Abstract
Various host-guest peptide series are used by experimentalists as reference conformational states. One such use is as a baseline for random-coil NMR chemical shifts. Comparison to this random-coil baseline, through secondary chemical shifts, is used to infer protein secondary structure. The use of these random-coil data sets rests on the perception that the reference chemical shifts arise from states where there is little or no conformational bias. However, there is growing evidence that the conformational composition of natively and nonnatively unfolded proteins fail to approach anything that can be construed as random coil. Here, we use molecular dynamics simulations of an alanine-based host-guest peptide series (AAXAA) as a model of unfolded and denatured states to examine the intrinsic propensities of the amino acids. We produced ensembles that are in good agreement with the experimental NMR chemical shifts and confirm that the sampling of the 20 natural amino acids in this peptide series is be far from random. Preferences toward certain regions of conformational space were both present and dependent upon the environment when compared under conditions typically used to denature proteins, i.e., thermal and chemical denaturation. Moreover, the simulations allowed us to examine the conformational makeup of the underlying ensembles giving rise to the ensemble-averaged chemical shifts. We present these data as an intrinsic backbone propensity library that forms part of our Structural Library of Intrinsic Residue Propensities to inform model building, to aid in interpretation of experiment, and for structure prediction of natively and nonnatively unfolded states.
Introduction
Unfolded and unstructured protein states have long been of interest, because the residual structure that resides within such states potentially guides the early stages of protein folding (1). The exact nature of this residual structure remains elusive, and its characterization has been the goal of many experiments (1). An emerging facet of structural biology is the prevalence of intrinsically disordered proteins (IDPs) (2). This native disorder appears to confer a structural plasticity that expands the functionality of the protein universe beyond one in which each protein has a single binding partner, function, and structure (3, 4). One of the best examples of this expansion of the protein structural universe and functionality is p53, which has disordered regions capable of binding hundreds of interaction partners, allowing it to function as a signaling hub (5). Unsurprisingly, understanding the nature of these disordered states has been as challenging as characterizing the conformational behavior and residual structure of denatured states.
A commonly accepted misconception is that denatured or unfolded protein states are random coils (6, 7). For our purposes, a random coil is defined as a peptide or protein where the orientation and sampling of each residue is random, with an absence of any preference to a given conformational region. Data that support the random-coil nature of unfolded states, such as intrinsic viscosity and radius of gyration (7, 8, 9, 10, 11, 12), are largely indiscriminate. Although the denatured or unfolded states of some proteins may conform to global descriptions of a random coil (9, 13), this does not conclusively preclude the presence of local structure or amino acid conformational preferences (6, 7, 9, 13, 14). The exact nature of these states at the microscopic level is often debated, usually within the context of the polyproline hypothesis that the left-handed polyproline helix (PIIL) is the dominant conformer in denatured and unfolded states (8, 9, 13, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25). Although some experimental evidence may support the conjecture that the PIIL conformation is prevalent in polyalanine and other peptides (18, 24, 26, 27, 28, 29, 30) and that PIIL content increases in urea (13, 14, 15, 19), other evidence refutes these ideas (6, 10, 11). In other contexts, such as the Δ131Δ fragment of staphylococcal nuclease, there is evidence that comparable residual and topological structural arrangements can exist in both 8 M urea and water (31). Furthermore, some proteins retain native-like topologies in their denatured states (32, 33, 34, 35). Although a bias toward defined local secondary structure conformations has been previously observed for other short peptides and host-guest peptide series (16, 30, 36), the details regarding these propensities and how they vary with environment is limited.
Knowing the nature of nascent structure in the early stages of protein folding could allow more accurate mechanisms to be developed (37), and since the acceptance of IDPs (38), there has been a reshaping of the protein structure universe to include disorder and structurally adaptable regions (39, 40). Further, given that many experimental measurements of these disordered or unfolded states are often analyzed relative to a control random-coil system, believed to be devoid of structure, knowledge of the conformational nature of such states, and of the control systems themselves, is of paramount importance. Hence, characterization of the intrinsic conformational propensities of the amino acids and their responses to different conditions is of great interest to better understand unfolded, denatured, and disordered states.
Molecular dynamics (MD) simulations are well suited to investigate the breadth of conformations present in these systems at the atomic level. Previously, we simulated the GGXGG host-guest peptides to survey the intrinsic conformational preferences for each of the 20 naturally occurring amino acids (36). However, the achirality of glycine and the unrealistic degrees of freedom due to the lack of side chains makes the GGXGG host a nonideal choice to model the unfolded states of proteins. Here, we reassess the intrinsic conformational propensities of amino acids within the alanine-based AAXAA pentapeptide host. Ala was selected as a more suitable host for two reasons. First, Ala is one of the most abundant amino acids in proteins (41), and second, the presence of the methyl side chain enforces backbone chirality while remaining small and relatively unobtrusive. Ala is notorious for its frequent occurrence in helical structures within globular proteins, but this proclivity for helical structure is dramatically reduced in short peptides. For example, helical content in the KKAnKKGY peptide series is only detectable by circular dichroism when n ≥ 9 (42). Hence, the host residues in our AAXAA peptides should not exert a great influence on the guest residues and should allow for a more realistic model of the conformational propensities in denatured or disordered states. A final advantage in selecting the Ala-based host system is that Prestegard et al. recently determined chemical shifts for this AAXAA series in 8 M urea to present an improved random-coil data set (43). Their data were used to validate the MD simulations, providing more confidence in the results to allow us to investigate and gather information at a level not accessible to experiment on the underlying conformational ensembles giving rise to the spectroscopic observables. The intrinsic propensities and dihedral angles for all the amino acids presented here are available as part of our Structural Library of Intrinsic Residue Propensities, available through our Dynameomics project website (http://dynameomics.org).
Materials and Methods
MD simulations of AAXAA pentapeptides
Pentapeptides for all 20 amino acids were built with extended conformations, with ϕ and ψ angles set to 180° and −180°, and simulated under three conditions: in pure water at 298 K; in 8 M urea at 298 K; and in pure water at 498 K. The microcanonical NVE ensemble (constant number of particles, volume, and energy) was employed for a variety of reasons, one being that it provides natural Boltzmann sampling without the need to artificially, and potentially arbitrarily, weight the sampled conformations. Where necessary, additional pentapeptides were generated with both neutral and acidic protonation states (Asp, Ash, Glu, and Glh). Three individual simulations of His were performed for each of the protonation states, Hid (δH), Hie (εH), and Hip (δH and εH). Cysteine was modeled in the reduced state with a protonated side chain (-CH2-SH, named Cyh in our force field). All of the peptides were explicitly modeled with N-acetylated and C-amidated blocked termini to minimize charged-end effects. Simulations were performed according to previously reported protocols (44) using the in lucem molecular mechanics (ilmm) package (45) with the Levitt et al. force field (46). Nonbonded interactions were treated with an 8-Å force-shifted cutoff (47), and explicit-solvent molecules were used for both the 8 M urea (48) and pure water simulations (49). Solvation in 8 M urea was achieved through random substitution of water molecules in a presolvated peptide system with urea molecules to give a final mole fraction of 0.186. Across all 8 M urea systems, the solvent box contained between 453 and 532 water molecules and between 107 and 126 urea molecules. Both pure water and 8 M urea solvent systems at 298 K were simulated with a box size that reproduced the experimental densities, 0.9970 g/mL and 0.7813 g/mL, respectively. For the simulations at 498 K, the density was set to the reduced density for that temperature, 0.829 g/mL (50). To assess convergence, multiple simulations were performed of the AAAAA, AAGAA, and AAWAA peptides. All simulations were performed for a minimum of time consistent with requirements to reach convergence: 1 μs in 8 M urea (1.5 μs for Ala, Tyr, and Gly); 600 ns in water at 298 K; and 100 ns in water at 498 K (200 ns for Ala), amounting to 53 μs of sampling.
Calculation of conformational propensities
Populations were calculated for the four quadrants of the conformational ψ/ϕ space and for specific conformational regions, defined as αR, −100° ≤ ϕ ≤ −30°, −80° ≤ ψ ≤ −5°; near-αR (nαR), −175° ≤ ϕ ≤ −100°, −55° ≤ ψ ≤ −5°; αL, 5° ≤ ϕ ≤ 75°, 25° ≤ ψ ≤ 120°; β, −180° ≤ ϕ ≤ −50°, 80° ≤ ψ ≤ −170°; and PIIL: −110° ≤ ϕ ≤ −50°, 120° ≤ ψ ≤ 180°; and PIR, −180° ≤ ϕ ≤ −115°, 50° ≤ ψ ≤ 100°. A second β region definition, termed nonpolyproline β (nPβ), excludes the areas that overlap with PIIL and PIR. Please note that both β region definitions include the portion of the β-basin that protrudes into the lower part of the quadrant containing the αR and nαR regions. Populations were calculated as percentages to account for the different trajectory lengths. Two-dimensional histograms with 5° × 5° ϕ/ψ bins were used to generate the Ramachandran distributions. Correlations between the ϕ/ψ frequency distributions were determined to generate similarity correlation matrices. The modes of the Ramachandran distributions, and of those within defined conformational regions, coincide with free-energy minima, and the modal angles were identified from the histograms to provide the most probable ϕ/ψ dihedral angles for each residue.
