Abstract
Autophagy, an important catabolic pathway involved in a broad spectrum of human diseases, implies the formation of double-membrane-bound structures called autophagosomes (AP), which engulf material to be degraded in lytic compartments. How APs form, especially how the membrane expands and eventually closes upon itself, is an area of intense research. Ubiquitin-like ATG8 has been related to both membrane expansion and membrane fusion, but the underlying molecular mechanisms are poorly understood. Here, we used two minimal reconstituted systems (enzymatic and chemical conjugation) to compare the ability of human ATG8 homologs (LC3, GABARAP, and GATE-16) to mediate membrane fusion. We found that both enzymatically and chemically lipidated forms of GATE-16 and GABARAP proteins promote extensive membrane tethering and fusion, whereas lipidated LC3 does so to a much lesser extent. Moreover, we characterize the GATE-16/GABARAP-mediated membrane fusion as a phenomenon of full membrane fusion, independently demonstrating vesicle aggregation, intervesicular lipid mixing, and intervesicular mixing of aqueous content, in the absence of vesicular content leakage. Multiple fusion events give rise to large vesicles, as seen by cryo-electron microscopy observations. We also show that both vesicle diameter and selected curvature-inducing lipids (cardiolipin, diacylglycerol, and lyso-phosphatidylcholine) can modulate the fusion process, smaller vesicle diameters and negative intrinsic curvature lipids (cardiolipin, diacylglycerol) facilitating fusion. These results strongly support the hypothesis of a highly bent structural fusion intermediate (stalk) during AP biogenesis and add to the growing body of evidence that identifies lipids as important regulators of autophagy.
Introduction
Macroautophagy, autophagy in the context of this work, is an intracellular degradation pathway conserved in all eukaryotes (1). This process does not only provide nutrients to maintain vital cellular functions under amino acid-limiting conditions, but also plays an important role in a wide range of physiological processes (2). Not surprisingly, impaired autophagic function seems to underlie a wide range of pathological conditions (3). During autophagy portions of the cytosol and even entire organelles are sequestered by double-membrane structures called autophagosomes (AP). Eventually, AP fuse with the lysosomal system where the inner membrane and the cargo are degraded. One outstanding question is how the AP membrane is formed and elongated. The most widely accepted hypotheses propose the formation of a core preautophagosomal structure, named isolation membrane or phagophore, upon which adhesion and fusion of further membrane structures would occur (4, 5, 6). To date, at least 35 autophagy-related genes (ATG) have been identified and related to the process (7, 8). Among them, the ATG8 family of ubiquitin-like proteins is among the late autophagosome-specific proteins to bind the AP (9). Conjugation of ATG8 to phosphatidylethanolamine (PE) at the autophagosomal membrane is essential for AP elongation (10, 11) but the underlying mechanism of AP growth remains essentially unknown.
Although most studies have been focused on the ATG8 conjugation system derived from Saccharomyces cerevisiae, the role of ATG8 homologs in higher eukaryotes is less well characterized (12). Remarkably, whereas yeast has a single ATG8 gene, mammals have up to seven homologs (13), which can be divided into LC3 and GABARAP subfamilies. Members of both subfamilies have overall similar structures (14, 15) (Fig.1 A), are localized to autophagosomal membranes (16, 17), and are all essential for the autophagic process (18, 19). However, far from being simply redundant members of a protein family, specific members are suggested to act at different stages of AP biogenesis (19).
Figure 1.
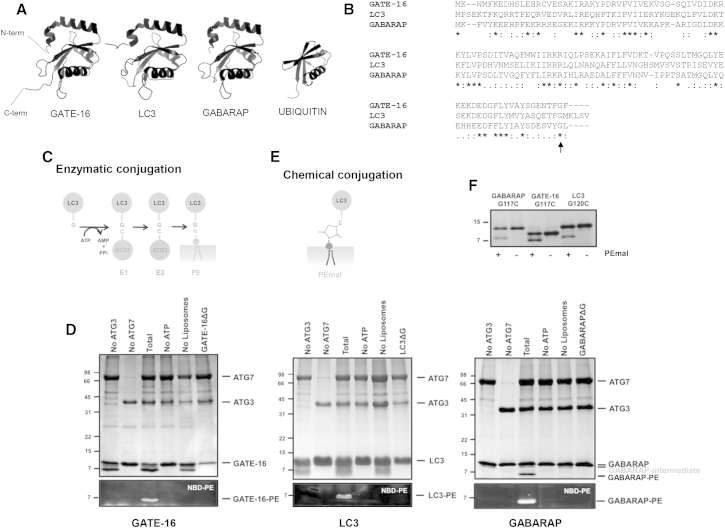
In vitro reconstitution of the lipidation reaction. (A) Crystal structures of the indicated human ATG8 family members and ubiquitin. All ATG8S share a similar ubiquitin-like fold with two additional amino-terminal α-helices. GATE-16 (Protein Data Bank (PDB): 1EO6), LC3B (PDB: 1V49), GABARAP (PDB: 1KOT), and ubiquitin (PDB: 1UBI). Helix shape represents an α-helix; arrow shape represents a β-sheet; arrowhead represents the carboxyl termini of β sheets. (B) ClustalW sequence alignment of human GATE-16 (database accession number: P60520), LC3B (Q9GZQ8), and GABARAP (Q6IAW1). The black arrow indicates the conserved PE-conjugating glycine residue present at the C-terminus of the processed forms of the ATG8-like proteins. (C) Outline representation of the enzymatic conjugation of LC3/GABARAP to PE. (D) Conjugation of recombinant LC3, GATE-16, and GABARAP with PE. Reaction media contain 1 μM ATG7, 1 μM ATG3, 10 μM ATG8 homologs, 1 mM DTT, 5 mM ATP, and 0.4 mM LUV [PC/DOPE/bl-PI/NBD-PE (35/45/10/10 mol ratio)]. Each reaction mixture was incubated at 37°C for 90 min, run on 12.5% SDS-PAGE gels, and visualized by CBB-staining (top panels) or using a VersaDoc MP 4000 Imaging System (bottom panels). In CBB-stained gels lipidated forms of GATE-16 and LC3 are undistinguishable from the adenylated intermediates (compare lanes 1, 3, and 5 from the right). In the case of GABARAP, the adenylated intermediate appears immediately below the nonlipidated form (GABARAP-intermediate) and clearly distinguishable from the lipidated form (GABARAP-PE). Conjugations of proteins to NBD-PE results in the appearance of a fluorescent faster migrating band corresponding to the lipidated form of the protein. Molecular masses are shown in kDa on the left-hand side. For negative controls, ATG8 homologs lacking the reactive C-terminal glycine (ΔG) or reactions lacking either ATG7, ATG3, ATP, or LUVs were used. (E) Outline representation of the chemical conjugation of LC3/GABARAP to PEmal. (F) Chemical conjugation of recombinant LC3BG120C, GATE-16G117C, and GABARAPG117C. The indicated recombinant proteins (10 μM) were incubated at 37°C for 90 min with 0.4 mM LUV [PC/DOPE/PEmal/bl-PI (35/40/15/10 mol ratio)]. Cross-linking of the proteins to the maleimide-functionalized lipid results in the appearance of a faster migrating band corresponding to the lipidated form of the protein. For negative control liposomes lacking PEmal were used. Molecular masses are shown in kDa on the left-hand side.
