Main Text
For over 50 years, bacteriophage have stood as some of the most informative systems used to investigate the fundamental aspects of macromolecular interaction and allostery, yielding insight into how complex structures form. The tractable nature of these systems presents both the simplicity needed to rapidly adopt and develop new experimental technologies and a level of complexity valued in revealing insights that reside at the leading edge of our understanding of biological molecular processes. Of the events that comprise phage assembly, the DNA packaging step that occurs in the dsDNA phages remains one of the most dramatic and complex. During packaging, phage DNA is translocated into a preformed capsid, or prohead, to crystalline density and the molecular machine that drives this process is necessarily one of the most powerful described (1). Fueled by ATP binding and hydrolysis, packaging is catalyzed by a transiently attached pentameric ring-ATPase docked to an elegant dodecameric portal connector structure (Fig. 1) embedded in the capsid shell.
Figure 1.
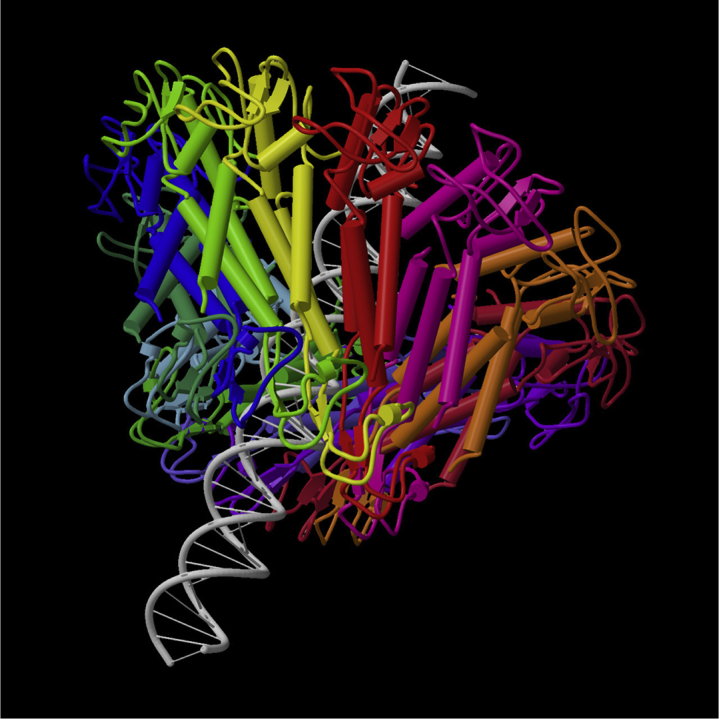
Crystal structure of the bacteriophage ϕ29 connector. DNA is modeled for perspective. Reproduced with permission of the International Union of Crystallography (12). To see this figure in color, go online.
Studies of the mechanism of the DNA translocation process have both matured and diverged over the past decade. With advanced genetic, structural, biophysical, and modeling approaches have come specific observations and interpretations as to how the energy of ATP binding and hydrolysis is harnessed to move DNA. Recent work on bacteriophage ϕ29 indicated the mechanism of translocation is complex: rather than a simple ratchet, with each ATPase subunit pushing DNA incrementally through the connector into the head, packaging entails a highly coordinated dwell-burst structure where the ATPase pentamer functions in a completely asymmetric fashion (2). A role for the connector in driving or regulating this process has been discussed for decades, with models ranging from those that presume the connector is a passive channel (3) to ones where the connector is active, dynamic, and even capable of generating the force required to move DNA (4).
The work of Kumar and Grubmüller (5) that appears in this issue addresses the connector question in a new and valuable way. Here the authors begin to rein in some of the models of DNA packaging by subjecting them to a feasibility study based on the inherent physical structure of the connector itself. Using advanced and detailed modeling and simulation, these authors take a different tack: rather than asking what role the connector plays during DNA packaging, they ask what role it can play based on the connector’s structure and dynamics.