Comparison with NMR
The experimental chemical shifts for the 15NH and 1HN resonances for AAXAA were acquired in 8 M urea at 298 K at pH 2.5 (43). As experimental chemical shifts of the other nuclei for these peptides are not available, comparisons were made with data for the GGXGG host-guest peptides in 8 M urea and AAAAA in water at 300 K (51, 52). The HN, Hα, Hβ, NH, Cα, Cβ, and C′ chemical shifts for the simulations were calculated using SHIFTX2 (53) with a 1% selection of Protein Data Bank (PDB) structures (5000 structures) randomly selected from the 500 ns production dynamics portion of each 8 M urea trajectory. These 1% subsets were determined to be highly representative of the full ensemble of conformations generated (Fig. S1 in the Supporting Material), and the computed chemical shifts were assessed for convergence (R > 0.93) against additional subsets taken from earlier portions of the trajectories (Table S1). As no reasonable homology models exist for these model peptides, the SHIFTX+ component of SHIFTX2 was used in isolation. Scalar couplings, 3JHNHα, were computed using the Karplus equation (Eq. 1) with the Schmidt et al. parameters, where A = 7.90, B = 1.05, and C = 0.65 (54).
| (1) |
Results and Discussion
We performed MD simulations of the AAXAA peptides with each of the 20 amino acids in the guest position to model control native conditions (water at 298 K), thermal denaturation (water at 498 K), and chemical denaturation (8 M urea at 298 K). Different amino acid protonation states were simulated, giving a total of 24 AAXAA peptides that included the protonated forms of Asp and Glu (Ash and Glh), and Hid, Hie, and Hip for the neutral δ and ε protonated and positively charged doubly protonated forms of His. Details are provided below, but the simulations typically converged in 50 ns (498 K) to 500 ns (8 M urea), and the ensembles obtained after convergence were used to assess the sampling of the central guest residues and the modulation of this sampling under different conditions. A total of 53 μs of simulation time was required to obtain meaningful statistics for the conformational propensities, which were validated against experiment.
Sampling and experimental validation of AAXAA simulations
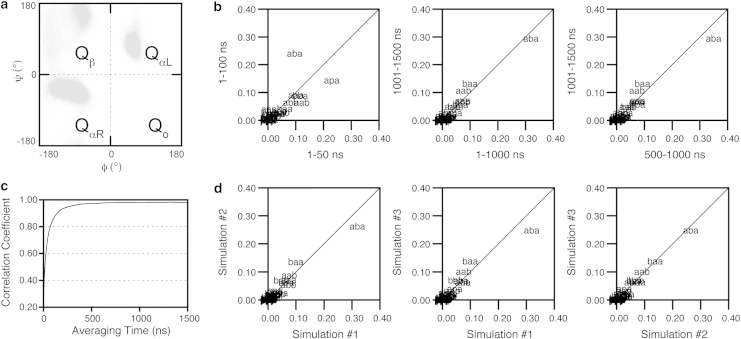
The sampling of the central guest residue, X, and the two neighboring host Ala residues within each peptide was used to assess trajectory convergence. Each pair of backbone ϕ/ψ dihedrals can inhabit one of the four quadrants of the Ramachandran plot: Qβ, QαR, QαL, Qo (Fig. 1 a). For the three central residues, AXA, this provides 64 potential substates, i.e., 34 conformational possibilities for each of the ϕ/ψ pairs. Assessing sampling in terms of the fractional population of these 64 substates increases the sensitivity with which variations and convergence can be detected over that from examination of the central residue alone. The terminal residues and end-capping groups were neglected due to their ability to more extensively sample conformational space and potentially mask any nonconvergence of the guest residues.
Figure 1.
Convergence of AAAAA in 8 M urea at 298 K assessed from the conformational sampling of the three central residues. (a) Quadrants of the Ramachandran plot used for defining the 64 states. Abbreviations used in (b) and (d): a (QαR) −ψ,−ϕ; b (Qβ) +ψ,−ϕ; p (QαL) +ψ,+ϕ; o (Qo) −ψ,+ϕ, where QαR, is the right-handed α-helical, QαL the left-handed α-helical, Qβ, the β, and Qo other. Occupation of a single quadrant by each of the three residues results in a maximum of 34, i.e., 64, possible states that can be populated. (b) Comparison of the fractional population of the 64 states sampled over different sections of the trajectory for run no. 1 of AAAAA. (c) Correlation coefficient for run no. 1 of AAAAA. (d) Comparison of the fractional populations sampled by all three AAAAA simulations.
Convergence was observed by comparing different time periods within the same simulation (Fig. 1 b) and populations compared over different averaging times were correlated (Fig. 1 c). Convergence was also obtained between three independent AAAAA simulations in 8 M urea (Fig. 1 d). Further details of the convergence of AAAAA in pure water are given in Fig. S2. In addition, the results for the AAGAA and AAWAA simulations are also provided, as AAGAA exhibited the greatest extent of conformational sampling, whereas AAWAA was one of the slowest members in the series to converge (Fig. S3). Compared to the sampling in 8 M urea, convergence was achieved significantly faster in pure water both at 298 K and at 498 K. Further analysis of the conformational propensities described below was made using the latter parts of the trajectories after the point of convergence: the last 300 ns in water at 298 K; the last 500 ns in 8 M urea at 298 K; and the last 50 ns in water at 498 K. The correlations over different portions of the production dynamics used in the analysis of all simulations were on average ≥0.93 (Table S2).
As our simulation protocols provide natural Boltzmann sampling and they had converged, the populations after convergence reflect free energy of the states relative to each other within a simulation and between simulations. Further, the water dynamics agreed with experimental values. The self-diffusion coefficients of bulk water by MD at 298 K was 0.25 vs. 0.23–0.25 Å2 ps−1 from experiment, and 2.00 Å2 ps−1 by MD at 498 K compared with the experimental value of 1.91 Å2 ps−1 (49, 55, 56, 57).
Amino acids exhibit conformational preferences
On average, the amino acid residues sampled ∼50% of ϕ/ψ space, with the exception of Gly and Pro, which covered ∼80% and ∼20%, respectively, of ϕ/ψ space (Table 1). Between 20 and 30% of the Ramachandran plot becomes closed off once heavy-atom side chains are present on the guest residue compared to the Gly in the AAGAA peptide. And if the residue is β-branched, or if Ash or Asp pair, the coverage of the ϕ/ψ area is reduced by a further 10% (Table 1). For our peptides to be considered random coils they must sample all sterically accessible conformational regions within these areas with equal probability (58, 59, 60). However, random, uniform sampling was not exhibited by any of the guest residues, even when chemical or thermal denaturation was simulated (Fig. 2). Gly approached a uniform sampling of conformational space, yet it still demonstrated some conformational bias (Tables S3–S5). Although >60% of the Gly ensemble sampled outside of the well-defined secondary structure regions (Table S3), on average 50% more than the other guest residues, there were distinct preferences toward the αR and αL regions. And although Gly has more freedom to breach the boundaries of the defined regions, only 10% visits Qo, which coincides with a narrow area attributed to γ-turns (61); for the other residues, sampling of this area is negligible (<1%).
Table 1.
Coverage of φ/ψ Space by Ala-Based Pentapeptide Systems in Control and Denaturing Conditions
| Water 298 K (%) | 8 M Urea 298 K (%) | Water 498 K (%) | |
|---|---|---|---|
| Residue | |||
| Ala | 61a | 63 ± 2a | 62a |
| Arg | 50 | 50 | 53 |
| Asn | 50 | 49 | 53 |
| Asp | 37 | 36 | 52 |
| Ash | 44 | 46 | 50 |
| Cys | 56 | 53 | 53 |
| Gln | 50 | 50 | 53 |
| Glu | 50 | 49 | 51 |
| Glh | 48 | 53 | 52 |
| Gly | 81 | 81a | 79 |
| Hid | 50 | 51 | 55 |
| Hie | 51 | 52 | 54 |
| Hip | 52 | 53 | 50 |
| Ile | 39 | 39 | 39 |
| Leu | 50 | 51 | 52 |
| Lys | 50 | 48 | 52 |
| Met | 51 | 49 | 53 |
| Phe | 51 | 49 | 50 |
| Pro | 13 | 15 | 18 |
| Ser | 51 | 55 | 54 |
| Thr | 39 | 36 | 42 |
| Trp | 50 | 53 ± 1a | 53 |
| Tyr | 53 | 44 | 53 |
| Val | 38 | 40 | 40 |
| Meanb | 49 ± 11 | 52 ± 13 | 52 ± 10 |
Averages across triplicate simulations; standard deviations are omitted where they were <0.5%.
Mean coverage is calculated excluding glycine and proline values.
Figure 2.
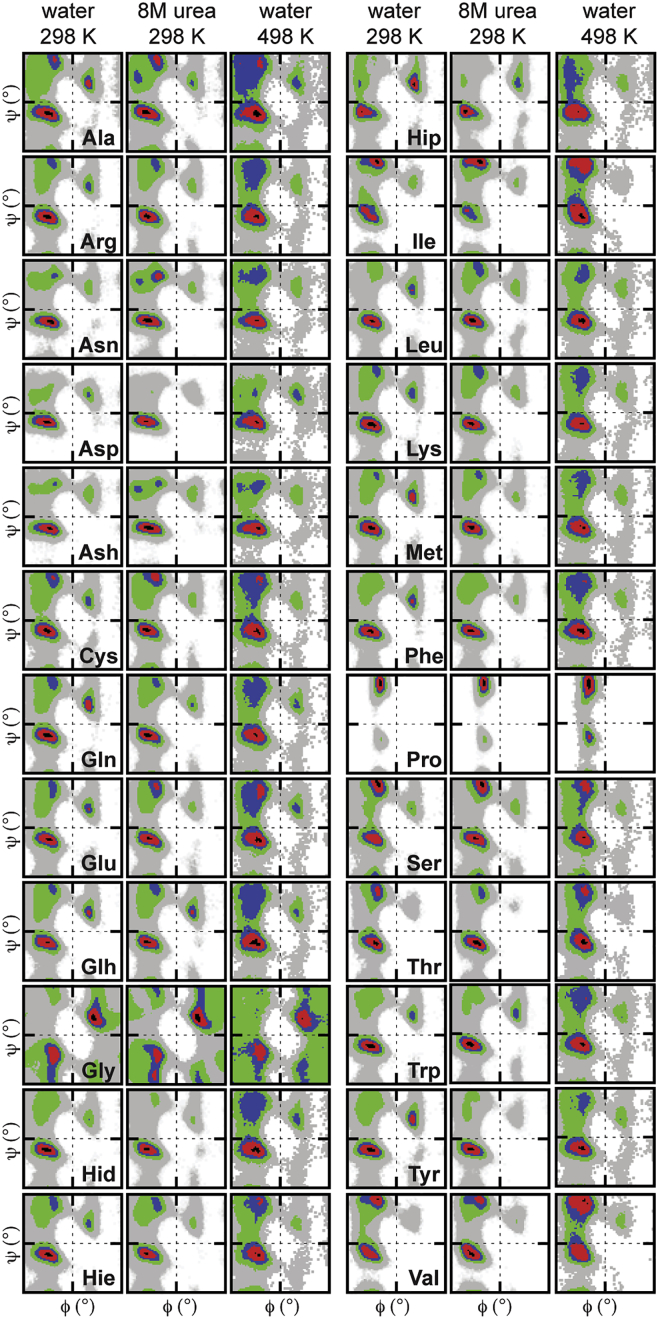
Ramachandran plots of the conformational populations of the guest (X) residues. Plots are provided for the peptides in different environments: pure water at 298 K, 8 M urea at 298 K, and pure water at 498 K. The conformational regions are colored by increasing percentage population from gray to black: 0 = white; 0 < gray < 0.05; 0.05 ≤ green < 0.2; 0.2 ≤ blue < 0.4; 0.4 ≤ red < 0.8; 0.8 ≤ black. To see this figure in color, go online.
