Abstract
Metabolic syndrome (MS) is a combination of factors that increases the risk of cardiovascular atherosclerotic diseases including diabetes, obesity, dyslipidemia, and high blood pressure. Cardiovascular diseases are one of the leading causes of death in the adult Saudi population where the increase in cardiovascular-related mortality is augmented by the rise in the prevalence of MS. Metabolic syndrome is a multi-factorial disorder influenced by interactions between genetic and environmental components. This review aims to provide a comprehensive assessment of studied environmental and genetic factors explaining the prevalence of MS in the Kingdom of Saudi Arabia. Additionally, this review aims to illustrate factors related to the population genetics of Saudi Arabia, which might explain a proportion of the prevalence of MS.
Genetic epidemiology
The determinants of any living creature are a combination of their inner propensity to survive and an outer surrounding environment, which decides their chance of thriving. Humans, as any living creature have an inner factor, which determines most, if not all of their characteristics. These internal characteristics are usually determined by - but not limited to - the transmission of genetic materials from parents to offspring. Genetic epidemiology has been defined as the ‘study of joint action of genes and environment in causing disease in human populations and their patterns of inheritance in families’.1 The genetic impact on illnesses and phenotypes has a wide range of variations. On certain occasions, a change of a single nucleotide can result in a disease, such as sickle cell anemia. Alternatively, genetic determinants may not be enough to cause disease and environmental components are deemed essential. Metabolic syndrome (MS) is an example of diseases where the genetic determinants are not solely responsible for the development of the disease, and the augmenting environmental causes are important. Earlier genetic investigations were able to detect the genetic factors associated with the strong effect on monogenic conditions on a small number of recruited subjects. However, during the last decade, advancements in molecular measurement have enabled the detection of genetic variants with modest effects on polygenic and complex diseases. Genome-wide association studies (GWAS) are able to scan the whole genome and recruit larger numbers of individuals. Having a larger number of scanned single nucleotide polymorphisms (SNPs), and the ability to compare unrelated cases with controls has increased the power to detect variants with modest effect on phenotypes.
The MS
Metabolic syndrome is defined as a combination of factors that increase the risk of cardiovascular atherosclerotic diseases including diabetes, obesity, dyslipidemia, and high blood pressure.2,3 According to the International Diabetes Federation (IDF), a quarter of the adult population worldwide suffer from MS.2 A literature review was conducted to investigate the prevalence of MS in the Kingdom of Saudi Arabia (KSA).4 The review indicated a variance of MS between 13.6 and 57%.4 This variation in prevalence is explained by the variation of targeted populations, age groups, gender, and the criteria used to define MS. However, it is noticeable that the prevalence of MS is higher among females compared with males, and the increment was associated with aging.4 The largest cross-sectional survey using a sample of 17,293 participants reported an overall MS prevalence of 39.3% among Saudis aged between 30 and 70 years.5
Review scope
The increased prevalence of MS in a population can be partially explained by environmental factors relating to socioeconomic transition, imbalanced eating habits and higher levels of physical inactivity. Similarly, it is possible to argue that the genetic components of a population might have an effect on incurring a higher population risk of MS. This review aims to investigate factors influencing the prevalence of MS in KSA through several steps. Firstly, environmental factors related to eating behavior, levels of physical activity, and gender variations in KSA were explored. Secondly, genetic investigations conducted in KSA concerning MS, diabetes, hypertension, obesity, and dyslipidemia are studied. Thirdly, this review explains biochemical effects of detected genetic variants on the increasing risks of MS in the Saudi population. Finally, the review illustrates factors related to the population genetics of KSA, which might further explain the prevalence of MS in the population.