Several authors have approached autophagy, and particularly the fusion processes leading to autophagosomal growth, using model membrane systems consisting of vesicles of defined lipid compositions and purified proteins. Nakatogawa et al. (20) showed in this kind of system that yeast ATG8 can trigger liposome tethering and hemifusion, whereas Weidberg et al. (21) found that mammalian orthologues of ATG8 have the same activities. Furthermore, Nair and co-workers (22) described how the in vitro fusion activity of ATG8-like proteins depended strongly on the presence of high amounts of PE (in the 50% mol range). These studies have elicited the interest of investigators on the role of specific lipids and their corresponding synthesizing enzymes in the process of AP formation. Because of the important role of PI3P in autophagy (23, 24), this area of research has recently attracted considerable attention and a variety of other lipids have been involved (25, 26, 27). However, how the various membrane lipid species modulate AP biogenesis, particularly at the level of autophagic membrane elongation, remains largely unexplored. The ever-increasing number of ATG proteins and the potential combinatorial interactions among them and with cellular factors make the autophagy network very complex and difficult to dissect and analyze in intact cells. In this regard, compositionally defined in vitro reconstituted systems are increasingly being used to reveal fundamental information concerning the mechanism of action of individual autophagy proteins and their regulatory factors (28). Here, we used two minimal reconstituted systems, based on a minimal set of recombinant proteins together with synthetic vesicles of defined lipid compositions, to gain further insight into the ability of human ATG8 homologs (LC3, GABARAP, and GATE-16) to mediate membrane fusion. Our in vitro studies shed light on the role of the various human ATG8 proteins during AP elongation and identify selected lipids as potent modulators of the process.
Materials and Methods
Materials
Egg phosphatidylcholine (PC), bovine liver phosphatidylinositol (bl-PI), heart cardiolipin (CL), 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (DOPE), lysophosphatidylcholine (LPC), egg diacylglycerol (DAG), 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine-N-(lissamine rhodamine B sulfonyl) (Rho-PE), 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine-p-maleimidomethyl-cyclohexanecarboxamide (PEmal), and 1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphatidylethanolamine (NBD-PE) were purchased from Avanti Polar Lipids (Alabaster, AL). NaCl, Tris Base, MgCl2, Triton X-100, and n-octyl-β-D-glucopyranoside were obtained from Calbiochem (Darmstadt, Germany). N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadecanoyl-sn-glycero-3-Phosphatidylethanolamine (NBD-PE), (1,3,6 aminonaphtalene-trisulfonate (ANTS), and p-xilene-bisdipicolinic acid (DPX) were purchased from Molecular Probes (Eugene, OR). Dithiothreitol (DTT) and tris(2-carboxyethyl)phosphine (TCEP) were purchased from Sigma-Aldrich (St. Louis, MO).
DNA constructs and site-directed mutagenesis
Plasmids for expression of human ATG7, human ATG3, and the various homologs of ATG8 (human LC3B, human GATE-16, and human GABARAP) were kindly provided by Dr. Isei Tanida (National Institute of Infectious Diseases, Tokyo, Japan). Note that each ATG8 homolog was a truncated form ending in the reactive C-terminal glycine such that no ATG4-mediated preprocessing was necessary. Wild-type plasmids were mutated by site-directed mutagenesis to obtain LC3G120C, GATE-16C15SG117C (GATEG117C for simplicity), and GABARAPG117C by TopGeneTech (Montreal, Canada) and confirmed by sequencing.
Expression and purification of recombinant proteins
Proteins were purified from soluble fractions of bacterial extracts obtained in the absence of detergents, and were >90% pure as evaluated by Coomassie-stained sodium didecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). ATG7 was expressed by baculoviral infection of HighFive (H5) insect cells. ATG7 in pFASTBAC HTa plasmid DNA was used to transform DH10Bac Escherichia coli for transposition into the bacmid. We used blue/white colony selection to identify colonies containing the recombinant bacmid and they were confirmed by polymerase chain reaction. The recombinant bacmid was then isolated and purified using the Macherey Nagel endotoxin-free DNA purification kit. H5 insect cells were transfected with purified bacmid using Lipofectamine from Invitrogen (Waltham, MA) in TC-100 insect media (Sigma-Aldrich) supplemented with the appropriate antibiotics. When the transfected cells demonstrated signs of late stage infection (typically around 72 h), the medium containing the free virus was collected. We repeated cycles of transfection and virus collection to amplify the viral stock. Cells were collected after 48 h infection followed by centrifugation at 5000 × g for 10 min. The pellet was resuspended and sonicated in a breaking buffer consisting of 50 mM Tris pH 8, 150 mM NaCl, and freshly prepared 1 mM TCEP and protease inhibitors. The lysate was cleared by 30 min centrifugation at 30,000 × g and loaded on cobalt resin (Clontech, Mountain View, CA) for 3 h at 4°C. Protein was eluted with 500 mM imidazole, concentrated up to 500 μl using YM-30 microcons (Millipore, Darmstadt, Germany) and loaded onto a Superdex TM200HR 10/30 size exclusion column (GE Healthcare, Buckinghamshire, UK) equilibrated in 50 mM Tris pH 8, 150 mM NaCl, supplemented with freshly added 1 mM DTT and protease inhibitors. Purified protein was kept at 4°C. Plasmids of ATG3, LC3, GATE-16, GABARAP, and their corresponding mutant forms were transformed onto E. coli BL21 (λDE3) cells. Cells were grown to OD600 = 0.8 and induced with 0.5 mM isopropyl-b-D-thiogalactoside for 3 h at 37°C in the case of GATE-16 and LC3, 4 h at 20°C in the case of GABARAP, and 18 h at 18°C in the case of ATG3. Following centrifugation at 5000 × g for 10 min, the pellet was resuspended and sonicated in breaking buffer (phosphate buffered saline with protease inhibitors mixture and 1 mM DTT). The lysate was cleared by 30 min centrifugation at 30,000 × g, loaded on glutathione-agarose (Sigma-Aldrich) and incubated for 3 h at 4°C. Proteins were eluted by cleavage with PreScission Protease (GE Healthcare) in Precission Buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM EDTA) with freshly added 1 mM DTT, for 4 h at 4°C. In the case of cysteine C-terminal mutants 1 mM TCEP was used instead of DTT to avoid interference with the thiol-maleimide coupling. Proteins were aliquoted, flash-frozen and stored at −80°C until further use.
Liposome preparation
The appropriate lipids were mixed in organic solution, and the solvent was evaporated to dryness under a stream of N2. The sample was then kept under vacuum for 2 h to remove solvent traces. The lipids were swollen in appropriate buffers. Small unilamellar vesicles (SUVs) were prepared from the swollen lipids by sonication with a probe tip sonicator (MSE Soniprep 150 (MSE, UK)) for 10 min (10 s on and off cycles) at 10–20 μm amplitude. The vials were kept on ice during the process to avoid overheating. When large unilamellar vesicles (LUVs) were required, swollen lipids were subjected to 10 freeze/thaw cycles and then extruded using 0.05-μm pore-size Nuclepore (San Diego, CA) filters, as described by Mayer et al (29). Vesicle size was checked by quasielastic light scattering, using a Malvern Zeta-Sizer 4 spectrometer (Malvern Instruments, Worcestershire, UK). LUVs had an average diameter of 80 nm and SUVs had an average diameter of 50 nm. Lipid concentration was determined by phosphate analysis (30).
Dynamic light scattering measurements
To determine size-distribution profiles of the liposomes, the samples were appropriately diluted with the conjugation reaction buffer and subjected to dynamic light scattering (DLS) measurements at 37°C using a Zetasizer Nano-S system (Malvern Instruments).
In vitro enzymatic lipidation reaction
To recreate PE-conjugation in vitro, purified ATG7 (1 μM), ATG3 (1 μM), and LC3 or one of its homologs (10 μM) were mixed with liposomes (0.4 mM total lipid) in the presence of 1 mM DTT and 5 mM ATP in conjugation buffer (50 mM Tris pH 7.5, 150 mM NaCl, and 1 mM MgCl2) and incubated at 37°C for 90 min. The reaction was stopped with 6× SDS-PAGE sample buffer and heated at 90°C for 5 min. Samples were run on SDS-PAGE gels and visualized with Coomassie Blue stain (Thermo Scientific; Waltham, MA).