The sampling bias was more pronounced for the other non-Gly guest residues. In the control 298 K water simulations, the majority of residues preferentially populated the QαR quadrant, whereas residues with smaller side chains, Ala, Cys, and Ser, had almost equivalent populations in the QαR and Qβ quadrants. In contrast, the β-branched residues, Ile and Val, preferentially sampled the Qβ quadrants (Fig. 2). Although correlations between the ϕ/ψ distributions of the amino acids showed that many exhibit similar sampling, distinct categories were present (Table S6): β-branched, linear chains, aromatics, and charged residues. Of the 24 amino acid side chains considered here, 15 had a maximal population in the broadly defined β-region (Fig. 3 a) due to this area spanning over twice that of the other defined regions (Fig. 3 b). However, partitioning of this β-region into three smaller areas that more finely reflect the range of conformations in this region, PIIL, PIR, and non-polyproline β (nPβ), revealed the majority of residues to preferentially cluster in αR (Fig. 3 c and Table S3). Again, residues that did not exhibit this bias toward αR were Ser and the β-branched Ile and Val residues. Thr exhibited a slight preference for αR but had PIIL content (26%) comparable to the other β-branched residues. Ser and Thr, which share similar chemistry, stand out as being very similar to one another, Thr being the least similar of the β-branched residues. Interestingly, the residues with ϕ/ψ frequency distributions most similar to Thr (Cys, Ser, and Hie) are found in enzyme active sites (Table S6). Cys, Ser, and Thr, in particular, all have been identified as playing the role of nucleophile in catalytic triads (63, 64, 65). Residues with long, linear side chains also exhibit very similar behaviors. Charge can have a big effect, which is exhibited by the three protonated forms of His as well as Ash, which is more similar to its homologous amidated form, Asn, than to its own charged form, Asp (Tables S3 and S6). And, as expected, proline, constrained by its cyclic nature, sampled the smallest area of ϕ/ψ space, being confined to the PIIL region (Fig. 2).
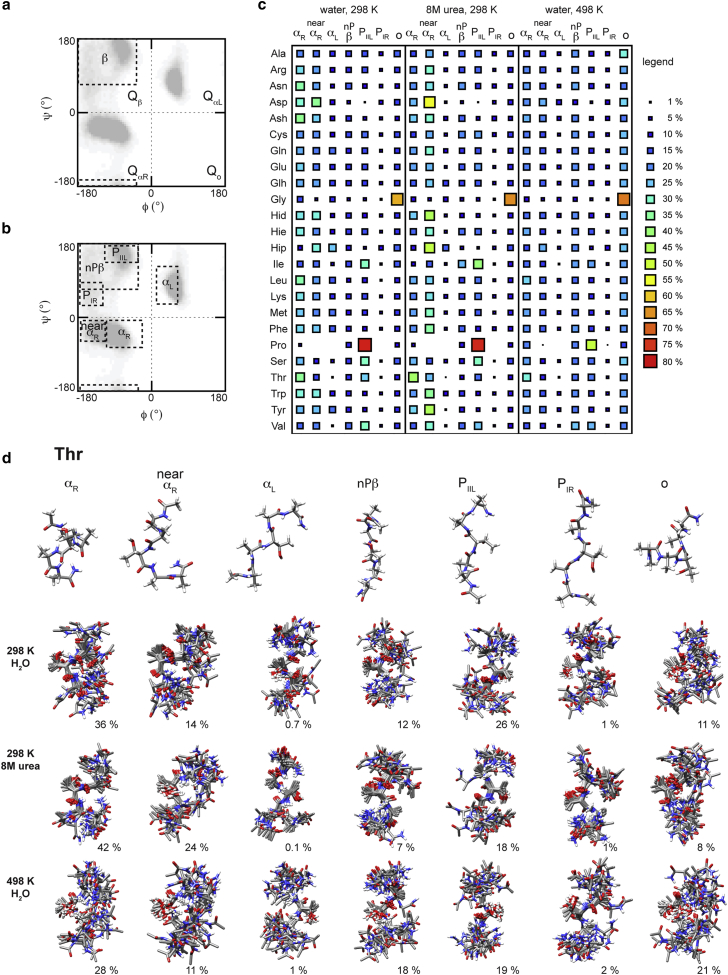
Figure 3.
Changes in the percentage populations of secondary structure conformations related to denaturing environment. (a) Four quadrants of the Ramachandran plot showing the broadly defined β-region. (b) Regions within the quadrants pertaining to specific conformations: right-handed α-helix (αR), left-handed α-helix (αL), polyproline type II left-handed helix (PIIL), polyproline type I right-handed helix (PIR), and the remaining nonpolyproline β-region (nPβ). Please note that both β and nPβ regions include a small area where the β-basin is shown to protrude into the bottom of the QαR quadrant. (c) Percentage populations of conformational regions for guest (X) residues in pure water at 298 K, 8 M urea at 298 K, and pure water at 498 K. (d) Ensembles of 20 structures randomly selected from each of the conformation regions for Thr from all three conditions. UCSF Chimera from the University of California, San Francisco (supported by NIH grant P41 RR-01081) was used to produce structural images (62).To see this figure in color, go online.
This grouping of residues into those that are β-branched and those that are not is a result of the increased steric constraints at the Cβ atom and is consistent with the known predilection of β-branched residues for β-sheet structure (66, 67, 68, 69). However, in general, a strong correlation between the conformational preference for β-regions and previously reported β-sheet propensity of an amino acid was not observed. For example, Tyr has high β-sheet propensity (66), yet this residue preferentially sampled the αR quadrant under both native control and denaturing conditions (Fig. 2). This mercurial behavior was not unexpected, given that there are known cases of residues in β-sheet regions within native states manifesting nonnative helical structure in isolation or in intermediate or denatured states (70, 71, 72, 73, 74, 75). However, there is some correlation between the ranking of the residues by their αR propensities and helix propensity observed in some experiments (76). One observation that was unexpected, that Asn and Ash showed greater helical propensities than Ala, is echoed here (76). Furthermore, the observed preference and range of αR, αL, and PIIL populations for the guest residues are in line with experimental estimates of the structural content in the disordered solution state of α-synuclein (77). And although there is concern that MD force fields induce excessive helicity, the average helix content for the blocked AAAAA peptide simulations presented here (19.4%) is also in closer agreement with experimental estimates, which are ∼20% for Ala3-, Ala4-, and Ala5-based peptides (42, 78), than are many other force fields, which, aside from GROMOS, have unrealistically high helical contents: 13.1% (GROMOS53a6), 57.5% (CHARMM27 with CMAP); 62.3% (AMBER03); 94.2% (AMBER99); and 97.6% (AMBER94) (79).
Some of these general trends in the intrinsic behaviors of the amino acids in water at 298 K are similar to those previously reported by us for the GGXGG series (36). For example, trends in the ϕ/ψ coverage are similar to that of the amino acids in the GGXGG series where β-branched residues, Asp and Pro, covered less area (mean coverage of 45% for GGXGG and 49% for AAXAA). Unfortunately, the previous data were limited to water at 298 K and did not include different protonation states, so despite Asp in GGDGG showing the same reduced sampling of ϕ/ψ space (31% for GGDGG and 37% for AADAA), it is unknown if the effects of protonation are isolated to the AAXAA series. It was noted that within the achiral glycine host, the amino acids sampled QαR to a greater extent, with reduced sampling in the Qβ and Qo quadrants. The greater Qβ sampling in the AAXAA series coincided with increases in PII populations (PIIL + PIR) for many of the residues, with an average increase of 6%, with Pro showing the greatest difference (e.g., 58% for GGPGG versus 78% for AAPAA). This increase in PIIL observed for the AAXAA series is in closer agreement with experimental estimates (80). The most notable differences between the GGXGG and AAXAA data sets were the concomitant increases in the AAXAA β propensities (average 8%), with decreased nαR populations (average 8%). However, using these differences to draw conclusions, such as that the chiral nature of Ala hosts may favor PIIL formation, is problematic, as it is possible that the differences originate from the sampling discrepancy between the 100 ns GGXGG and 600 ns AAXAA data sets (36). Due to an interest in the sequence context on the intrinsic propensities, we have now extended our previous GGXGG simulations and performed new simulations under the denaturing conditions presented here to better compare the two systems, which reflect different aspects of the intrinsic propensities of the amino acids (M.C. Childers, C.-L.T., and V.D., unpublished data). Here, we focus on the intrinsic AAXAA propensities in response to different denaturing environments.
Denatured state propensities differ
Except for the Ash/Asp pair, where increases in ϕ/ψ coverage of up to 15% were observed, the environment did not significantly alter the area of ϕ/ψ space sampled with only an average change of 2.5% (Table 1). The charge on the Asp side chain reduces the area sampled compared to the protonated form, Ash, in pure water and 8M urea where long water residence times around the COO- group of 46 ps and 58 ps, respectively, appear to be responsible for the decreased coverage of ϕ/ψ space. At high temperature, the solvent contacts formed by Asp that limit its sampling of ϕ/ψ space are weaker due to the increase in kinetic energy, with water residence times substantially reduced to 7 ps, so that the sampling of the charged Asp becomes more like that of the protonated Ash residue (Fig. 3 c and Tables S5 and S7). Although Glu shares the same functional group as Asp and has a similar water residence time, the Glu/Glh pair did not exhibit this behavior. The greater separation between the Glu side chain terminus and the main chain reduces the effect of the solvent interactions with the COO- group on the main-chain mobility; the intervening CH2 group has short water residence times that are unchanged by the environment.