Environmental components of MS
Non-genetic factors, which might increase the risk of MS could be related to higher consumption of foods rich in fat, carbohydrates and salt, lower consumption of fruits and vegetables, and lower levels of physical activities. Several investigations conducted in KSA highlighted that eating and physical activity habits in the population provide an explanation for the increased prevalence of MS. The Saudi Health Interview Survey (SHIS) is a recent national investigation, which measured several health indicators in 13 administrative regions in KSA.6 The survey, which included 10,735 participants aged between 15 and 65 years, reported that only 6.7% of the participants had an average of more than 5 servings of fruit and vegetables on a daily basis. Similarly, a nationwide investigation targeting 10,500 military personnel revealed that most participants (87.6%) consumed fast food on a weekly basis compared with 44.8% who reported fruit consumption, and 32.6% reporting vegetable consumption on a weekly basis.7 Additionally, 2 studies8,9 concerning adolescents reported a similar trend of high intake of carbohydrates and fats. Although several investigations indicated the significant contribution of eating habits in increasing levels of MS in the Saudi population, the magnitude of salt consumption is a less studied risk factor for MS. A limited study of 87 subjects indicated that the daily salt intake of the studied sample exceeded the recommended daily intake of sodium.10 High levels of fast food consumption might indicate a higher level of salt consumption, as fast food products have been reported to have elevated levels of salt.11 Physical inactivity is another contributor to the increased prevalence of MS in Saudis.12-16
A cross-cultural study17 compared levels of physical activity in male and female adolescents aged between 15 and 17 years. Three groups were selected from Birmingham and Coventry in UK, and a third one from the Al-Ahsa region in KSA. This study showed that the proportion of adolescents with low levels of physical activity in KSA was higher than those reported in the UK.17 Additionally, the proportion of individuals engaged in physical activities in the UK was higher compared with Saudi adolescents.17 This study17 signifies the effect of cultural attitudes toward reduced physical activity observed in KSA. Saudi men are more likely to engage in physical activities than women.18 Amin et al19 compared patterns of leisure and physical activity between males and females, where 6 out of 12 types of sports and leisure activities were not even practised by females. Another study9 including adult subjects confirmed this association, indicating that females were more likely to exhibit a sedentary lifestyle compared with males (p<0.001).20 Additionally, a similar trend was observed in adolescent subjects.9 This could be a result of the cultural background in KSA, which limits the physical activity of females. Furthermore, lack of resources can be another barrier leading to both men and women engaging in less physical activities.13,21,22 It appears that the exposure of Saudis to elevated levels of physical inactivity, higher consumption of fast foods, and lower consumption of vegetables and fruits are important explanations for the increased prevalence of MS in the country. Additionally, the reported lower levels of physical activities make an important contribution to the increased prevalence of MS among Saudi females compared with Saudi males.
Genetic components of MS
Determinants can be driven by variants that contribute to a higher risk of developing traits related to MS, such as increasing the risk of elevated blood sugar level, elevated blood pressure, dyslipidemia, and obesity. Genome-wide investigations were able to find several genetic variants increasing the risk of MS that were mainly related to the genetic components affecting lipid metabolism, glucose sensing, insulin signalling, and appetite control.23 However, only one study24 was conducted in KSA to investigate genetic components of MS where measured variants in the vitamin D receptor (VDR) gene were studied. This study24 recruited 285 cases with MS, and these were compared with 285 healthy control subjects. It was hypothesized in this study that variants related to the VDR gene can be associated with the control of lipid metabolism, and subsequently influence the risk of MS. Nonetheless, this study was not able to find any statistically significant effects of the 4 measured variants near the VDR genes on increasing the risk of MS.24
Genetic components of diabetes
Genome-wide associated studies have been conducted in several populations in order to indicate the genetic components of diabetes. A literature review25 revealed that approximately 60 loci have been reported to be associated with type 2 diabetes in the European and Asian populations. The large number of genetic variants proves the multiple genes’ influence on type 2 diabetes. However, these loci only explain approximately 10% of type 2 diabetes heritability.26 In reviewing the efforts conducted in KSA to replicate the genetic associations detected in other populations, we were able to find only 5 studies.24,27-30 No studies targeted screening for novel association, and all of the investigations were replication studies. Fifty SNPs incurring a higher risk of type 2 diabetes have been investigated in the Saudi populations. However, only 12 SNPs were successfully replicated in the Saudi population as risk factors for type 2 diabetes (Table 1). Failure to replicate the remaining associations could be mainly explained by the small sample size of the studies. Among the genetic investigations concerning diabetes, the sample size varied between 185 and 2,401 subjects. Only one study by AlDaghri et al29 recruited a relatively large sample size (2,401 subjects), and was able to detect 8 associations out of 38 measured genetic variants. As shown in Table 1, the effect size of variants incurring a higher risk of type 2 diabetes in the Saudi population is witnessing a wide range of variation, which is likely to be affected by factors related to the sampling method and sample size. All of the detected associations indicate a higher risk of developing type 2 diabetes when inheriting the risk allele, except for one variant near the VDR gene, which seems to have a protective effect against type 2 diabetes. However, the protective effect is contradictory to other studies, indicating an increased risk of diabetes when inheriting the risk allele.31,32
Table 1.