Lipidation of LC3 and homologs with PEmal
To reconstitute LC3-PE conjugation in vitro without the use of ATG7, ATG3, or PE, purified LC3G120C, GATE-16G117C, or GABARAPG117C (10 μM each) were incubated with 15% PEmal-containing liposomes at a final lipid concentration of 0.4 mM in conjugation buffer. The reaction mixture was incubated at 37°C for 90 min and visualized by SDS-PAGE.
Aggregation assays
Liposome aggregation was monitored in an Uvikon 922 (Kontron, Augsburg, Germany) spectrophotometer as an increase in turbidity (absorbance at 400 nm) of the sample. All assays were carried out at 37°C with continuous stirring.
Lipid mixing and aqueous contents mixing assays in large unilamellar vesicles
A fluorescence resonance energy transfer assay was used to monitor intervesicular membrane lipid mixing (31). The appropriate LUVs containing 1.5 mol % NBD-PE and 1.5 mol % Rh-PE were mixed with ninefold excess of unlabeled LUVs. NBD-PE emission was monitored in a QuantaMaster spectrofluorometer (Photon Technology International, Birmingham, New Jersey) in a thermostatically controlled 1-cm path length cuvette with constant stirring at 37°C. NBD emission was monitored at 530 nm with the excitation wavelength set at 467 nm (slits, 2 nm). A 515 nm cut-off filter was placed between the sample and the emission monochromator to avoid scattering interference. Inner monolayer lipid mixing was measured using asymmetrically labeled membrane vesicles produced by the quenching of the outer leaflet NBD-PE fluorescence upon addition of sodium dithionite (see (32)). Dithionite was removed by gel filtration in Sephadex G-25M, using conjugation buffer for elution.
Intervesicular content mixing was monitored by the ANTS/DPX assay (33). ANTS emission was monitored at 530 nm with the excitation wavelength set at 360 nm (slits, 2 nm). A 470 nm cut-off filter was placed between the sample and the emission monochromator to avoid scattering interference. 0% vesicle content mixing was set by using a 1:1 mixture of ANTS and DPX liposomes. 100% mixing of contents corresponded to the fluorescence of the vesicles containing coencapsulated ANTS and DPX. Details for the intervesicular lipid mixing and contents mixing assays can be found in Goñi et al. (34).
Vesicle contents leakage assay
Leakage of vesicle contents was monitored by the ANTS/DPX assay (29). ANTS emission was monitored at 530 nm with the excitation wavelength set at 360 nm (slits, 2 nm). A 470 nm cut-off filter was placed between the sample and the emission monochromator to avoid scattering interference. To establish the 100% leakage signal, Triton X-100 was added to a concentration of 1%. Details for the vesicle contents leakage assay can be found in Goñi et al. (34).
Cryo-electron microscopy
For the cryoelectron microscopy (cryo-EM) studies, conjugation reactions were performed as explained previously. Grids were prepared following standard procedures and observed at liquid nitrogen temperatures in a JEM-2200FS/CR transmission electron microscope (JEOL Europe, Croissy-sur-Seine, France) operated at 200 kV. An in-column omega energy filter helped to record images with improved signal/noise ratio by zero-loss filtering. The energy selecting slit width was set at 9 eV. Digital images were recorded on an UltraScan4000 CCD camera under low-dose conditions at a magnification of 55,058 obtaining a final pixel size of 2.7 Å/pixel.
Results
Reconstitution of the lipidation reaction
After activation by ATG4B (Fig. 1 B), covalent attachment of ATG8 to PE is mediated by a ubiquitin-like chain of enzymatic steps involving the E1-like ATG7 and the E2-like ATG3 (Fig. 1 C). These reactions can be reconstituted in vitro, using recombinant purified proteins, liposomes and ATP (35, 36). To study the role of these protein-lipid complexes in membrane tethering and fusion processes, we first tried to reconstitute the enzymatically driven lipidation reaction of the human ATG8 proteins, LC3, GABARAP, or GATE-16. Reaction systems including ATG7, ATG3, ATP, and liposomes led to the formation of a faster migrating band readily visualized by Coomasie brilliant blue (CBB) staining (Fig. 1 D, upper panels). The latter was particularly well-resolved for the GABARAP case. However, for LC3 and GATE-16 this band was undistinguishable from the presumably adenylated form of the protein that appears in the presence of ATP and ATG7 (37). To confirm the lipidation reaction, conjugation reactions were prepared with liposomes containing 10% NBD-PE. In each case, reactions led to the formation of a fluorescent faster migrating band representing the lipidated products of LC3/GABARAP subfamilies (Fig. 1 D, lower panels).
Alternatively to the enzyme-driven reaction LC3 homologs can also be chemically lipidated (32, 38) (Fig. 1 E). Here, the C-terminal glycine of the proteins is replaced by a cysteine residue and then linked to PE carrying a reactive maleimide on its headgroup (PEmal). Structurally, this linkage differs from the native coupling by the addition of the maleimide ring; however, these maleimide-anchored conjugates have been reported to be functionally active and are generally considered as useful mimetic analogs to the naturally coupled proteins. Conjugation of the corresponding C-terminal mutants to PEmal-containing liposomes caused the appearance of a faster migrating band in SDS-PAGE, similar to the lipidated forms obtained using the enzymatic-conjugation machinery (Fig. 1 F).Taken together these experiments indicate that we can detect the production of the lipidated products for the three proteins used in this study, both in the context of the whole conjugation enzymatic system and using a chemically modified lipid.
Effect of human LC3 homologs on liposome clustering in vitro
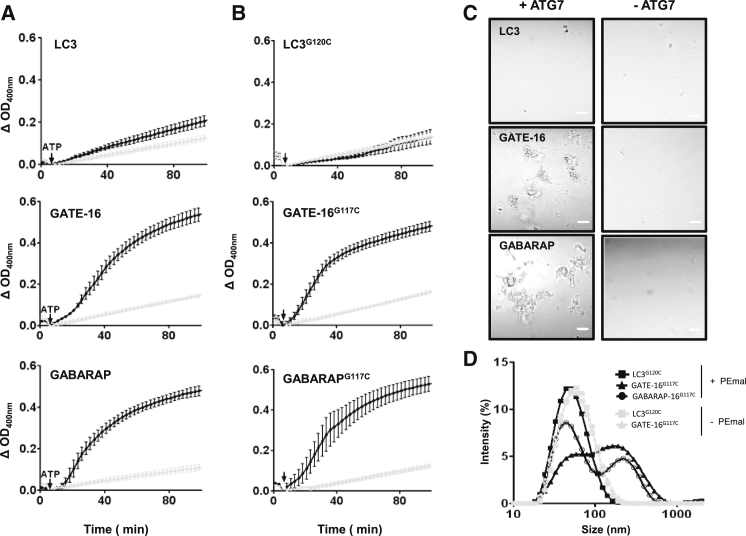
In the course of autophagosome maturation membrane tethering should be essential both in the recruitment of vesicles targeting the expanding phagophore and in the closure of the autophagosome itself. In fact, yeast ATG8-PE was proven to induce liposome aggregation in vitro (20), shedding some light on the possible role of this protein during AP biogenesis. As an initial test for analyzing the functional activity of LC3 homologs, we evaluated their ability to induce liposome aggregation in vitro. As shown in Fig. 2, of the three proteins tested, GATE-16 and GABARAP induced a marked liposome aggregation, measured as an increase in absorbance at 400 nm (Fig. 2 A) and by visual observation under the light microscope (see Fig. 2 C). This effect was not observed in the absence of ATG7. LC3 had but a small effect under the same conditions. We also evaluated the capacity of the PEmal-anchored homologs to induce liposome aggregation. Again only GATE-16-PEmal and GABARAP-PEmal were able to induce efficient liposome aggregation according to either turbidimetric (Fig. 2 B) or DLS measurements (Fig. 2 D). Interestingly, PEmal conjugates induce liposome aggregation at similar rates than their enzymatically conjugated counterparts, supporting the reliable use of this chemical model. These results are consistent with the growing number of evidences indicating a membrane tethering activity for autophagy-related ubiquitin molecules and might represent the first evidence of such an in vitro activity for the GABARAP homolog. LC3-PEmal was shown to efficiently induce membrane tethering in vitro (21). The fact that in our system this protein lacked this activity may result from differences in the experimental system, particularly differences in the lipidic composition of the LUVs that included phosphatidylserine and cholesterol in their case.