Although the area of ϕ/ψ space sampled by the majority of amino acids did not differ substantially, both the preference for a given quadrant and the propensities within defined conformational regions were dependent on the denaturing conditions (Tables S3–S5). Similarity in the amino acid sampling increased at high temperature but remained more distinct in the presence of urea (Tables S6–S8). The majority of residues dominated the QαΡ quadrant under both native control conditions and in 8 M urea (Fig. 2 and Table S4). However, at high temperature, there was a distinct shift away from QαΡ to the Qβ quadrant, resulting in evening out of these populations (Table S5). The only exception where sampling of the Qβ quadrant at high temperature did not increase was Pro. The differential sampling exhibited by the two denaturing conditions is more apparent in the rearrangement of populations across the specific conformational regions (Fig. 3). For example, the conformational sampling of Pro was affected differently by high temperature and urea, although its preference for the Qβ quadrant remained unchanged (Fig. 3 and Tables S4 and S5). The sampling was indistinguishable at 298 K in water and in 8 M urea (Fig. 3); 88% of the population within the β-region was confined to the PIIL region. However, at high temperature, there was a marked movement of sampling outside of the β-region (Fig. 2 and Table S5).
For most residues, more expanded structures within QαΡ were sampled in urea, where large increases in nαΡ occurred (Table S4). At high temperature, the preferences across the specific conformational regions were diminished overall. Most residues showed increased sampling of the more extended β-region, resulting in the shift to Qβ. Moreover, the percentage of these dominant populations in the β-region was almost invariant with respect to amino acid identity, ranging from 28% to 37%, except for the β-branched residues, Ile and Val, and Pro, which sampled this region to a greater extent (>45%). Only Gly (12%) and Asp (15%) had notably low populations in this region.
This divergent sampling for the amino acids under different denaturing conditions, specifically changes in the PIIL populations, is in agreement with recent experiments (15). Many expect, as initially hypothesized by Tiffany and Krimm (18), that elevated populations of PIIL will be observed when proteins are placed in urea (15, 19). Although we observed an increase in the PIIL populations in urea for some residues, the PIIL conformation was not dominant under denaturing or native control conditions, except for five residues, Pro, Ser, and the β-branched residues Ile and Val, and to a lesser extent Thr, which we attribute to the larger steric constraints at Cβ (Fig. 3 c). In 8 M urea, the PIIL content of Ile increased by 8%, whereas Val and Thr had reduced PIIL, with population shifts toward the α-helical regions, possibly related to the difference in the side-chain length. For most of the non-β-branched residues, however, there was no significant increase in the PIIL population in 8 M urea (Table S4). The marked increase in the population of the β-region in urea (Fig. 3 and Table S4) was actually due to an increase in extended, non-PIIL conformations, and this was outweighed by the larger shift toward the nαR region.
In contrast, larger modulations of the PIIL populations were observed at high temperature. Except for Pro, which retained a preference for PIIL conformations, there was no considerable shift toward the PIIL region. Instead, more extended β conformations were populated (Fig. 3 c). Elam et al. detected a similar contrast in the modulation of PIIL propensity in urea and at high temperatures for the Src-homology domain (15). Further, as observed for the XAO peptide (10), we also find that although PIIL is present, it is not as substantial as some have inferred from experiment and reported elsewhere (15, 19, 21). Further, the increase in more extended β conformations and decreases in PIIL and αR seen here at high temperature are also in agreement with other experimental observations and simulations (21, 24, 81). Hence, while distinct preferences are present at 298 K in both urea and water, at 498 K, there is a reduction in the intrinsic propensities resulting from the increased dynamics. Although sampling of the conformational regions varied with the conditions, ensembles taken from within each conformational region were equivalent, as shown for Thr (see the populations of the different conformations in Fig. 3 d). At 298 K, in both water and urea, some of the ensembles from each conformational region were well defined, with secondary structures involving more than the central residue. However, at high temperature, the structure around the central guest residue was more variable, resulting in broader conformational ensembles.
Many experimental observations, for example, the helical content and the offset between β and decreasing PIIL content with increasing temperature, are reproduced here (21, 22, 52). However, the dominance and increase of PIIL content in 8M urea that some expect was not observed; here, the PIIL content ranges are comparable in water at 298 K and 8 M urea (Tables S3 and S4). Consideration of trends in the PIIL content for the different amino acids against those previously reported reinforces the known inconsistency of PIIL propensity scales (80). Across all environments, Pro, the β-branched residues, and Ser exhibited the highest PIIL propensities, in agreement with experimental observations by Shi et al. (21) but not with those of Rucker et al. (82) and Brown and Zondlo (23). In 8 M urea, Glu, Ala, and Cys moved toward the top, somewhat in line with experimental reports that Ala has high PIIL propensity (19, 23). Gly, Hip, and Asp were consistently at the bottom of the scale. The aromatic residues also exhibited low PIIL propensities, lowered further in 8 M urea, in agreement with the Brown and Zondlo scale (23). Except for Pro, the PIIL contents were closer to other theoretical scales (14) but lower than experimental estimates using the PPXPP and GGXGG series (21, 82).
The disagreement with other PIIL scales is especially apparent for the charged residues, which we find to have lower propensities than other scales might suggest. However, PIIL has been shown to be sensitive to sample conditions (temperature, charge, and ions) (22). The sensitivity of PIIL to sample conditions in conjunction with probable experimental overestimations, due to the assumptions required to estimate PIIL from spectroscopic data, may explain the inconsistency in the PIIL scales reported (80). Further, given that the PIIL scales determined thus far have used different hosts (23, 80, 82), including Pro-rich hosts, they are reflecting the extent to which the guest residue propensities can be modulated by the different neighboring residues rather than providing a measure of intrinsic PIIL propensity. Hence, the disagreement with the experimental PIIL scales, especially for the charged residues, should not be seen as discouraging. And, interestingly, although at odds with some scales, the high and low PIIL propensities we observed for Ser and Asp, respectively, correlate with their frequency in Pro-rich regions of proteins (23).
Transition pathways and left-handed helix propensities
Although left-handed helices are scarcely found in protein structures, they can appear in functional regions that form part of an active site or are involved in ligand binding (83). Previous surveys of the PDB have tried to resolve the true nature of αL structures and establish propensity scales for use in engineering molecules, sometimes using D-amino acids, to enforce similar functional structure (83, 84). We, too, have been interested in such propensities to inform our development of heterochiral peptide inhibitors of amyloidosis (85, 86). The αL propensities captured here support these earlier PDB-based observations (83). We find that all amino acids except Pro will assume αL ϕ/ψ angles (Figs. 2 and 3 c; Table S3). All but Hip prefer αR; Hip is unusual in being the only residue to have a substantially larger population in αL (25%) than αR (9%). Interestingly, Novotny and Kelywegt (83) reported that the His-containing sequence HAGEGG formed the longest αL-helix found in the PDB, which was crystallized using a slightly acidic solution and coordinated with Zn2+ (87). This may have made sampling of αL more favorable, as observed here for the protonated form. The β-branched residues and Ser all have the lowest propensities for αL (<5%), with Thr the least likely to sample αL (Table S3). Encouragingly, these observations agree with the prevalence of certain amino acid types found in αL helices or αL-turns in experimental structures. It has been hypothesized that amyloidogenic regions may form soluble intermediates that contain α-sheet stretches where the residues assume alternating αR and αL ϕ/ψ angles (86, 88, 89, 90). Interestingly, it is residues at either end of the αL propensity scale, Ile, Val, Leu, Phe, and Tyr, that are most frequently found in amyloidogenic regions and are predicted to have the greatest amyloid propensities (91, 92).