Genetic variants incurring higher risk of type 2 diabetes in the Saudi population as found in different studies.
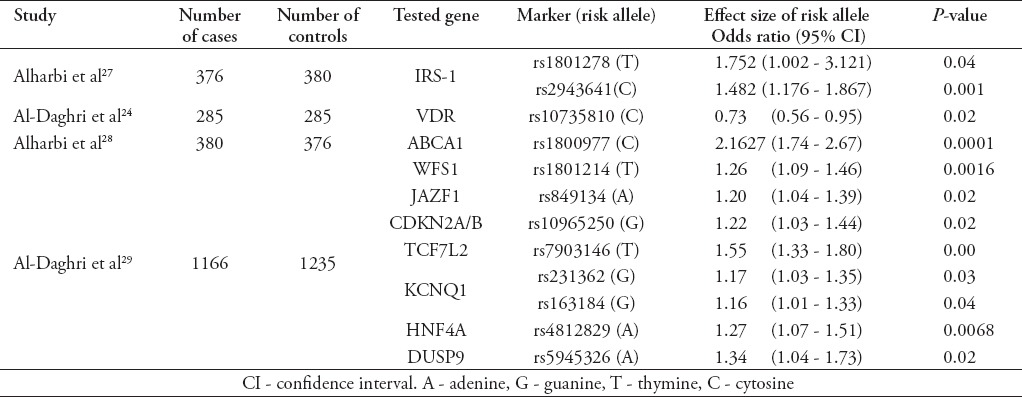
The Rs7903146 variant near transcription factor 7-like 2 (TCF7L2) showed the highest level of statistical significance among all of the detected variants in the Saudi population (1.13x10-8). This finding is consistent with the findings in other populations. The TCF7L2 was replicated by association studies in the European, Asian, and African ethnicities,33 and was found to have the strongest effect on the type 2 diabetes risk.34 The detected genetic associations in the Saudi population increased the risk of diabetes through several mechanisms. Juxtaposed with another zinc finger (JAZF1)35 have been reported to affect the development of pancreatic beta cells, which are responsible for insulin production. The TCF7L2,36 Wolfram syndrome 1 gene (WFS1),37 and hepatocyte nuclear factor 4 alpha (HNF4A)38 affect insulin processing and maturation in the pancreatic beta cells. Dual specificity phosphatase 9 (DUSP9) has been suggested to affect insulin signalling and stimulation.39 The ATP-binding cassette transporters A1 (ABCA1),40 cyclin-dependent kinase inhibitor 2B (CDKN2A/B),41 and potassium voltage-gated channel (KCNQ1)42,43 are associated with the secretion of insulin from the pancreatic cells. While all other detected variants affect maturation of beta cells and signalling and production of insulin, insulin receptor substrate 1 (IRS-1) is associated with modulating insulin action in target cells.44
Genetic components of hypertension
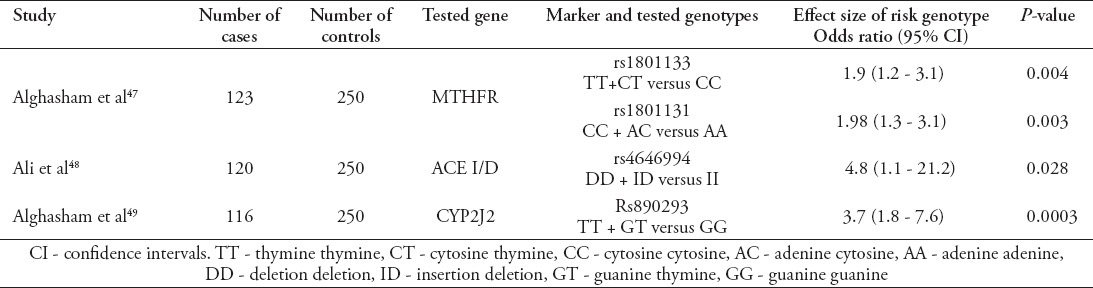
Similar to diabetes, hypertension is a multi-factorial condition requiring both environmental and genetic factors of the disease to develop. However, the number of genetic variants detected to increase the risk of hypertension is lower compared with diabetes. Only 32 SNPS are detected that are so far associated with hypertension.45 It has been speculated that the investigation of the genetic components of hypertension is less fruitful compared with other chronic conditions.46 Only 3 studies have been conducted in KSA to investigate the genetic components of hypertension.47-49 Six genetic variants incurring the higher risk of hypertension were studied in the Saudi population, although only 4 were found to be statistically associated with hypertension.