Figure 2.
Membrane tethering activities by human autophagic proteins. (A and C) Enzymatic conjugation reactions (maleimide-free) were performed as described in Fig. 1C followed by changes in absorbance at 400 nm, 37°C and observation under a light microscope (bar = 10μm). Liposomes were composed of PC/DOPE/bl-PI (35/55/10 mol ratio). For a negative control, ATG7 was excluded from the reaction (gray trace). (B and D) Chemical conjugation to PEmal was performed as described in Fig. 1D followed by measurements of changes in the absorbance at 400 nm, 37°C. Aggregate size distribution was measured by DLS at 37°C. In (B) and (D) liposomes were composed of PC/DOPE/PEmal/bl-PI (35/40/15/10 mol ratio). For negative controls, liposomes lacking PEmal were used (gray traces).
Effect of human LC3 homologs on membrane fusion
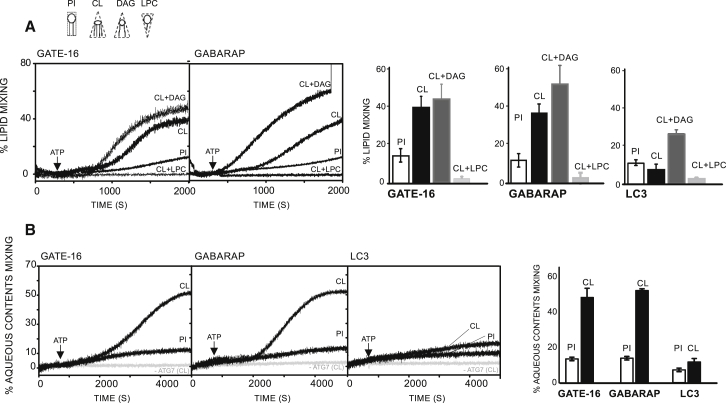
Liposome aggregation could simply represent membrane tethering, but it could also be an indication of hemifusion (i.e., fusion of the outer leaflets of adjacent membranes, whereas inner leaflets remain intact) or even of complete fusion (i.e., the merger of both inner and outer leaflets) of apposed membranes (38). Previous studies with an in vitro reconstituted system similar to ours reported that yeast ATG8 mediates membrane hemifusion, but not complete fusion (20). In a different study, mammalian forms of LC3 and GATE-16, artificially linked to PEmal, were shown to induce full membrane fusion (21) (lipid mixing). To clarify the situation, we initially used a lipid-mixing assay based on fluorescence resonance energy transfer from NBD-PE to Rho-PE (31). When the two dyes are present at an appropriate concentration in the same liposome, the fluorescence of NBD is quenched by rhodamine. Upon fusion of these LUVs with unlabeled LUVs and subsequent probe dilution in the bilayer the distance between the two dyes increases, resulting in a dequenching of NBD fluorescence. As shown in Fig. 3 A, when the two LUV populations were subjected to the conjugation reaction, a significant increase in fluorescence was observed with GATE-16-PE and GABARAP-PE proteins. Of importance, lipid mixing was not observed in the absence of ATG7. In agreement with the inefficient capacity of LC3-PE to tether liposomes, only a small increase in NBD fluorescence was recorded in this case.
Figure 3.
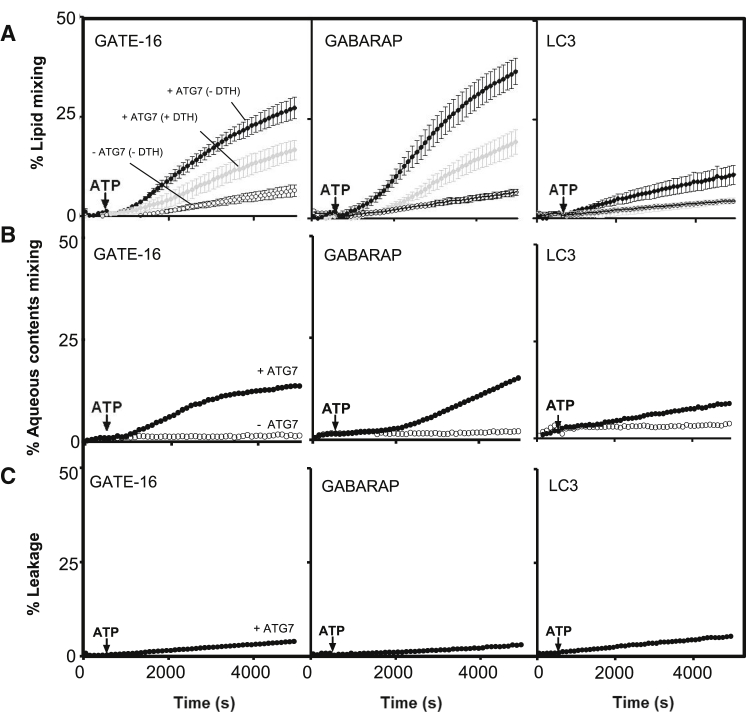
Membrane fusion induced by enzymatically conjugated LC3/GABARAP-PE. (A) Time courses of total (black circles) and inner lipid (black circles) mixing induced by ATG7 (1 μM), ATG3 (1 μM), and the indicated ATG8 human homologs (10 μM) in liposomes (0.4 mM) composed of PC/DOPE/bl-PI (35/55/10 mol ratio). Lipid mixing was monitored by the NBD/Rhodamine lipid dilution assay. For inner monolayer lipid mixing NBD + Rho-liposomes were pretreated with the appropriate amounts of sodium dithionite to quench NBD fluorescence of the outer leaflet. Unlabeled and (NBD + Rho)-labeled liposomes (9:1) were mixed and incubated for 5 min before ATP addition. The extent of lipid mixing was quantified on a percentage basis according to the equation: (Ft-F0/F100-F0) × 100 where Ft is the measured fluorescence of protein-treated LUVs at time t, F0 is the initial fluorescence of the LUV suspension before protein addition, and F100 is the fluorescence value after complete disruption of LUVs by addition of 30 mM β-octylglucoside Each curve represents the average and SE of three independent experiments. For negative controls, ATG7 was excluded from the reactions (open circles). (B) Time course of intervesicular aqueous content mixing. Proteins and lipids as in (A). Content mixing (black circles) was monitored by the ANTS/DPX mixing assay; 100% mixing was determined using LUVs containing coencapsulated ANTS and DPX. ATG7 was excluded from the reactions for negative controls (open circles). (C) Time courses of fusion-related vesicle content leakage. Proteins and lipids as in (A). Vesicle leakage (black circles) was monitored by the ANTS/DPX leakage assay; 100% leakage signal was obtained by adding 1% Triton X-100.