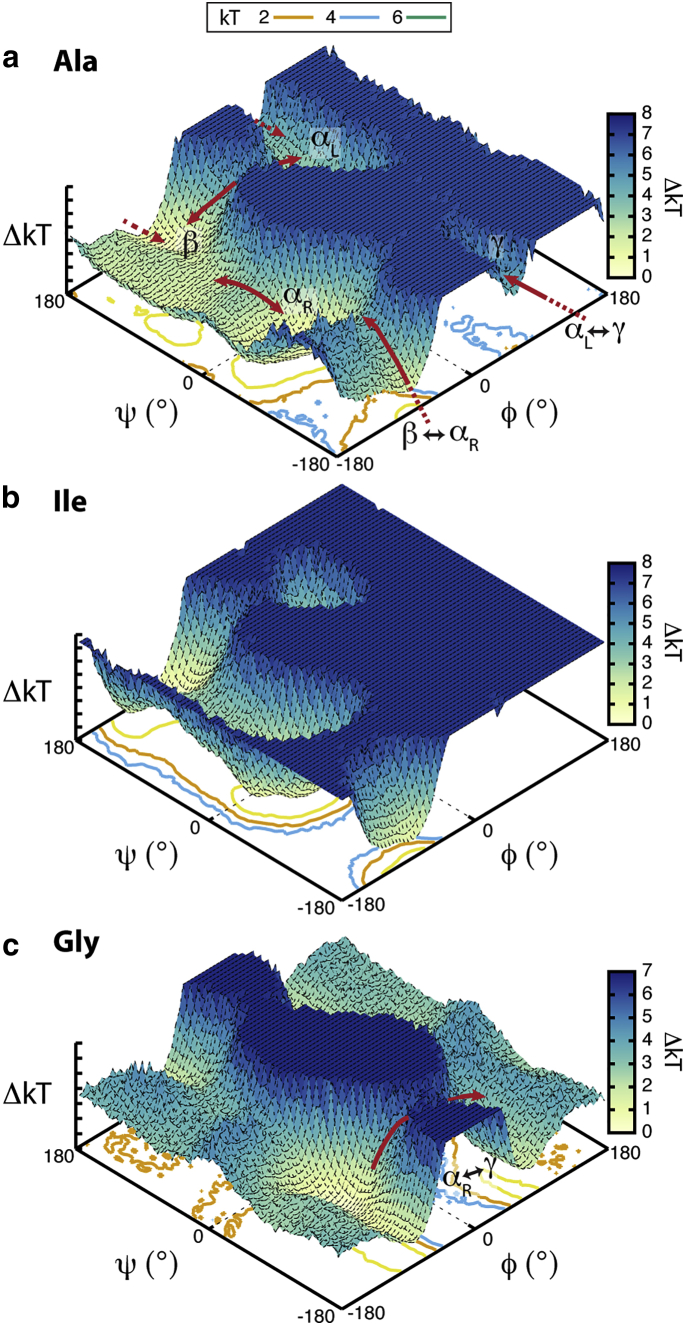
Although all non-Pro residues sample the αL region, transitions to αL are rare. There are five possible pathways across ϕ/ψ space linking the conformational regions, the most expansive being between the αR/nαR and β regions (Fig. 4). Most residues access αL and Qo via a narrow path between β and αL and, although diminished for β-branched residues (Fig. 4 b), the γ-turn region is another one rarely sampled, as it is reached through transitions from αL. Only for Gly is a αR↔γ path available in addition to the greater access between all regions due to the significantly diminished barrier where ϕ approaches 180° (Fig. 4 c). The common transitions are between αR↔nαR and β↔PIIL, with the most frequent occurring between nαR and αR, except in the cases of Ile, Val, Ser, and Pro. Except for Gly, transitions involving the αL region are rarely observed. In any one trajectory, there are only ∼1000–2000 transitions involving αL, where sampling remained within QαL or traveled back and forth between αL and the small channel in Qo (Fig. 4 a). Transition between αL and the other defined conformational regions was minimal (<40). The small populations that appear in αL reach that region via transitions through the β region, specifically PIIL, or from outside the defined secondary structure areas where lower energy barriers exist (Fig. 4 b). This pathway was previously observed for a similar peptide plane flip to αL within an amyloidogenic region of transthyretin (90). The barriers to other conformations are sufficient enough that once an amino acid assumes the αL backbone conformation, it will reside in the αL region for longer periods of time than any other region (Table S9). This trapping of the αL conformation is what accounts for the αL propensities observed here. Correspondingly, the regions where the landscape between them is easier to traverse, e.g., nαR and β, have shorter residence times and a greater number of rapid transitions (Table S9).
Figure 4.
Accessible pathways across the ϕ/ψ free-energy landscapes of (a) alanine, (b) isoleucine, and (c) glycine. To see this figure in color, go online.
Experimental validation and underlying conformational ensembles
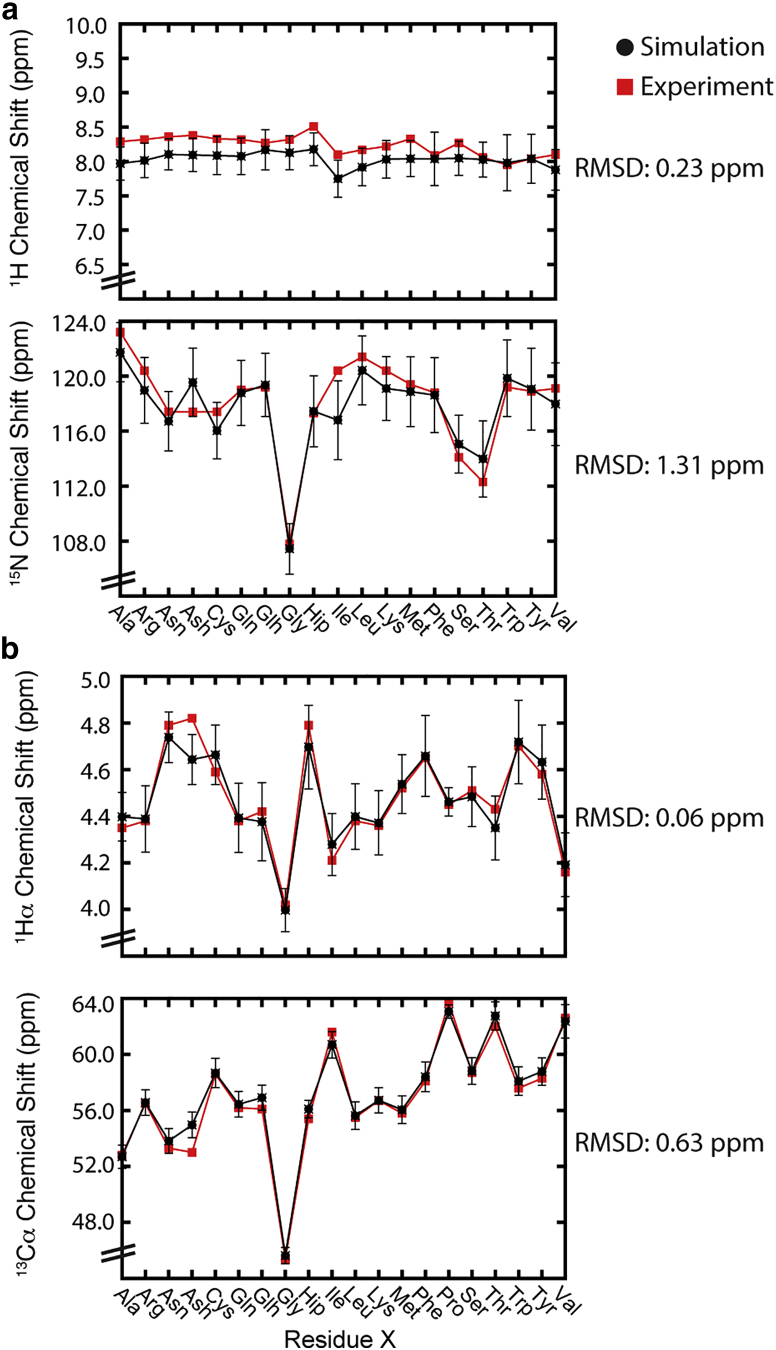
There was good agreement between the experimental spectroscopic observables and those calculated from our MD simulations (Fig. 5; Tables 2 and S10). The calculated 1HN chemical shifts fall slightly upfield, except for Tyr, in line with the error expected from SHIFTX2 (Fig. 5 a). The root mean-square deviations (RMSDs) between the experimental and calculated shifts was 0.23 ppm, but the low dispersion in both the experimental and calculated 1HN chemical shifts produced a correlation coefficient of 0.63. There was greater variation in the 15NH shifts, and the experimental and calculated values were strongly correlated (R = 0.93, with RMSD = 1.31 ppm), with no consistent offset (Fig. 5 a). Again, the 15NH shifts were within experimental error, with remarkable agreement in some cases (Fig. 5 a). For the remaining nuclei, highly satisfactory correlations (R ≥ 0.95) were obtained (Tables 2 and S10), and for all nuclei, the RMSDs between the experimental and calculated values ranged from 0.06 (1Hα) to 1.31 (15NH) ppm and were comparable to those expected using SHIFTX2 (Fig. 5) (53). There is also general agreement between the calculated 3JHNHα coupling constant, 6.96 ± 2.12 Hz, and experiment, 5.98 Hz, for the central Ala in the AAAAA peptide (52). For the other residues, the coupling constants fell in the range 7.03–7.25 Hz, which were within the range typical of many residues in an IDP or such a rapidly fluctuating ensemble of short peptides as studied here (77).
Figure 5.
Experimental and simulated chemical shifts for the guest (X) residues. (a) The 1HN and 15NH chemical shifts of the main-chain NH groups. (b) The 1Hα and 13Cα chemical shifts of main-chain α-CH groups. To see this figure in color, go online.
Table 2.
Correlations between Experimental and Calculated NMR Chemical Shifts
| Nucleus | R | n |
|---|---|---|
| HN | 0.63 | 19 |
| Hα | 0.96 | 20 |
| Hβ1 | 1.00 | 4 |
| Hβ2 | 0.99 | 16 |
| Hβ3 | 0.99 | 16 |
| NH | 0.93 | 19 |
| Cα | 0.99 | 20 |
| Cβ | 1.00 | 19 |
| C′ | 0.95 | 20 |
| Overall | 1.00 | 153 |
Experimental data for the AAXAA peptides generated by Prestegard et al. was for the 1HN and 15NH nuclei in 8 M urea at pH 2.5 (43). For the other nuclei, the data of Schwarzinger et al. for GGXGG peptides in 8 M urea at pH 2.3 were used (51). Calculated NMR chemical shifts were for the 8 M urea MD simulations.