As indicated in Table 2, the sample sizes of the included studies are smaller than those used to investigate the genetic components of diabetes in the Saudi population. All detected studies did not use additive models by investigating the effect of a single risk allele. Instead, dominance models were used by comparing genotypes rather than comparing single alleles. Although it is possible to assume that the non-additive effect of the measured genetic variants can be found in Saudi subjects by observing the small sample sizes of the studies, it is possible to argue that fewer studies would have been able to detect significant associations if an additive model were to be used. Additionally, the same genetic variants were measured in other populations and were found to have an additive effect on incurring a higher risk of hypertension in a study targeting Indian population,50 and of marginal significance in a Chinese population.51 Statistically significant associations of genetic variants incurring a higher risk of hypertension in the Saudi populations were detected near genes influencing blood pressure level through several mechanisms. The methylenetetrahydrofolate reductase (MTHFR) gene has been reported to increase the level of blood homocysteine in individuals with homozygote mutations in the gene.52 The link between homocysteine level and blood pressure increase has been postulated to be due to increasing arteriolar constriction, increasing sodium renal reabsorption.53 Similarly, the angiotensin-converting enzyme (ACE) gene is reported to increase blood pressure though the vasoconstrictive effect of angiotensin II.54 The cytochrome P450 (CYP2J2) gene increases the risk of hypertension through the regulation of renal of sodium transportation.55
Table 2.
Genetic variants associated with increased risk of hypertension in the Saudi population according to findings in this study.
Genetic components of obesity
Investigating the genetic components of obesity has further dimensions of complexities compared with assessing the risk of diabetes or hypertension. Obesity is a risk factor for type 2 diabetes where shared genetic components between both conditions are possible. This notion was reflected on how genetic components were investigated in KSA. This review was able to detect 3 genetic studies that assessed the effect of 48 SNPs in terms of increasing the risk of obesity in KSA.56-58 Only 4 SNPs were reported to increase the risk of obesity with statistical significance (Table 3). In both studies, which were able to find statistically significant effects, several models were used to measure these effects. However, only the additive model is reported in Table 3. Additionally, it can be seen that both studies have an under-presentation of lean controls, which could indicate a difficulty in recruiting lean subjects in KSA.