We then investigated whether GATE-16/GABARAP-PE-induced lipid mixing resulted from complete membrane fusion or membrane hemifusion. To this aim, we prepared asymmetrically labeled liposomes by adding the membrane-impermeable reductant sodium dithionite to the liposome suspension to selectively quench the fluorescence of NBD in the outer leaflet (32). Thus, any observed lipid mixing arises from intervesicular mixing of inner monolayer lipids, i.e., from full membrane fusion. Both GATE-16-PE and GABARAP-PE stimulated NBD fluorescence of dithionite-treated liposomes in a time-dependent manner, indicating that fusion of the inner leaflets of apposed membranes, i.e., complete fusion of vesicles, had occurred (Fig. 3 A). As expected, the intensity of the fluorescence signal corresponding to fusion of inner monolayer lipids is about one-half of the one indicating total lipid mixing. Note that fusion of inner monolayers exhibits a longer lag time than total lipid mixing. This is particularly clear for GABARAP. The phenomenon occurs because total lipid mixing includes hemifusion, which is an obligatory step previous to inner monolayer mixing, i.e., complete fusion (39, 40, 41). These results indicate that the observed membrane fusion elicited by human GATE-16 and by GABARAP in vitro, in the context of a minimal human conjugation machinery, consists of full membrane fusion events. Because in the process of vesicle-vesicle fusion a pore connecting the vesicle inner spaces is opened, intervesicular mixing of aqueous contents should occur. This was tested, and confirmed, using the ANTS/DPX fluorescence quenching couple (Fig. 3 B). Note that the extent of contents mixing corresponds roughly with that of inner monolayer lipid mixing (Fig. 3 A) at a given time. Most biological fusion events occur in such a way that vesicle contents are not leaked out. This is also the case in our model fusion system (Fig. 3 C).
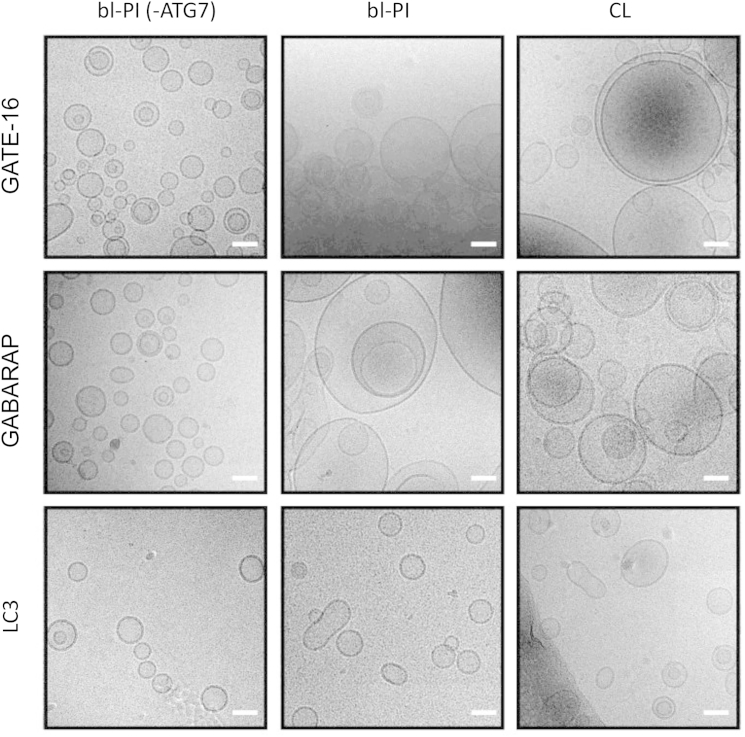
To obtain a morphological evidence of fusion, we analyzed the morphology of liposomes by cryo-EM. As shown in Fig. 4, in the case of conjugation reactions including GATE-16 and GABARAP homologs, formation of much larger vesicles was observed at the end of the fusion reaction, clearly suggesting repetitive fusion events. These results strongly support our conclusion that GATE-16-PE and GABARAP-PE cause fusion of liposomes. In control experiments, no increase in liposome size was observed in the absence of ATG7. Moreover, consistent with the biochemical results suggesting that LC3 is a weaker inducer of membrane fusion in the in vitro system used here, liposome size increased less markedly. However, a significant number of liposomes lost their spherical shape and appeared as elongated structures that may derive from either deformation of single vesicles and/or fusion events between no more than 2–3 vesicles.
Figure 4.
Fusion-dependent increase in vesicle size analyzed by cryo-EM. Typical cryo-EM images of liposomes treated with human autophagic proteins. Enzymatic conjugation reactions including the indicated LC3 homologs (30 μM), ATG3 (3 μM), ATG7 (3 μM), and 3 mM ATP were performed in conjugation buffer for 1 h at 37°C and the products examined by cryo-EM. Liposome (LUVs) compositions: PC/DOPE/bl-PI (35/55/10 mol ratio), column 1 and 2, or PC/DOPE/CL (35/55/10 mol ratio), column 3. In all cases, liposome concentration was 1.2 mM. In control experiments (column 1) ATG7 was excluded from the reaction. The same scale (bar = 50 nm) was used in all images.
The role of specific lipids in LC3- and GATE-16/GABARAP-promoted fusion
According to the widely accepted stalk-pore fusion model (40, 41, 42, 43, 44, 45), fusion is thought to start with the formation of a nonlamellar lipidic stalk, a local connection between the contacting monolayers of two membranes. The stalk then extends connecting the facing monolayers (hemifusion) before pore formation (fusion) occurs. The model predicts that addition of inverted cone-shaped lipids (i.e., positive curvature-inducing lipids) such as lyso-phosphatidylcholine (LPC) to contacting membrane leaflets should prevent formation of hemifused intermediates, whereas cone-shaped lipids such as DAG or CL (i.e., lipids with a negative intrinsic curvature) should promote formation of hemifusion intermediates (45). As shown in Fig. 5 A, substituting bl-PI for CL (10 mol %) strongly promoted GATE/GABARAP-PE-induced lipid mixing. Addition of DAG (10 mol %) further promoted intervesicular lipid mixing and LPC (10%) totally inhibited it. Furthermore, fusion pore formation and content mixing occurred as well with lipidated GATE-16 and GABARAP proteins. Consistent with what was observed in lipid mixing experiments, CL strongly potentiates mixing of aqueous contents (Fig. 5 B). Again, vesicle fusion was accompanied by only a residual leakage of vesicular content (data not shown). For all lipid composition tested vesicle size was centered around 80 nm as assessed by DLS measurements (data not shown).
Figure 5.
Effect of lipids modifying the membrane intrinsic curvature on membrane fusion induced by LC3/GABARAP-PE conjugates. (A) Representative time courses (left panels) and comparison at t = 2000 s (right panels) of intervesicular total lipid mixing induced by ATG7 (1 μM), ATG3 (1 μM), and the indicated ATG8 human homologs (10 μM) in liposomes (0.4 mM) with the basic lipid composition PC/DOPE (35/55 mol ratio) to which either bl-PI (+10 mol ratio), CL (+10 mol ratio), CL+DAG (+10 +10 mol ratio), or CL+LPC (+10+10 mol ratio) was added. (B) Representative time courses (left panels) and comparison at t = 2000 s (right panel) of aqueous contents mixing for the same reactions as in (A). Lipid mixing and content mixing assays were performed as described in Fig. 3. ATG7 was excluded from the reaction in control experiments (gray traces). Liposomes and proteins were incubated for 5 min before ATP addition. Each curve represents the average and SE of three independent experiments.