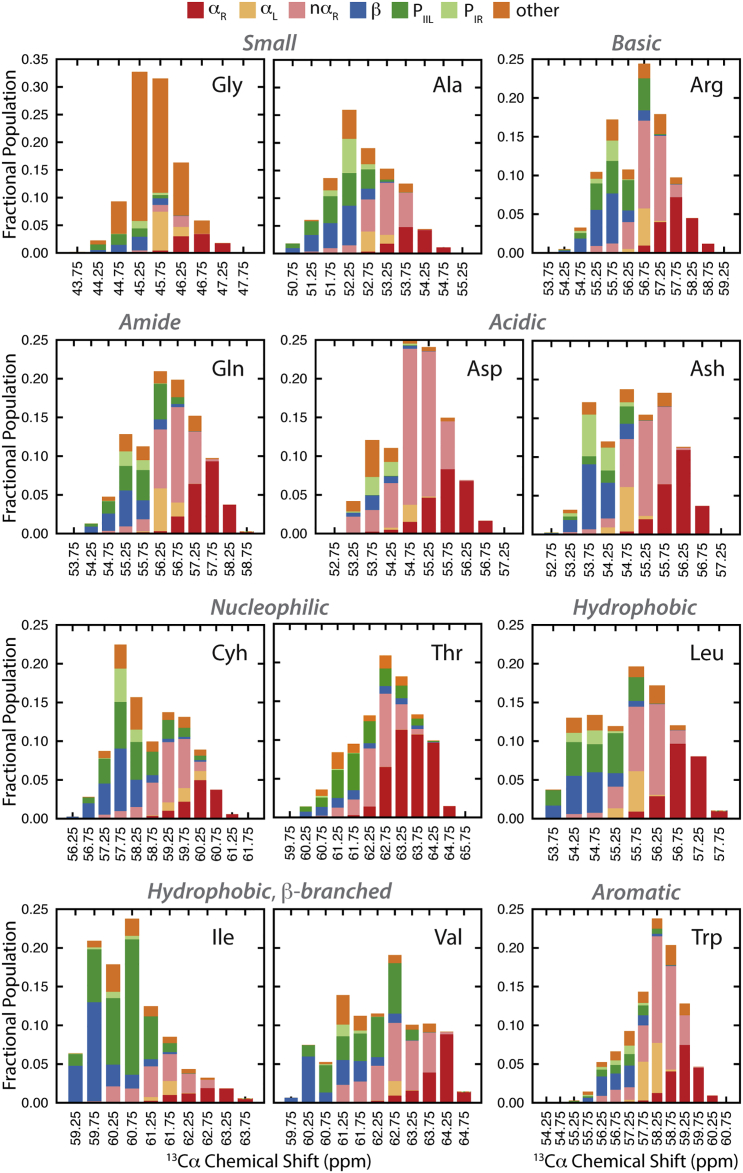
Spectroscopic measures, such as chemical shifts, are a property of the ensemble, and as unfolded states are heterogeneous, the true distribution of conformational states between which rapid interconversion is occurring is difficult to ascertain from the experimentally observed ensemble average. Examination of the underlying ensembles giving rise to the chemical shifts allowed us to investigate the conformational makeup of these systems. Here, because of their sensitivity to secondary structure and side-chain chemistry, we focus on 13Cα shifts (93). Fig. 6 shows the population distribution of chemical shifts in our MD ensembles colored by the originating conformations. Within the ensembles, conformations where the central residue was in the β-region (blue) and the polyproline PIIL (green) and PIR (pale green) regions have chemical shifts upfield of the ensemble average (Fig. 6). Shifts arising from conformations where the guest residue is in αR are downfield of the ensemble average. This is consistent with what is expected from empirical assessments of secondary chemical shifts (93). Further, the chemical-shift differences between the average of the α and β distributions, isolated here, are of similar magnitude to empirically determined secondary shifts. When transitioning from αR to β structure, residues commonly traverse a pathway through nαR (Fig. 4 a). Correspondingly, conformations from the nαR region have shifts that overlay those from both αR and β regions, indicating a gradual upfield shift that occurs as helical turns become more extended and assume β conformations (Fig. 6). Interestingly, when residues are in αL, their chemical-shift populations, although overlapping, are skewed slightly upfield compared to when the guest residue is in αR.
Figure 6.
Conformational makeup of the MD ensembles giving rise to the empirically validated ensemble averaged 13Cα chemical shifts. Histograms show the fractional population of the ensemble at a given chemical shift for a guest amino acid and the proportion of that population where the residue inhabits a given conformational region. The legend for the conformational regions is above the histograms. To see this figure in color, go online.
Hence, the relationship between chemical shift and conformation is not distinct, nor separable, for all residues. The chemical-shift distributions from the different ϕ/ψ regions overlap, creating multimodal profiles (Fig. 6). This degeneracy is not altogether unheard of (94); however, being able to examine the ensembles in great detail beyond just considering αR and β structure indicates a greater degree of overlap and complexity in the relationship between the local conformation of a residue and the chemical shift. For example, some residues in β (as well as PIIL and PIR) have chemical shifts in the same range as those arising from αR and αL. This ambiguous relationship between chemical shift and conformation is demonstrated most significantly by the Thr ensemble (Fig. 6). Further, these data indicate that substantial conformational populations can exist and, although the ensemble-average may move in a direction corresponding to coalescence of a given structure, that shift may only be detectable once the population redistribution has been considerable.
Backbone conformational propensity library for unfolded states
Having achieved Boltzmann sampling of conformational space, we are able to provide details on the relative conformational populations and generate an intrinsic backbone propensity library of highly probable dihedral angles (Table S11). The mode of the local and global ϕ/ψ population distributions, which would be coincident with the local and global minima on a free-energy landscape, were taken to obtain a library of ϕ/ψ angles for each amino acid residue. Although ideal angles for secondary structure elements are well known (61, 95), we observed that while capturing the intrinsic nature of the amino acids, the most probable dihedral angles within a conformational region did not necessarily cluster around typical secondary structure values and instead varied with amino acid identity. For example, the most probable ϕ angle in αR ranges from -80° to −96°, with the largest variation in both ϕ and ψ angles observed for residues in the β-region (Table S11). Within the αR region, only Pro and Gly have a ϕ-angle in the vicinity of that expected for an ideal α-helix (−60° to −65°). This amino acid dependence is noteworthy and should be of value for those interested in building natively or nonnatively unfolded models, such as proteins with known disordered regions.
Conclusions
The MD simulations presented here of the AAXAA peptides under three different conditions show that amino acids have intrinsic conformational preferences and that they can be modulated by solvent and temperature. Although preferences observed under native conditions, water at 298 K, are largely retained in urea, the differences in the conformational makeup of the ensembles are diminished at high temperature. However, for most residues the coverage of ϕ/ψ space was relatively invariant with environment. The MD-generated ensembles reproduced the spectroscopic observables and are in excellent agreement with experimental chemical shifts. Although more detailed structural information from experiment is unavailable to compare with our ensembles, our force field and methods provide excellent agreement with experiment under numerous denaturing conditions elsewhere, including the accurate prediction of protein structures and properties during folding ahead of experiment (96, 97, 98, 99). Our simulations also reproduce many other known tendencies of the amino acids in globular proteins, indicating that some of the structural preferences observed for globular proteins are related to the intrinsic propensities of the amino acids.
Small host-guest systems are often used to generate sets of random-coil chemical shifts to be used as a reference for estimating secondary structure content. Recently, Prestegard et al. (43) published a set of chemical shifts for the AAXAA peptide series. As conventional structure determination methods struggle to simultaneously capture the dynamics and atomic-level detail (100), the conformational makeup giving rise to an experimental, yet ensemble-averaged, measurement in these reference systems is unknown. We probed these experimentally intractable details and observed the same relationship between conformation and chemical shift as determined empirically. However, the overlapping mixture of conformations at a given chemical shift confirmed the inherent difficulty in estimating residual or secondary structure in unfolded states. The conformational makeup of the ensembles giving rise to random-coil chemical shifts is reported here to enable more accurate estimation of the extent of residual structure in denatured and unstructured states. Moreover, by performing exhaustive sampling of ϕ/ψ space we have been able to construct a set of intrinsic backbone angles. The backbone propensities and angles presented here form part of our Structural Library of Residue Propensities for use in the modeling or structure prediction of natively and nonnatively unfolded systems. This library is available through our Dynameomics site (http://dynameomics.org/SLIRP).
Author Contributions
V.D. and J. Vondrasek designed and supervised the research; C-L.T. and J. Vymetal performed research and analyzed data; and C-L.T. and V.D. wrote the manuscript. All authors contributed to manuscript revisions.
Acknowledgments
We thank Dr. James Prestegard for providing chemical shift values ahead of publication for experimental validation of our simulations. We are grateful for computing resources provided by the National Energy Research Scientific Computing Center, supported by the Department of Energy Office of Biological Research, which is supported by the U.S. Department of Energy under contract no. DE-AC02-05CH11231 (to V.D.).
This work was supported by National Institutes of Health grant GM 50789 (to V.D.). J. Vymetal was supported by grant no. LH11020 from the Ministry of Education, Youth and Sports (MSMT) of the Czech Republic and, in part, by a development of research organization grant (RVO:61388963, to J. Vondrasek and V.D.). The UCSF Chimera from the University of California, San Francisco (supported by National Institutes of Health grant P41 RR-01081) was used to produce structural images (62).
Editor: Rohit Pappu.
Footnotes
Three figures and eleven tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(15)04748-7.
Supporting Citations
References (43, 51, 52) appear in the Supporting Material.