Table 3.
Genetic variants associated with increased risk of obesity in the Saudi population according to findings in this study.
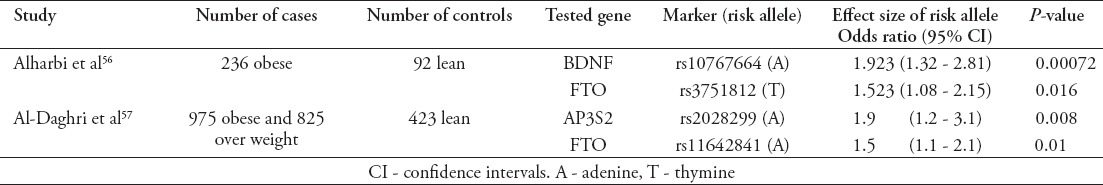
A study by Daghestani et al58 investigated the association between the Arg16Gly polymorphism of the β2-adrenergic receptor gene and the risk of obesity in Saudi subjects. This study58 indicated that the frequency of the risk allele was higher in obese and overweight subjects compared with lean controls. Additionally, this study conducted genotypes and allele effect estimations. However, the statistical method of calculating the odds ratio was not clearly indicated in the study. Additionally, conflicting findings were observed especially when comparing the odds of having obesity in controls compared with overweight subjects using the genotype effect (3.38, 95% confidence interval [CI]: 1.456 - 7.822) compared with using the allelic effect (0.60, 95% CI: 0.46 - 0.78) of the same measured variant. This finding suggests that the genotype effect could increase the risk of obesity while the allelic measurement indicates a protective effect. While the study by Alharbi et al56 investigated 11 variants which were reported to increase the risk of obesity in other populations, the study by Al-Dhagri et al57 investigated the association between 36 type 2 diabetes related SNPs with the risk of obesity in a Saudi sample. However, Al-Dhagri’s et al57 study included subjects with type 2 diabetes. Inclusion of cases with diabetes to investigate the association of genetic variants with obesity could be biased, as several factors are likely to influence body weight in patients with diabetes, such as lifestyle modification and diabetes medications. When this study stratified the sample into type 2 diabetes patients and healthy subjects, the detected effects of the 2 variants (rs2028299, rs11642841) were only observable in healthy subjects, and were not significant in type 2 diabetes patients. Additionally, these SNPs were not found to have an influence of type 2 diabetes risks in Saudi subjects.29
Clathrin-associated adaptor complex (AP3S2) has been speculated to regulate the transportation of intracellular vesicles in several tissues including the pancreatic cells.59 It has been reported to be associated with type 2 diabetes in the south Asian population.60 Although the variant rs2028299 near gene AP3S2 was not found to be associated with risk of diabetes in Saudis, it was found to have a significant additive effect on the obesity risk in Saudis. Given that there is limited evidence concerning the effect of AP3S2 on obesity traits in other populations, the detection of an effect on obesity but not diabetes in a Saudi population remains a mystery.