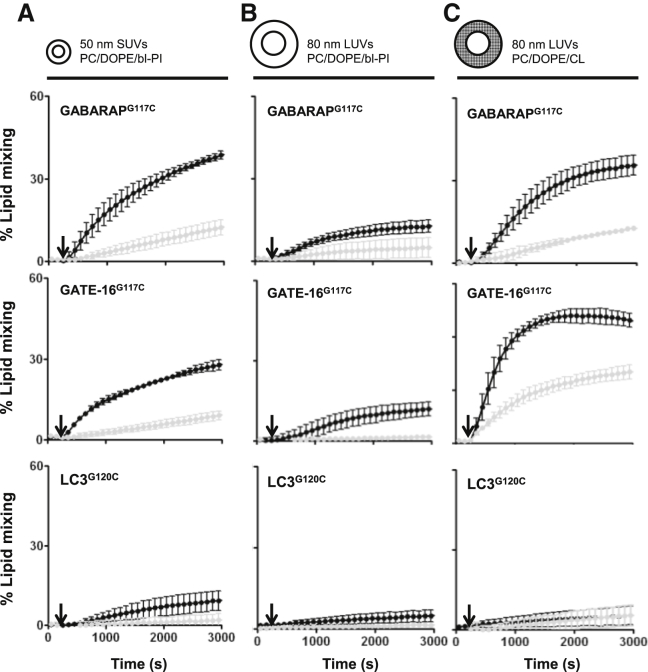
Vesicle radius has been proven to modulate LC3/GABARAP lipidation in vitro (37). In their study, Melia and co-workers showed that ATG3 contains a membrane inserting, curvature-sensitive domain and that the conjugation of the ATG8-family proteins occurs preferentially on highly curved membranes in vivo. In addition to promoting the lipidation process, membrane curvature may also facilitate the fusion process itself (46, 47), inducing autophagosome elongation on the curved edge of the growing isolation membrane. To address the effect of vesicle radius on the process of membrane fusion, we chose the maleimide system, as it allows the evaluation of membrane fusion elicited by LC3 homologs independently of ATG3. To this aim, labeled and nonlabeled PC/DOPE/bl-PI liposomes containing 15% PE-mal were prepared either by sonication (SUVs) or by extrusion through polycarbonate membranes 0.05-μm pore size (LUVs). As shown in Fig. 6, decreasing liposome membrane radius from 40 to 25 nm markedly stimulates total lipid mixing induced by artificially coupled GATE-16-PEmal and GABARAP-PEmal (compare Fig. 6, A and B). Hardly a stimulatory effect was observed in the case of LC3-PEmal, although it was efficiently conjugated to PEmal (Fig. 1 F). Consistent with what was observed previously for enzymatically conjugated GATE-16 and GABARAP, substitution of PI by CL strongly enhances lipid-mixing induced by GATE-16-PEmal and GABARAP-PEmal (compare Fig. 6, B and C). Again, inclusion of CL did not affect particle size (data not shown). These results add to the growing body of evidence supporting an important role of both vesicle radius and membrane intrinsic curvature during autophagosome biogenesis (48, 49, 50).
Figure 6.
Effect of vesicle diameter and lipid composition on membrane fusion elicited by LC3/GABARAP-PEmal conjugates. Time courses of total (black circles); and inner (black circles); lipid mixing induced by the indicated proteins (10 μM) in liposomes (total lipid 0.4 m) of different sizes (sonicated (A), or extruded, (B) and (C)) and different lipid compositions: PC/DOPE/PEmal/bl-PI (35/40/15/10 mol ratio) (A and B) or PC/DOPE/PEmal/CL (35/40/15/10 mol ratio) (C). The arrow marks the moment of protein addition. Each curve represents the average and SE of three independent experiments.
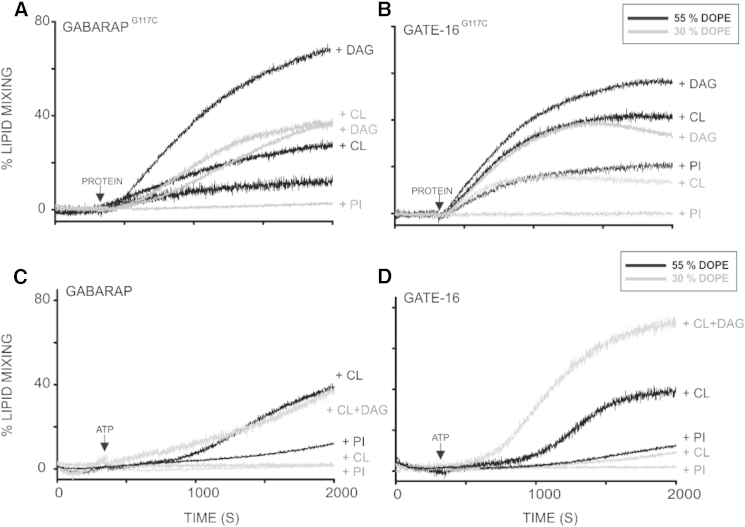
The fusogenic potential of ATG8-PEmal has been shown to be highly dependent on the concentration of PE in the target membrane (22). In that study, a reduction of PE concentration from 55% to 30% made ATG8-PEmal unable to fuse liposomes, although coupling of ATG8 to maleimide was equally efficient in both cases. We evaluated whether reducing the PE content in our liposomes had an impact on GABA/GATE-PE-dependent vesicle fusion. In agreement with the previous report, GATE-PEmal and GABA-PEmal were hardly able to fuse liposomes containing 30% PE (Fig. 7, A and B). However, substitution of bl-PI by cone-shaped lipid molecules such as CL or DAG promoted vesicle fusion, nearly to the extent observed with 55% DOPE. With enzymatically conjugated GATE and GABARAP proteins (Fig.7, C and D) vesicle fusion is significantly impaired by reduction of DOPE to 30%, probably due to a drop in the efficiency of the lipidation reaction. However, incorporation of CL and DAG into the vesicles sustained potent vesicle fusion, even in the context of a limiting DOPE concentration.
Figure 7.
CL- and DAG-sustained membrane fusion under PE limiting concentrations. Time courses of total lipid mixing induced by the indicated proteins (10 μM) in liposomes (0.4 mM) of different lipid compositions. (A and B) (chemical conjugation): with the basic lipid composition PC/DOPE/PEmal (45/30/15 mol ratio, gray lines) to which either bl-PI (+10 mol ratio), CL (+10 mol ratio), or DAG (+10 mol ratio) were used; or basic lipid composition of PC/DOPE/PEmal (35/40/15 mol ratio, black lines) to which either bl-PI (+10 mol ratio), CL (+10 mol ratio), or DAG (+10 mol ratio) were used. (C and D) (enzymatic conjugation): with the basic lipid composition of PC/DOPE (60/30, gray lines) to which either bl-PI (+10 mol ratio), CL (+10 mol ratio,), and CL+DAG (+10+10 mol ratio) were used or the basic lipid composition of PC/DOPE (35/55, black lines) to which either bl-PI (+10 mol ratio,) or CL (+10 mol ratio) were added. Liposomes and proteins were incubated for 5 min before ATP addition. Traces are representative of at least three independent experiments.
Discussion
The ATG8 ubiquitin-like conjugation system plays a central role in autophagy. However, despite its likely importance during autophagosome formation, its mode of action is only incompletely understood. This is particularly true in the case of higher eukaryotes and information regarding mammalian LC3/GABARAP subfamilies is just recently starting to emerge (12, 51, 52, 53). In this study, we used two reconstituted liposomal systems to learn more about the molecular mechanisms by which human LC3 and GABARAP subfamilies trigger AP biogenesis. Using these simplified and biochemically accessible model systems, we systematically explored the membrane activities of human ATG8 homologs in the context of different lipid compositions. Our data provide to our knowledge, new insight into several aspects of the human autophagy machinery, namely on the nature of the fusion process leading to AP elongation, and the role of lipid intrinsic curvature in the process. From the methodological point of view, our results fully validate the use of chemically lipidated ATG8 homologs.
The fusion mechanism
According to the current view, phagophore expansion requires the supply and subsequent fusion of membranal material of yet uncharacterized origin. In this regard, several studies have involved both yeast ATG8 (20) and human LC3 and GATE-16 (21) in the fusion processes underlying phagophore expansion. However, the molecular mechanism is still obscure. The liposomal assays applied here allowed us to explicitly test the capacity of the different homologs for inducing membrane fusion in vitro, in the absence of other cellular proteins.
