Raderer M, Chott A, Streubel B. Mucosa-associated lymphoid tissue lymphoma: novel translocations including rearrangements of ODZ2, JMJD2C, and CNN3. Clin Cancer Res. 2008;14(20):6426–6431. doi: 10.1158/1078-0432.CCR-08-0702. [DOI] [PubMed] [Google Scholar]
- 15.Italiano A, Attias R, Aurias A, et al. Molecular cytogenetic characterization of a metastatic lung sarcomatoid carcinoma: 9p23 neocentromere and 9p23–p24 amplification including JAK2 and JMJD2C. Cancer Genet Cytogenet. 2006;167(2):122–130. doi: 10.1016/j.cancergencyto.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Suikki HE, Kujala PM, Tammela TL, van Weerden WM, Vessella RL, Visakorpi T. Genetic alterations and changes in expression of histone demethylases in prostate cancer. Prostate. 2010;70(8):889–898. doi: 10.1002/pros.21123. [DOI] [PubMed] [Google Scholar]
- 17.Liu G, Bollig-Fischer A, Kreike B, et al. Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene. 2009;28(50):4491–4500. doi: 10.1038/onc.2009.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rui L, Emre NC, Kruhlak MJ, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo W, Chang R, Zhong J, Pandey A, Semenza GL. Histone demethylase JMJD2C is a coactivator for hypoxia-inducible factor 1 that is required for breast cancer progression. Proc Natl Acad Sci U S A. 2012;109(49):E3367–3376. doi: 10.1073/pnas.1217394109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang ZQ, Imoto I, Fukuda Y, et al. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60(17):4735–4739. [PubMed] [Google Scholar]
- 21.Hou J, Wu J, Dombkowski A, et al. Genomic amplification and a role in drug-resistance for the KDM5A histone demethylase in breast cancer. American journal of translational research. 2012;4(3):247–256. [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young LC, McDonald DW, Hendzel MJ. Kdm4b Histone Demethylase Is a DNA Damage Response Protein and Confers a Survival Advantage following γ-Irradiation. J Biol Chem. 2013;288(29):21376–21388. doi: 10.1074/jbc.M113.491514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black JC, Manning AL, Van Rechem C, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154(3):541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young LC, Hendzel MJ. The oncogenic potential of Jumonji D2 (JMJD2/KDM4) histone demethylase overexpression1. Biochemistry and Cell Biology. 2012;91(6):369–377. doi: 10.1139/bcb-2012-0054. [DOI] [PubMed] [Google Scholar]
- 26.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Research. 2006;66(24):11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 27.Forozan F, Veldman R, Ammerman CA, et al. Molecular cytogenetic analysis of 11 new breast cancer cell lines. British journal of cancer. 1999;81(8):1328–1334. doi: 10.1038/sj.bjc.6695007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao JJ, Aksoy BA, Dogrusoz U, et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Science signaling. 2013;6(269) doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22(8):447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 31.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34(4):369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 32.Kaneko S, Bonasio R, Saldana-Meyer R, et al. Interactions between JARID2 and Noncoding RNAs Regulate PRC2 Recruitment to Chromatin. Molecular Cell. 2014;53(2):290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prat A, Karginova O, Parker JS, et al. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast cancer research and treatment. 2013;142(2):237–255. doi: 10.1007/s10549-013-2743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adelaide J, Finetti P, Bekhouche I, et al. Integrated profiling of basal and luminal breast cancers. Cancer Res. 2007;67(24):11565–11575. doi: 10.1158/0008-5472.CAN-07-2536. [DOI] [PubMed] [Google Scholar]
- 35.Crowder RJ, Phommaly C, Tao Y, et al. PIK3CA and PIK3CB Inhibition Produce Synthetic Lethality when Combined with Estrogen Deprivation in Estrogen Receptor-Positive Breast Cancer. Cancer Research. 2009;69(9):3955–3962. doi: 10.1158/0008-5472.CAN-08-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldman M, Craft B, Swatloski T, et al. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Research. 2013;41(D1):D949–D954. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gatza ML, Silva GO, Parker JS, Fan C, Perou CM. An integrated genomics approach identifies drivers of proliferation in luminal-subtype human breast cancer. Nat Genet. 2014;46(10):1051–1059. doi: 10.1038/ng.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang R, Barbera AJ, Xu Y, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39(2):222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katz TA, Vasilatos SN, Harrington E, Oesterreich S, Davidson NE, Huang Y. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast cancer research and treatment. 2014;146(1):99–108. doi: 10.1007/s10549-014-3012-9. [DOI] [PubMed] [Google Scholar]