The brain-derived neurotrophic factor (BDNF) gene, and the fat mass and obesity associated (FTO) gene were reported to affect eating behaviors in the European population.61 Unlike variants near the BDNF gene, the FTO gene was speculated to influence the risk of both type 2 diabetes and obesity traits. As indicated in Table 3, 2 tested variants are located near the FTO gene. The conflicting evidence of the FTO gene on the risk of diabetes and obesity in the Saudi population has been observed in other European populations. Variants near the FTO gene are presumed to increase the risk of type 2 diabetes. The odds of having type 2 diabetes increases by 1.15 (95% CI: 1.09 - 1.22) when inheriting the risk allele of rs8050136 near the FTO gene.62 However, the effect of the FTO gene on type 2 diabetes has been argued to be mediated through its effect on body mass index (BMI).63
Genetic components of dyslipidemia
Genetic polymorphisms influencing lipid metabolism are related to genes influencing the levels of high-density lipoprotein cholesterol (HDL-C), high serum triglycerides (TG), and small low-density lipoprotein cholesterol (LDL-C). A systematic review64 revealed that approximately 13 genetic regions are detected so far as being involved in the regulation of lipid metabolism using the candidate gene approach, and more new novel associations were detected using the GWAS approach. Two studies highlighted the genetic components of lipid metabolism in a Saudi population where 5 polymorphisms were measured.24,58 The studies used different genetic statistical methods to measure the effect size. Additionally, the measured polymorphisms in these samples were reported to increase the risk of type 2 diabetes or obesity in other populations, but were not specifically related to lipid metabolism.
The study by Daghestani et al58 investigated the association between the Arg16Gly polymorphism of the β2-adrenergic receptor gene and HDL, LDL, and TG by simply looking at the presence of a statistically significant difference of the levels of HDL, LDL, and TG depending on the detected genotype, and no effect size were calculated. It was able to find that carriers of identical copies of the risk alleles had higher levels of LDL and TG but not HDL.58 The study by Al-Dhagri et al57 measured the association between VDR gene polymorphisms and HDL. It used odds ratios as a measurement of the effect size of a measured genetic variant with the risk of having a low level of HDL. None of the measured 4 risk alleles were found to be significantly associated with the risk of having low level of HDL.24
Saudi population genetics of MS
The Hardy-Weinberg law states that allelic frequencies are constant when genes are passed from one generation to another, as long as mating is carried out randomly in a large population with no selection, migration, or mutations.65 The Saudi population has been affected by historical factors that might have deviated the overall allelic frequencies from Hardy-Weinberg equilibrium. Kingdom of Saudi Arabia is a relatively new country and was established approximately 8 decades ago. It is dominated by deserts and upon its establishment, it suffered from major lack of resources, and was one of the poorest countries in the world. Having had this environmental condition, this might indicate that natural selection could have been an important force that favored individuals who were able to survive in this environment. Over the last 3-4 decades, KSA has witnessed an economic surge caused by the discovery of the world’s largest reserve of oil in the eastern parts of the country. This change was associated with an increased availability of food, and more exposure of Saudis to the western lifestyle. Subsequently, this change in lifestyle was associated with the increased prevalence of overweight and obese populations, which could be partially explained by the thrifty genotype hypothesis. It has been suggested that natural selection is one of the forces that may inflict the risk of type 2 diabetes on certain populations. Natural selection influences allelic frequencies based on the ability of individuals with certain genotypes to survive in certain environments. When there is a genotype, which has been selectively favored, its frequency increases. If a person is carrying an unfavored genotype, that person is more likely to have reduced viability and fertility, and consequently may be less able to pass their unfavored genotypes to the next generation.65 The thrifty genotype hypothesis has been postulated to explain the variation in the incidence of obesity and diabetes among ethnicities. This hypothesis claims that certain genotypes were advantageous during periods of starvation and low food availability in the past. Natural selection might have led to an increased frequency of favored genotypes, which later became risky genotypes in periods when food was abundant.66
Another well studied force affecting the genetic structure on Saudi Arabian populations is inbreeding. The percentage of consanguineous marriages ranged between 54 and 57%, and most of these marriages were between first-degree cousins.67-69 The prevalence of consanguineous marriages was also found to be higher in rural areas.69 The prevalence of consanguinity depends on several traditional and social factors. Families in KSA are adherent to cultural behaviors, and marriage between first-degree cousins is almost a norm. It has been argued that marriage between relatives is more likely to be easily mediated; hence, mating individuals are brought up together in the same environment. Furthermore, such arrangements may ease spousal adjustment after marriage, and enhance the stability of marriages. However, other economic reasons have been suggested, since marriage between relatives is a method of keeping fortunes and properties within the same family or tribe.67 Inbreeding may lead to the aggregation of type 2 diabetes risk alleles. Three epidemiological studies70-72 have suggested a synergism between type 2 diabetes and consanguineous marriages. The increased degree of consanguinity increases the inbreeding coefficient, and consequently increases the risk of inheriting a common allele shared by a common ancestor. On certain occasions, the shared allele could be a type 2 diabetes risk allele.