Supporting Material
References
- 1.Bowler B.E. Residual structure in unfolded proteins. Curr. Opin. Struct. Biol. 2012;22:4–13. doi: 10.1016/j.sbi.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oldfield C.J., Dunker A.K. Intrinsically disordered proteins and intrinsically disordered protein regions. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- 3.Uversky V.N. Flexible nets of malleable guardians: intrinsically disordered chaperones in neurodegenerative diseases. Chem. Rev. 2011;111:1134–1166. doi: 10.1021/cr100186d. [DOI] [PubMed] [Google Scholar]
- 4.Hsu W.-L., Oldfield C.J., Dunker A.K. Exploring the binding diversity of intrinsically disordered proteins involved in one-to-many binding. Protein Sci. 2013;22:258–273. doi: 10.1002/pro.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oldfield C.J., Meng J., Dunker A.K. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding F., Jha R.K., Dokholyan N.V. Scaling behavior and structure of denatured proteins. Structure. 2005;13:1047–1054. doi: 10.1016/j.str.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Smith L.J., Fiebig K.M., Dobson C.M. The concept of a random coil. Residual structure in peptides and denatured proteins. Fold. Des. 1996;1:R95–R106. doi: 10.1016/S1359-0278(96)00046-6. [DOI] [PubMed] [Google Scholar]
- 8.Tanford C., Kawahara K., Lapanje S. Proteins in 6-M guanidine hydrochloride. Demonstration of random coil behavior. J. Biol. Chem. 1966;241:1921–1923. [PubMed] [Google Scholar]
- 9.McCarney E.R., Kohn J.E., Plaxco K.W. Is there or isn’t there? The case for (and against) residual structure in chemically denatured proteins. Crit. Rev. Biochem. Mol. Biol. 2005;40:181–189. doi: 10.1080/10409230591008143. [DOI] [PubMed] [Google Scholar]
- 10.Makowska J., Rodziewicz-Motowidlo S., Scheraga H.A. Further evidence for the absence of polyproline II stretch in the XAO peptide. Biophys. J. 2007;92:2904–2917. doi: 10.1529/biophysj.106.097550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller W.G., Goebel C.V. Dimensions of protein random coils. Biochemistry. 1968;7:3925–3935. doi: 10.1021/bi00851a021. [DOI] [PubMed] [Google Scholar]
- 12.Kohn J.E., Millett I.S., Plaxco K.W. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzkee N.C., Rose G.D. Reassessing random-coil statistics in unfolded proteins. Proc. Natl. Acad. Sci. USA. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran H.T., Wang X., Pappu R.V. Reconciling observations of sequence-specific conformational propensities with the generic polymeric behavior of denatured proteins. Biochemistry. 2005;44:11369–11380. doi: 10.1021/bi050196l. [DOI] [PubMed] [Google Scholar]
- 15.Elam W.A., Schrank T.P., Hilser V.J. Temperature and urea have opposing impacts on polyproline II conformational bias. Biochemistry. 2013;52:949–958. doi: 10.1021/bi301435p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oh K.-I., Jung Y.-S., Cho M. Conformational distributions of denatured and unstructured proteins are similar to those of 20 × 20 blocked dipeptides. J. Biomol. NMR. 2012;53:25–41. doi: 10.1007/s10858-012-9618-5. [DOI] [PubMed] [Google Scholar]
- 17.Oh K.-I., Lee K.-K., Cho M. A comprehensive library of blocked dipeptides reveals intrinsic backbone conformational propensities of unfolded proteins. Proteins. 2012;80:977–990. doi: 10.1002/prot.24000. [DOI] [PubMed] [Google Scholar]
- 18.Tiffany M.L., Krimm S. New chain conformations of poly(glutamic acid) and polylysine. Biopolymers. 1968;6:1379–1382. doi: 10.1002/bip.1968.360060911. [DOI] [PubMed] [Google Scholar]
- 19.Whittington S.J., Chellgren B.W., Creamer T.P. Urea promotes polyproline II helix formation: implications for protein denatured states. Biochemistry. 2005;44:6269–6275. doi: 10.1021/bi050124u. [DOI] [PubMed] [Google Scholar]
- 20.Mezei M., Fleming P.J., Rose G.D. Polyproline II helix is the preferred conformation for unfolded polyalanine in water. Proteins. 2004;55:502–507. doi: 10.1002/prot.20050. [DOI] [PubMed] [Google Scholar]
- 21.Shi Z., Chen K., Kallenbach N.R. Polyproline II propensities from GGXGG peptides reveal an anticorrelation with β-sheet scales. Proc. Natl. Acad. Sci. USA. 2005;102:17964–17968. doi: 10.1073/pnas.0507124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makowska J., Rodziewicz-Motowidło S., Scheraga H.A. Polyproline II conformation is one of many local conformational states and is not an overall conformation of unfolded peptides and proteins. Proc. Natl. Acad. Sci. USA. 2006;103:1744–1749. doi: 10.1073/pnas.0510549103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown A.M., Zondlo N.J. A propensity scale for type II polyproline helices (PPII): aromatic amino acids in proline-rich sequences strongly disfavor PPII due to proline-aromatic interactions. Biochemistry. 2012;51:5041–5051. doi: 10.1021/bi3002924. [DOI] [PubMed] [Google Scholar]
- 24.Gokce I., Woody R.W., Lakey J.H. Single peptide bonds exhibit poly(pro)II (“random coil”) circular dichroism spectra. J. Am. Chem. Soc. 2005;127:9700–9701. doi: 10.1021/ja052632x. [DOI] [PubMed] [Google Scholar]
- 25.Adzhubei A.A., Sternberg M.J.E., Makarov A.A. Polyproline-II helix in proteins: structure and function. J. Mol. Biol. 2013;425:2100–2132. doi: 10.1016/j.jmb.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 26.Shi Z., Chen K., Kallenbach N.R. Conformation of the backbone in unfolded proteins. Chem. Rev. 2006;106:1877–1897. doi: 10.1021/cr040433a. [DOI] [PubMed] [Google Scholar]
- 27.Shi Z., Woody R.W., Kallenbach N.R. Is polyproline II a major backbone conformation in unfolded proteins? Adv. Protein Chem. 2002;62:163–240. doi: 10.1016/s0065-3233(02)62008-x. [DOI] [PubMed] [Google Scholar]
- 28.Shi Z., Olson C.A., Kallenbach N.R. Polyproline II structure in a sequence of seven alanine residues. Proc. Natl. Acad. Sci. USA. 2002;99:9190–9195. doi: 10.1073/pnas.112193999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eker F., Griebenow K., Schweitzer-Stenner R. Preferred peptide backbone conformations in the unfolded state revealed by the structure analysis of alanine-based (AXA) tripeptides in aqueous solution. Proc. Natl. Acad. Sci. USA. 2004;101:10054–10059. doi: 10.1073/pnas.0402623101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eker F., Griebenow K., Schweitzer-Stenner R. Tripeptides with ionizable side chains adopt a perturbed polyproline II structure in water. Biochemistry. 2004;43:613–621. doi: 10.1021/bi035740+. [DOI] [PubMed] [Google Scholar]
- 31.Shortle D., Ackerman M.S. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 32.Lietzow M.A., Jamin M., Wright P.E. Mapping long-range contacts in a highly unfolded protein. J. Mol. Biol. 2002;322:655–662. doi: 10.1016/s0022-2836(02)00847-1. [DOI] [PubMed] [Google Scholar]
- 33.Kristjansdottir S., Lindorff-Larsen K., Poulsen F.M. Formation of native and non-native interactions in ensembles of denatured ACBP molecules from paramagnetic relaxation enhancement studies. J. Mol. Biol. 2005;347:1053–1062. doi: 10.1016/j.jmb.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Arcus V.L., Vuilleumier S., Fersht A.R. A comparison of the pH, urea, and temperature-denatured states of barnase by heteronuclear NMR: implications for the initiation of protein folding. J. Mol. Biol. 1995;254:305–321. doi: 10.1006/jmbi.1995.0618. [DOI] [PubMed] [Google Scholar]
- 35.Sari N., Alexander P., Orban J. Structure and dynamics of an acid-denatured protein G mutant. Biochemistry. 2000;39:965–977. doi: 10.1021/bi9920230. [DOI] [PubMed] [Google Scholar]
- 36.Beck D.A.C., Alonso D.O.V., Daggett V. The intrinsic conformational propensities of the 20 naturally occurring amino acids and reflection of these propensities in proteins. Proc. Natl. Acad. Sci. USA. 2008;105:12259–12264. doi: 10.1073/pnas.0706527105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wright P.E., Dyson H.J., Lerner R.A. Conformation of peptide fragments of proteins in aqueous solution: implications for initiation of protein folding. Biochemistry. 1988;27:7167–7175. doi: 10.1021/bi00419a001. [DOI] [PubMed] [Google Scholar]
- 38.Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 39.Uversky V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J. 2015;282:1182–1189. doi: 10.1111/febs.13202. [DOI] [PubMed] [Google Scholar]
- 40.Dyson H.J. Expanding the proteome: disordered and alternatively folded proteins. Q. Rev. Biophys. 2011;44:467–518. doi: 10.1017/S0033583511000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pe’er I., Felder C.E., Beckmann J.S. Proteomic signatures: amino acid and oligopeptide compositions differentiate among phyla. Proteins. 2004;54:20–40. doi: 10.1002/prot.10559. [DOI] [PubMed] [Google Scholar]
- 42.Firestine A.M., Chellgren V.M., Creamer T.P. Conformational properties of a peptide model for unfolded α-helices. Biochemistry. 2008;47:3216–3224. doi: 10.1021/bi702474k. [DOI] [PubMed] [Google Scholar]
- 43.Prestegard J.H., Sahu S.C., Gruta C. Chemical shift prediction for denatured proteins. J. Biomol. NMR. 2013;55:201–209. doi: 10.1007/s10858-012-9702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beck D.A.C., Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34:112–120. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Beck D.A.C., McCully M.E., Daggett V. University of Washington; Seattle, WA: 2000–2015. In lucem molecular mechanics (ilmm) [Google Scholar]
- 46.Levitt M., Hirshberg M., Daggett V. Potential energy function and parameters for simulations of the molecular dynamics of proteins and nucleic acids in solution. Comput. Phys. Commun. 1995;91:215–231. [Google Scholar]
- 47.Beck D.A.C., Armen R.S., Daggett V. Cutoff size need not strongly influence molecular dynamics results for solvated polypeptides. Biochemistry. 2005;44:609–616. doi: 10.1021/bi0486381. [DOI] [PubMed] [Google Scholar]
- 48.Zou Q., Bennion B.J., Murphy K.P. The molecular mechanism of stabilization of proteins by TMAO and its ability to counteract the effects of urea. J. Am. Chem. Soc. 2002;124:1192–1202. doi: 10.1021/ja004206b. [DOI] [PubMed] [Google Scholar]
- 49.Levitt M., Hirshberg M., Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. J. Phys. Chem. B. 1997;101:5051–5061. [Google Scholar]
- 50.Kell G.S. Precise representation of volume properties of water at one atmosphere. J. Chem. Eng. Data. 1967;12:66–69. [Google Scholar]