The ensemble of the data are compatible with a fusion mechanism based on the stalk hypothesis (32, 38, 39, 40, 41, 42, 43, 44, 45), according to which the formation of a lipidic intermediate, the stalk, with a nonlamellar structure, is the limiting step in the fusion event. The canonical form of stalk-mediated fusion requires the independent demonstration of vesicle aggregation, intervesicular mixing of total membrane lipids, mixing of inner monolayer lipids, and mixing of aqueous contents in the absence of vesicular content leakage or spill out. Ideally, aggregation, total lipid mixing, inner lipid mixing, and contents mixing should start in the said order, i.e., should exhibit increasing lag times. All these predictions have been demonstrated in our case, particularly for GATE-16 and GABARAP (Figs. 2 and 3). Moreover, the proposed stalk architecture demands that lipids with an intrinsic negative curvature (e.g., DAG, CL) must facilitate stalk formation, thus membrane fusion (43, 44). This is also amply demonstrated by our data, as well as the opposite, inhibitory effect of positive intrinsic-curvature lipids (e.g., lyso PC) (Fig. 5). The same considerations on intrinsic lipid curvature apply to the requirement of high proportions of PE in the vesicle composition (Fig. 7). We can thus conclude that, at least in the in vitro situation, ATG8 proteins induce AP growth via stalk-mediated fusion events.
In the cell environment, the AP is seen to grow by elongation, i.e., along a main axis. In our case the most effective ATG8 analogs, namely GATE-16 and GABARAP, give rise to a spectacular growth of vesicles, leading to roughly spherical structures (Fig. 4), whereas LC3, a much less efficient fusogen, gave rise to elongated, peanut-shaped structures, reminiscent of the growing AP in cell images. The reason why LC3 is less effective than the other two, and why it originates elongated vesicles deserves further study. At least part of the answer will rely in the yet unknown mechanisms of AP growth regulation (22, 54). Currently, we cannot rule out the possibility that, being less effective, LC3 is more physiologically relevant in AP elongation.
Proteins and lipids
Both SNAREs and viral fusion proteins contain a transmembrane domain, which is critical for their fusion activity (32, 55, 56). ATG8 homologs do not present such a transmembrane domain and yet are capable of inducing full membrane fusion when anchored only to one membrane leaflet via PE. Several models have been proposed including that self-oligomerization of ATG8-PE molecules would drive membrane tethering and hemifusion through a yet uncharacterized mechanism (20). Alternatively, it has been shown that peptides representing the N-terminus of LC3 and GATE are sufficient to drive full membrane fusion when artificially linked to PEmal (21). An additional, nonexclusive possibility is that the asymmetric insertion of this N-terminal region would promote positive curvature and tension in the outer leaflet of the membrane thus promoting hemifusion and eventually fusion (57). Amphipathic proteins devoid of transmembrane domains can induce vesicle fusion, e.g., muscle glyceraldehyde 3-phosphate dehydrogenase (58). Determining membrane structures of LC3/GABARAP proteins, particularly at the fusogenic state will likely broaden our understanding of ATG8-driven membrane fusion.
According to a growing body of evidence, AP biogenesis is a lipid-modulated process (25). However, with the exception of the relatively well-characterized role of PI3P, little is known about the specific role that each lipid species plays during the different steps of AP biogenesis. Here, we obtained direct evidence that intrinsic-curvature modifying lipids (DAG, LPC, and CL) act in concert with autophagic proteins to modulate the fusion of vesicles in vitro. This observation is compatible with a model in which the local generation or transfer of these lipids from the ER or mitochondria to the isolation membrane may regulate phagophore expansion in vivo. Interestingly the PLD1 pathway, which produces phosphatidic acid, a cone-shaped lipid, has been reported to promote autophagy (59). Furthermore, another study showed that DAG generation is required for the initiation of Salmonella-induced autophagy (60). In both cases, however, the underlying mechanism is not known. What still needs to be determined is the lipid composition of the phagophore as well as the lipid changes that accompany AP biogenesis in vivo. In this regard, reliable autophagosome purification procedures in conjunction with lipidomic approaches should help understand the specific roles of lipids in autophagy.
In agreement with a recent report (37), we observe that both human LC3 and GABARAP subfamilies can be enzymatically lipidated in vitro (Fig. 1 A), even in the absence of the ATG12-ATG5-ATG16L1 complex. However, both systems are essential for proper AP biogenesis in vivo, and several in vitro studies (61, 62) indicated that the ATG12-ATG5-ATG16L1 complex functions, at least in part, facilitating the transfer of ATG8/LC3 from ATG3 to PE. Whether human ATG12-ATG5-ATG16L1 complex promotes lipidation of the GABARAP subfamily in a similar manner to LC3 needs to be tested and will be addressed in future studies.
As an alternative to the enzymatically driven reaction the chemical conjugation approach enables direct conjugation of proteins to PE lipids (Fig. 1 B), thus bypassing the requirement for the conjugation machinery. Although the enzymatic conjugation is more physiologically relevant, chemically conjugated versions of LC3/GABARAP subfamilies are easily prepared and are increasingly being used in both mechanistic (35, 36) and structural (63) studies. Here, we report that PEmal conjugates faithfully recapitulate key features of their naturally conjugated homologs, supporting the reliable use of these artificial complexes. In fact, PEmal conjugates show faster aggregation and fusion kinetics than the enzymatically conjugated proteins, consistent with the fact that they are not conjugated trough a relatively complex ubiquitin-like reaction comprising multiple steps (compare lag times between enzymatic and chemical conjugation reactions).
Author Contributions
A.L. and A.A. conceived and coordinated the study. A.L. wrote the article. F.M.G. and L.R.M. helped in coordinating the study, designed experiments, and edited the article. A.L., J.H.H., and Z.A. designed, performed, and analyzed most of the experiments. D.J. and M.V. designed, performed, and analyzed the experiments shown in Fig. 4. J.F.R. helped with the design of protein expression and purification. All authors analyzed the results and approved the final version of the article.
Acknowledgments
This article is based upon work from COST Action (PROTEOSTASIS BM1307), supported by COST (European Cooperation in Science and Technology). The authors thank Dr. Isei Tanida (National Institute of Infectious Diseases, Tokyo, Japan) for providing human ATG7, ATG3, LC3, GATE-16, and GABARAP plasmids.
This work was supported in part by grants from the Spanish Ministry of Economy and FEDER (BFU 2011-28566, BFU 2012-36241, AGL2011-24758) and the Basque Government (IT838-13, IT849-13). J.H.H. and Z.A. were predoctoral students supported by the University of the Basque Country.
Editor: Joseph Falke.
Footnotes
Ane Landajuela and Javier H. Hervás contributed equally to this work.