Conclusion and recommendations
Environmental components explain a large proportion of the increased prevalence of MS in KSA. Genetic components of MS and contributing factors in the Saudi population are similar to those discovered in other populations. However, most of the studied polymorphisms failed to be replicated in Saudi subjects due to the small study sizes compared with other studies. The current genetic evidence does not explain the prevalence of MS in Saudis due to the small detected effect sizes, and due to an inability to replicate most of the discovered association in other populations. The Saudi population genetics explain an additional contributing factor to the increased prevalence of obesity and type 2 diabetes, where the thrifty genotype hypothesis and high inbreeding levels are major explanatory genetic forces.
The findings of this review necessitate the need for larger epidemiological investigations capable of detection of variants with smaller effects. Similarly, as no GWAS have been conducted in KSA, it is possible to argue that performing a genome-wide investigation could lead to the detection of novel genetic variants in the Saudi population incurring a higher risk of MS. Additionally, statistical methods in genetic epidemiology investigations, such as issues with multiple testing, gene-environment interactions, and assessing genetic risk in cohorts with different levels of consanguinity might bring more insight into the genetic components of MS in the Saudi population.
Footnotes
Related Articles.
Mohamed S, Hamad MH, Abu-Amero KK. Identification of 2 novel homozygous mutations in the methylmalonyl-CoA mutase gene in Saudi patients. Saudi Med J 2015; 36: 1110-1114.
Liu C, Yang Y, Peng D, Chen L, Luo J. Hyperhomocysteinemia as a metabolic disorder parameter is independently associated with the severity of coronary heart disease. Saudi Med J 2015; 36: 839-846.
Li YQ, Zhao LQ, Liu XY, Wang HL, Wang XH, Li B, et al. Prevalence and distribution of metabolic syndrome in a southern Chinese population. Relation to exercise, smoking, and educational level. Saudi Med J 2013; 34: 929-936.
References
- 1.Thomas D. Statistical Methods in Genetic Epidemiology. New York (NY): Oxford University Press; 2004. [Google Scholar]
- 2.Federation ID. [cited 2015 April 23];IDF Worldwide Definition of the Metabolic Syndrome. Available from: http://www.idf.org/metabolic-syndrome . [Google Scholar]
- 3.Kassi E, Pervanidou P, Kaltsas G, Chrousos G. Metabolic syndrome: definitions and controversies. BMC Med. 2011;9:48. doi: 10.1186/1741-7015-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bahijri SM, Al Raddadi RM. The importance of local criteria in the diagnosis of metabolic syndrome in Saudi Arabia. Ther Adv Endocrinol Metab. 2013;4:51–59. doi: 10.1177/2042018813483165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Nozha M, Al-Khadra A, Arafah MR, Al-Maatouq MA, Khalil MZ, Khan NB, et al. Metabolic syndrome in Saudi Arabia. Saudi Med J. 2005;26:1918–1925. [PubMed] [Google Scholar]
- 6.Ministry of Health. [cited 2015 April 12];Survey of Health Information. 2013 Available from: http://www.moh.gov.sa/en/ministry/statistics/pages/healthinformatics.aspx . [Google Scholar]
- 7.Bin Horaib G, Al-Khashan HI, Mishriky AM, Selim MA, Alnowaiser N, Binsaeed AA, et al. Prevalence of obesity among military personnel in Saudi Arabia and associated risk factors. Saudi Med J. 2013;34:401–407. [PubMed] [Google Scholar]