- 51.Schwarzinger S., Kroon G.J.A., Dyson H.J. Random coil chemical shifts in acidic 8 M urea: implementation of random coil shift data in NMRView. J. Biomol. NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 52.Graf J., Nguyen P.H., Schwalbe H. Structure and dynamics of the homologous series of alanine peptides: a joint molecular dynamics/NMR study. J. Am. Chem. Soc. 2007;129:1179–1189. doi: 10.1021/ja0660406. [DOI] [PubMed] [Google Scholar]
- 53.Han B., Liu Y., Wishart D.S. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt J.M., Blümel M., Rüterjans H. Self-consistent 3J coupling analysis for the joint calibration of Karplus coefficients and evaluation of torsion angles. J. Biomol. NMR. 1999;14:1–12. doi: 10.1023/A:1008345303942. [DOI] [PubMed] [Google Scholar]
- 55.Beck D.A.C., Alonso D.O.V., Daggett V. A microscopic view of peptide and protein solvation. Biophys. Chem. 2003;100:221–237. doi: 10.1016/s0301-4622(02)00283-1. [DOI] [PubMed] [Google Scholar]
- 56.Bennion B.J., Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl. Acad. Sci. USA. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beck D.A.C., Bennion B.J., Daggett V. Simulations of macromolecules in protective and denaturing osmolytes: properties of mixed solvent systems and their effects on water and protein structure and dynamics. Methods Enzymol. 2007;428:373–396. doi: 10.1016/S0076-6879(07)28022-X. [DOI] [PubMed] [Google Scholar]
- 58.Ho B.K., Thomas A., Brasseur R. Revisiting the Ramachandran plot: hard-sphere repulsion, electrostatics, and H-bonding in the α-helix. Protein Sci. 2003;12:2508–2522. doi: 10.1110/ps.03235203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- 60.Baldwin R.L., Zimm B.H. Are denatured proteins ever random coils? Proc. Natl. Acad. Sci. USA. 2000;97:12391–12392. doi: 10.1073/pnas.97.23.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Richardson J.S. The anatomy and taxonomy of protein structure. Adv. Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- 62.Pettersen E.F., Goddard T.D., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 63.Dodson G., Wlodawer A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998;23:347–352. doi: 10.1016/s0968-0004(98)01254-7. [DOI] [PubMed] [Google Scholar]
- 64.Polgár L. The catalytic triad of serine peptidases. Cell. Mol. Life Sci. 2005;62:2161–2172. doi: 10.1007/s00018-005-5160-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ekici Ö.D., Paetzel M., Dalbey R.E. Unconventional serine proteases: variations on the catalytic Ser/His/Asp triad configuration. Protein Sci. 2008;17:2023–2037. doi: 10.1110/ps.035436.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith C.K., Withka J.M., Regan L. A thermodynamic scale for the β-sheet forming tendencies of the amino acids. Biochemistry. 1994;33:5510–5517. doi: 10.1021/bi00184a020. [DOI] [PubMed] [Google Scholar]
- 67.Minor D.L., Jr., Kim P.S. Measurement of the β-sheet-forming propensities of amino acids. Nature. 1994;367:660–663. doi: 10.1038/367660a0. [DOI] [PubMed] [Google Scholar]
- 68.Kim C.A., Berg J.M. Thermodynamic β-sheet propensities measured using a zinc-finger host peptide. Nature. 1993;362:267–270. doi: 10.1038/362267a0. [DOI] [PubMed] [Google Scholar]
- 69.Chou P.Y., Fasman G.D. Conformational parameters for amino acids in helical, β-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–222. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 70.Hamada D., Kuroda Y., Goto Y. High helical propensity of the peptide fragments derived from β-lactoglobulin, a predominantly β-sheet protein. J. Mol. Biol. 1995;254:737–746. doi: 10.1006/jmbi.1995.0651. [DOI] [PubMed] [Google Scholar]
- 71.Hamada D., Goto Y. The equilibrium intermediate of β-lactoglobulin with non-native α-helical structure. J. Mol. Biol. 1997;269:479–487. doi: 10.1006/jmbi.1997.1055. [DOI] [PubMed] [Google Scholar]
- 72.Hamada D., Segawa S., Goto Y. Non-native α-helical intermediate in the refolding of β-lactoglobulin, a predominantly β-sheet protein. Nat. Struct. Biol. 1996;3:868–873. doi: 10.1038/nsb1096-868. [DOI] [PubMed] [Google Scholar]
- 73.Liu Z.P., Rizo J., Gierasch L.M. Equilibrium folding studies of cellular retinoic acid binding protein, a predominantly β-sheet protein. Biochemistry. 1994;33:134–142. doi: 10.1021/bi00167a017. [DOI] [PubMed] [Google Scholar]
- 74.Morrone A., McCully M.E., Travaglini-Allocatelli C. The denatured state dictates the topology of two proteins with almost identical sequence but different native structure and function. J. Biol. Chem. 2011;286:3863–3872. doi: 10.1074/jbc.M110.155911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blanco F.J., Serrano L., Forman-Kay J.D. High populations of non-native structures in the denatured state are compatible with the formation of the native folded state. J. Mol. Biol. 1998;284:1153–1164. doi: 10.1006/jmbi.1998.2229. [DOI] [PubMed] [Google Scholar]
- 76.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mantsyzov A.B., Maltsev A.S., Bax A. A maximum entropy approach to the study of residue-specific backbone angle distributions in α-synuclein, an intrinsically disordered protein. Protein Sci. 2014;23:1275–1290. doi: 10.1002/pro.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jiang F., Han W., Wu Y.-D. The intrinsic conformational features of amino acids from a protein coil library and their applications in force field development. Phys. Chem. Chem. Phys. 2013;15:3413–3428. doi: 10.1039/c2cp43633g. [DOI] [PubMed] [Google Scholar]
- 79.Best R.B., Buchete N.-V., Hummer G. Are current molecular dynamics force fields too helical? Biophys. J. 2008;95:L07–L09. doi: 10.1529/biophysj.108.132696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Elam W.A., Schrank T.P., Hilser V.J. Experimental and computational studies of polyproline II propensity. In: Schweitzer-Stenner R., editor. Protein and Peptide Folding, Misfolding, and Non-folding. John Wiley & Sons; Hoboken, NJ: 2012. pp. 159–185. [Google Scholar]
- 81.García A.E. Characterization of non-α helical conformations in Ala peptides. Polymer (Guildf.) 2004;45:669–676. [Google Scholar]
- 82.Rucker A.L., Pager C.T., Creamer T.P. Host-guest scale of left-handed polyproline II helix formation. Proteins. 2003;53:68–75. doi: 10.1002/prot.10477. [DOI] [PubMed] [Google Scholar]
- 83.Novotny M., Kleywegt G.J. A survey of left-handed helices in protein structures. J. Mol. Biol. 2005;347:231–241. doi: 10.1016/j.jmb.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 84.Annavarapu S., Nanda V. Mirrors in the PDB: left-handed α-turns guide design with D-amino acids. BMC Struct. Biol. 2009;9:61. doi: 10.1186/1472-6807-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Towse C.-L., Hopping G., Daggett V. Nature versus design: the conformational propensities of D-amino acids and the importance of side chain chirality. Protein Eng. Des. Sel. 2014;27:447–455. doi: 10.1093/protein/gzu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hopping G., Kellock J., Daggett V. Designed α-sheet peptides inhibit amyloid formation by targeting toxic oligomers. eLife. 2014;3:e01681. doi: 10.7554/eLife.01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sivaraman J., Li Y., Cygler M. Crystal structure of Escherichia coli PdxA, an enzyme involved in the pyridoxal phosphate biosynthesis pathway. J. Biol. Chem. 2003;278:43682–43690. doi: 10.1074/jbc.M306344200. [DOI] [PubMed] [Google Scholar]
- 88.Armen R.S., Bernard B.M., Daggett V. Characterization of a possible amyloidogenic precursor in glutamine-repeat neurodegenerative diseases. Proc. Natl. Acad. Sci. USA. 2005;102:13433–13438. doi: 10.1073/pnas.0502068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Daggett V. α-sheet: The toxic conformer in amyloid diseases? Acc. Chem. Res. 2006;39:594–602. doi: 10.1021/ar0500719. [DOI] [PubMed] [Google Scholar]
- 90.Armen R.S., DeMarco M.L., Daggett V. Pauling and Corey’s α-pleated sheet structure may define the prefibrillar amyloidogenic intermediate in amyloid disease. Proc. Natl. Acad. Sci. USA. 2004;101:11622–11627. doi: 10.1073/pnas.0401781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Das S., Pal U., Maiti N.C. Sequence complexity of amyloidogenic regions in intrinsically disordered human proteins. PLoS One. 2014;9:e89781. doi: 10.1371/journal.pone.0089781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ahmed A.B., Kajava A.V. Breaking the amyloidogenicity code: methods to predict amyloids from amino acid sequence. FEBS Lett. 2013;587:1089–1095. doi: 10.1016/j.febslet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 93.Wishart D.S. Interpreting protein chemical shift data. Prog. Nucl. Magn. Reson. Spectrosc. 2011;58:62–87. doi: 10.1016/j.pnmrs.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 94.Kragelj J., Ozenne V., Jensen M.R. Conformational propensities of intrinsically disordered proteins from NMR chemical shifts. ChemPhysChem. 2013;14:3034–3045. doi: 10.1002/cphc.201300387. [DOI] [PubMed] [Google Scholar]
- 95.Hoffman-Ostenhof O., Cohn W.E., Whelan W.J. IUPAC-IUB Commission on Biochemical Nomenclature. Abbreviations and symbols for the description of the conformation of polypeptide chains. J. Mol. Biol. 1970;52:1–17. [PubMed] [Google Scholar]
- 96.McCully M.E., Beck D.A.C., Daggett V. Refolding the engrailed homeodomain: structural basis for the accumulation of a folding intermediate. Biophys. J. 2010;99:1628–1636. doi: 10.1016/j.bpj.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Day R., Bennion B.J., Daggett V. Increasing temperature accelerates protein unfolding without changing the pathway of unfolding. J. Mol. Biol. 2002;322:189–203. doi: 10.1016/s0022-2836(02)00672-1. [DOI] [PubMed] [Google Scholar]
- 98.Day R., Daggett V. All-atom simulations of protein folding and unfolding. Adv. Protein Chem. 2003;66:373–403. doi: 10.1016/s0065-3233(03)66009-2. [DOI] [PubMed] [Google Scholar]
- 99.Mayor U., Johnson C.M., Fersht A.R. Protein folding and unfolding in microseconds to nanoseconds by experiment and simulation. Proc. Natl. Acad. Sci. USA. 2000;97:13518–13522. doi: 10.1073/pnas.250473497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.van den Bedem H., Fraser J.S. Integrative, dynamic structural biology at atomic resolution--it’s about time. Nat. Methods. 2015;12:307–318. doi: 10.1038/nmeth.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.