This in silico litmus test for packaging models suggests that the connector does, indeed, present the possibility of conformational change that both responds to the DNA and might regulate the ATPase catalyst that powers the process. Mechanisms where the charged channel loops that line the lumen of the connector interact with DNA and restrain it enable a valvelike mechanism to survive such scrutiny (6). There are limits to the models of connector action, however, and the experiments of Kumar and Grubmüller reveal that some of the more speculative models exceed what the connector appears to be capable of doing. The role of the connector in the push-roll (7) or revolution-without-rotation (8) models is not supported. The simulations that entail whether the connector helps in either rotating or rotationally restraining the DNA as it passes through the channel indicate that these roles are beyond what the connector structure can support. The revolution-without-rotation model, in particular, was advanced without experimental support (9) and appears to have been dispatched by a more sophisticated analysis of what the connector can do based on a dynamic analysis of its structure. Lastly, the observation that DNA rotates slightly to continually align the ATPase ring with the DNA during translocation is tolerated, demanding that the check-valve interaction and this rotation event be considered together in future studies.
Finally, a most interesting prediction is that the DNA in the channel is itself deformed by its interaction with the connector. The idea that DNA is an active participant in its own packaging, whether through a recently proposed scrunching mechanism (10) or through the deformation induced by the protein components of a motor crunching mechanism (11), is intriguing. This supports the idea that in this, and other complex macromolecular systems, all components should be considered dynamic, responsive to change, regulatory, and perhaps active participants in the mechanical process underway. Adding a new active player to the game makes this more complex, but in the end illustrates an elegance of evolutionary design worthy of continued investigation and marvel.
Acknowledgments
The author thanks Dr. Shelley Grimes for her helpful comments in preparing this article.
This work was supported in part by grants from the National Institutes of Health (GM071552 and GM095516).
Editor: Fazoil Ataullakhanov.
References
- 1.Smith D.E., Tans S.J., Bustamante C. The bacteriophage straight ϕ29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 2.Moffitt J.R., Chemla Y.R., Bustamante C. Intersubunit coordination in a homomeric ring ATPase. Nature. 2009;457:446–450. doi: 10.1038/nature07637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun S., Kondabagil K., Rao V.B. The structure of the ATPase that powers DNA packaging into bacteriophage T4 procapsids. Mol. Cell. 2007;25:943–949. doi: 10.1016/j.molcel.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Lebedev A.A., Krause M.H., Antson A.A. Structural framework for DNA translocation via the viral portal protein. EMBO J. 2007;26:1984–1994. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R., Grubmüller H. The interaction between the ϕ29 connector and DNA governs DNA crunching and rotation, supporting the check-valve model. Biophys. J. 2016;110:455–469. doi: 10.1016/j.bpj.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimes S., Ma S., Jardine P.J. Role of ϕ29 connector channel loops in late-stage DNA packaging. J. Mol. Biol. 2011;410:50–59. doi: 10.1016/j.jmb.2011.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J., Moffitt J., Oster G. Mechanochemistry of a viral DNA packaging motor. J. Mol. Biol. 2010;400:186–203. doi: 10.1016/j.jmb.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Zhao Z., Khisamutdinov E., Guo P. Mechanism of one-way traffic of hexameric ϕ29 DNA packaging motor with four electropositive relaying layers facilitating antiparallel revolution. ACS Nano. 2013;7:4082–4092. doi: 10.1021/nn4002775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz C., De Donatis G.M., Guo P. Revolution rather than rotation of AAA+ hexameric ϕ29 nanomotor for viral dsDNA packaging without coiling. Virology. 2013;443:28–39. doi: 10.1016/j.virol.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harvey S.C. The scrunchworm hypothesis: transitions between A-DNA and B-DNA provide the driving force for genome packaging in double-stranded DNA bacteriophages. J. Struct. Biol. 2015;189:1–8. doi: 10.1016/j.jsb.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dixit A.B., Ray K., Black L.W. Compression of the DNA substrate by a viral packaging motor is supported by removal of intercalating dye during translocation. Proc. Natl. Acad. Sci. USA. 2012;109:20419–20424. doi: 10.1073/pnas.1214318109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson A.A., Leiman P.G., Rossmann M.G. Structure determination of the head-tail connector of bacteriophage ϕ29. Acta Crystallogr. D Biol. Crystallogr. 2001;57:1260–1269. doi: 10.1107/s0907444901010435. [DOI] [PubMed] [Google Scholar]