References
- 1.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Boya P., Reggiori F., Codogno P. Emerging regulation and functions of autophagy. Nat. Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi A.M.K., Ryter S.W., Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 4.Shibutani S.T., Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24:58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamasaki M., Shibutani S.T., Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr. Opin. Cell Biol. 2013;25:455–460. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Rubinsztein D.C., Shpilka T., Elazar Z. Mechanisms of autophagosome biogenesis. Curr. Biol. 2012;22:R29–R34. doi: 10.1016/j.cub.2011.11.034. [DOI] [PubMed] [Google Scholar]
- 7.Homma K., Suzuki K., Sugawara H. The Autophagy Database: an all-inclusive information resource on autophagy that provides nourishment for research. Nucleic Acids Res. 2011;39:D986–D990. doi: 10.1093/nar/gkq995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Z., Klionsky D.J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki K., Kirisako T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichimura Y., Kirisako T., Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z., Nair U., Klionsky D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z., Klionsky D.J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shpilka T., Weidberg H., Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouno T., Mizuguchi M., Kawano K. Solution structure of microtubule-associated protein light chain 3 and identification of its functional subdomains. J. Biol. Chem. 2005;280:24610–24617. doi: 10.1074/jbc.M413565200. [DOI] [PubMed] [Google Scholar]
- 15.Coyle J.E., Qamar S., Nikolov D.B. Structure of GABARAP in two conformations: implications for GABA(A) receptor localization and tubulin binding. Neuron. 2002;33:63–74. doi: 10.1016/s0896-6273(01)00558-x. [DOI] [PubMed] [Google Scholar]
- 16.Kabeya Y., Mizushima N., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanida I., Tanida-Miyake E., Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J. Biol. Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 18.Sou Y.S., Waguri S., Komatsu M. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weidberg H., Shvets E., Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Weidberg H., Shpilka T., Elazar Z. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Nair U., Jotwani A., Klionsky D.J. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda T., Matsunaga K., Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin. Cell Dev. Biol. 2010;21:671–676. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Dooley H.C., Razi M., Tooze S.A. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dall’Armi C., Devereaux K.A., Di Paolo G. The role of lipids in the control of autophagy. Curr. Biol. 2013;23:R33–R45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vicinanza M., Korolchuk V.I., Rubinsztein D.C. PI(5)P regulates autophagosome biogenesis. Mol. Cell. 2015;57:219–234. doi: 10.1016/j.molcel.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu C.T., Ji J., Kagan V.E. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat. Cell Biol. 2013;15:1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zens B., Sawa-Makarska J., Martens S. In vitro systems for Atg8 lipidation. Methods. 2015;75:37–43. doi: 10.1016/j.ymeth.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer L.D., Hope M.J., Cullis P.R. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta. 1986;858:161–168. doi: 10.1016/0005-2736(86)90302-0. [DOI] [PubMed] [Google Scholar]
- 30.Fiske C.H., Subbarow Y. The colorimetric determination of phosphorus. J. Biol. Chem. 1925;66:375–400. [Google Scholar]
- 31.Struck D.K., Hoekstra D., Pagano R.E. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–4099. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y., Zhang F., Shin Y.-K. Hemifusion in SNARE-mediated membrane fusion. Nat. Struct. Mol. Biol. 2005;12:417–422. doi: 10.1038/nsmb921. [DOI] [PubMed] [Google Scholar]
- 33.Ellens H., Bentz J., Szoka F.C.H. H+- and Ca2+-induced fusion and destabilization of liposomes. Biochemistry. 1985;24:3099–3106. doi: 10.1021/bi00334a005. [DOI] [PubMed] [Google Scholar]
- 34.Goñi F.M., Villar A.V., Alonso A. Interaction of phospholipases C and sphingomyelinase with liposomes. Methods Enzymol. 2003;372:3–19. doi: 10.1016/S0076-6879(03)72001-1. [DOI] [PubMed] [Google Scholar]
- 35.Ichimura Y., Imamura Y., Ohsumi Y. In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Bio. Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 36.Nakatogawa H., Ohsumi Y. SDS-PAGE techniques to study ubiquitin-like conjugation systems in yeast autophagy. Methods Mol. Biol. 2012;832:519–529. doi: 10.1007/978-1-61779-474-2_37. [DOI] [PubMed] [Google Scholar]
- 37.Nath S., Dancourt J., Melia T.J. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chernomordik L.V., Kozlov M.M. Mechanics of membrane fusion. Nat. Struct. Mol. Biol. 2008;15:675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Basáñez G., Nieva J.L., Alonso A. Origin of the lag period in the phospholipase C cleavage of phospholipids in membranes. Concomitant vesicle aggregation and enzyme activation. Biochemistry. 1996;35:15183–15187. doi: 10.1021/bi9616561. [DOI] [PubMed] [Google Scholar]
- 40.Villar A.V., Goñi F.M., Alonso A. Diacylglycerol effects on phosphatidylinositol-specific phospholipase C activity and vesicle fusion. FEBS Lett. 2001;494:117–120. doi: 10.1016/s0014-5793(01)02333-x. [DOI] [PubMed] [Google Scholar]
- 41.Ibarguren M., Bomans P.H., Goñi F.M. End-products diacylglycerol and ceramide modulate membrane fusion induced by a phospholipase C/sphingomyelinase from Pseudomonas aeruginosa. Biochim. Biophys. Acta. 2010;1798:59–64. doi: 10.1016/j.bbamem.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goñi F.M., Alonso A. Membrane fusion induced by phospholipase C and sphingomyelinases. Biosci. Rep. 2000;20:443–463. doi: 10.1023/a:1010450702670. [DOI] [PubMed] [Google Scholar]
- 43.Basáñez G., Goñi F.M., Alonso A. Effect of single chain lipids on phospholipase C-promoted vesicle fusion. A test for the stalk hypothesis of membrane fusion. Biochemistry. 1998;37:3901–3908. doi: 10.1021/bi9728497. [DOI] [PubMed] [Google Scholar]
- 44.Basanez G., Nieva J.L., Goni F.M. Diacylglycerol and the promotion of lamellar-hexagonal and lamellar-isotropic phase transitions in lipids: implications for membrane fusion. Biophys. J. 1996;70:2299–2306. doi: 10.1016/S0006-3495(96)79795-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chernomordik L., Chanturiya A., Zimmerberg J. The hemifusion intermediate and its conversion to complete fusion: regulation by membrane composition. Biophys. J. 1995;69:922–929. doi: 10.1016/S0006-3495(95)79966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahyayauch H., Villar A.V., Goñi F.M. Modulation of PI-specific phospholipase C by membrane curvature and molecular order. Biochemistry. 2005;44:11592–11600. doi: 10.1021/bi050715k. [DOI] [PubMed] [Google Scholar]
- 47.de Arcuri B.F., Vechetti G.F., Morero R.D. Protein-induced fusion of phospholipid vesicles of heterogeneous sizes. Biochem. Biophys. Res. Commun. 1999;262:586–590. doi: 10.1006/bbrc.1999.1243. [DOI] [PubMed] [Google Scholar]
- 48.Knorr R.L., Nakatogawa H., Dimova R. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids. PLoS One. 2014;9:e115357. doi: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan W., Nassiri A., Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc. Natl. Acad. Sci. USA. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Antonny B. Mechanisms of membrane curvature sensing. Annu. Rev. Biochem. 2011;80:101–123. doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 51.Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanida I., Nishitani T., Kominami E. Mammalian Apg12p, but not the Apg12p.Apg5p conjugate, facilitates LC3 processing. Biochem. Biophys. Res. Commun. 2002;296:1164–1170. doi: 10.1016/s0006-291x(02)02057-0. [DOI] [PubMed] [Google Scholar]
- 53.Mizushima N., Yoshimori T., Ohsumi Y. Role of the Apg12 conjugation system in mammalian autophagy. Int. J. Biochem. Cell Biol. 2003;35:553–561. doi: 10.1016/s1357-2725(02)00343-6. [DOI] [PubMed] [Google Scholar]
- 54.Moreau K., Ravikumar B., Rubinsztein D.C. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H., Zick M., Jun Y. A lipid-anchored SNARE supports membrane fusion. Proc. Natl. Acad. Sci. USA. 2011;108:17325–17330. doi: 10.1073/pnas.1113888108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langosch D., Hofmann M., Ungermann C. The role of transmembrane domains in membrane fusion. Cell. Mol. Life Sci. 2007;64:850–864. doi: 10.1007/s00018-007-6439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zimmerberg J., Kozlov M.M. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 58.Morero R.D., Viñals A.L., Farías R.N. Fusion of phospholipid vesicles induced by muscle glyceraldehyde-3-phosphate dehydrogenase in the absence of calcium. Biochemistry. 1985;24:1904–1909. doi: 10.1021/bi00329a015. [DOI] [PubMed] [Google Scholar]
- 59.Dall’Armi C., Hurtado-Lorenzo A., Di Paolo G. The phospholipase D1 pathway modulates macroautophagy. Nat. Commun. 2010;1:142. doi: 10.1038/ncomms1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shahnazari S., Yen W.L., Brumell J.H. A diacylglycerol-dependent signaling pathway contributes to regulation of antibacterial autophagy. Cell Host Microbe. 2010;8:137–146. doi: 10.1016/j.chom.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanada T., Noda N.N., Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 62.Sakoh-Nakatogawa M., Matoba K., Ohsumi Y. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 63.Ma P., Mohrlüder J., Willbold D. Preparation of a functional GABARAP-lipid conjugate in nanodiscs and its investigation by solution NMR spectroscopy. ChemBioChem. 2010;11:1967–1970. doi: 10.1002/cbic.201000354. [DOI] [PubMed] [Google Scholar]