- 40.Defeojones D, Huang PS, Jones RE, et al. Cloning of Cdnas for Cellular Proteins That Bind to the Retinoblastoma Gene-Product. Nature. 1991;352(6332):251–254. doi: 10.1038/352251a0. [DOI] [PubMed] [Google Scholar]
- 41.Lin W, Cao J, Liu J, et al. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci U S A. 2011;108(33):13379–13386. doi: 10.1073/pnas.1110104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teng YC, Lee CF, Li YS, et al. Histone Demethylase RBP2 Promotes Lung Tumorigenesis and Cancer Metastasis. Cancer Res. 2013;73(15):4711–4721. doi: 10.1158/0008-5472.CAN-12-3165. [DOI] [PubMed] [Google Scholar]
- 43.Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9(11):773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- 44.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20(10):1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 45.Ren G, Baritaki S, Marathe H, et al. Polycomb protein EZH2 regulates tumor invasion via the transcriptional repression of the metastasis suppressor RKIP in breast and prostate cancer. Cancer Res. 2012;72(12):3091–3104. doi: 10.1158/0008-5472.CAN-11-3546. [DOI] [PubMed] [Google Scholar]
- 46.Lu PJ, Sundquist K, Baeckstrom D, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274(22):15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- 47.Yamane K, Tateishi K, Klose RJ, et al. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25(6):801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 48.Catchpole S, Spencer-Dene B, Hall D, et al. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. International journal of oncology. 2011;38(5):1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto S, Wu Z, Russnes HG, et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell. 2014;25(6):762–777. doi: 10.1016/j.ccr.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riaz M, van Jaarsveld MT, Hollestelle A, et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013;15(2):R33. doi: 10.1186/bcr3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kawazu M, Saso K, Tong KI, et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6(3):e17830. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang J, Jubb AM, Pike L, et al. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010;70(16):6456–6466. doi: 10.1158/0008-5472.CAN-10-0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi L, Sun L, Li Q, et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc Natl Acad Sci U S A. 2011;108(18):7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73(10):2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38(2):179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 57.Cheng Z, Cheung P, Kuo AJ, et al. A molecular threading mechanism underlies Jumonji lysine demethylase KDM2A regulation of methylated H3K36. Genes Dev. 2014;28(16):1758–1771. doi: 10.1101/gad.246561.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu T, Jackson MW, Wang B, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci U S A. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu T, Jackson MW, Singhi AD, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci U S A. 2009;106(38):16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rizwani W, Schaal C, Kunigal S, Coppola D, Chellappan S. Mammalian lysine histone demethylase KDM2A regulates E2F1-mediated gene transcription in breast cancer cells. PLoS One. 2014;9(7):e100888. doi: 10.1371/journal.pone.0100888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wagner KW, Alam H, Dhar SS, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2013;123(12):5231–5246. doi: 10.1172/JCI68642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang Y, Liu Y, Yu L, et al. Histone demethylase KDM2A promotes tumor cell growth and migration in gastric cancer. Tumour Biol. 2014 doi: 10.1007/s13277-014-2630-5. [DOI] [PubMed] [Google Scholar]
- 63.Levi L, Lobo G, Doud MK, et al. Genetic ablation of the fatty acid-binding protein FABP5 suppresses HER2-induced mammary tumorigenesis. Cancer Res. 2013;73(15):4770–4780. doi: 10.1158/0008-5472.CAN-13-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tilch E, Seidens T, Cocciardi S, et al. Mutations in EGFR, BRAF and RAS are rare in triple-negative and basal-like breast cancers from Caucasian women. Breast cancer research and treatment. 2014;143(2):385–392. doi: 10.1007/s10549-013-2798-1. [DOI] [PubMed] [Google Scholar]
- 65.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14(7):455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Salhia B, Kiefer J, Ross JT, et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PLoS One. 2014;9(1):e85448. doi: 10.1371/journal.pone.0085448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature reviews Molecular cell biology. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer discovery. 2013;3(11):1228–1237. doi: 10.1158/2159-8290.CD-13-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.