- 8.Washi SA, Ageib MB. Poor diet quality and food habits are related to impaired nutritional status in 13- to 18-year-old adolescents in Jeddah. Nutr Res. 2010;30:527–534. doi: 10.1016/j.nutres.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Al-Hazzaa HM, Abahussain NA, Al-Sobayel HI, Qahwaji DM, Musaiger AO. Physical activity, sedentary behaviors and dietary habits among Saudi adolescents relative to age, gender and region. Int J Behav Nutr Phys Act. 2011;8:140. doi: 10.1186/1479-5868-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alkhunaizi AM, Al JH, Al SZ. Salt intake in Eastern Saudi Arabia. East Mediterr Health J. 2013;19:915–918. [PubMed] [Google Scholar]
- 11.Dunford E, Webster J, Woodward M, Czernichow S, Yuan WL, Jenner K, et al. The variability of reported salt levels in fast foods across six countries: opportunities for salt reduction. CMAJ. 2012;184:1023–1028. doi: 10.1503/cmaj.111895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al-Rukban MO. Obesity among Saudi male adolescents in Riyadh, Saudi Arabia. Saudi Med J. 2003;24:27–33. [PubMed] [Google Scholar]
- 13.AlQuaiz AM, Tayel SA. Barriers to a healthy lifestyle among patients attending primary care clinics at a university hospital in Riyadh. Ann Saudi Med. 2009;29:30–35. doi: 10.4103/0256-4947.51818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Hazzaa H, Sulaimani R. Maximal oxygen uptake and daily physical activity in 7-to-12 year-old boys. Pediatric Exercise Science. 1993;5:357–366. [Google Scholar]
- 15.Al-Rafaee SA, Al-Hazzaa HM. Physical activity profile of adult males in Riyadh City. Saudi Med J. 2001;22:784–789. [PubMed] [Google Scholar]
- 16.Al-Hazzaa HM. Physical Activity Profiles of College Male Subjects. Journal of King Saud University. 1990:2. [Google Scholar]
- 17.Al-Nakeeb Y, Lyons M, Collins P, Al-Nuaim A, Al-Hazzaa H, Duncan MJ, et al. Obesity, physical activity and sedentary behavior amongst British and Saudi youth: a cross-cultural study. Int J Environ Res Public Health. 2012;9:1490–1506. doi: 10.3390/ijerph9041490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taha AZ. Self-reported knowledge and pattern of physical activity among school students in Al Khobar, Saudi Arabia. East Mediterr Health J. 2008;14:344–355. [PubMed] [Google Scholar]
- 19.Amin TT, Al Khoudair AS, Al Harbi MA, Al Ali AR. Leisure time physical activity in Saudi Arabia: prevalence, pattern and determining factors. Asian Pac J Cancer Prev. 2012;13:351–360. doi: 10.7314/apjcp.2012.13.1.351. [DOI] [PubMed] [Google Scholar]
- 20.Al-Nozha MM, Al-Hazzaa HM, Arafah MR, Al-Khadra A, Al-Mazrou YY, Al-Maatouq MA, et al. Prevalence of physical activity and inactivity among Saudis aged 30-70 years. A population-based cross-sectional study. Saudi Med J. 2007;28:559–568. [PubMed] [Google Scholar]
- 21.Sidawi B, Al-Hariri MT. The impact of built environment on diabetic patients: the case of Eastern Province, Kingdom of Saudi Arabia. Glob J Health Sci. 2012;4:126–138. doi: 10.5539/gjhs.v4n4p126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almajwal AM. Correlations of Physical Activity, Body Mass Index, Shift Duty, and Selected Eating Habits among Nurses in Riyadh, Saudi Arabia. Ecol Food Nutr. 2015;54:397–417. doi: 10.1080/03670244.2015.1004400. [DOI] [PubMed] [Google Scholar]
- 23.Chang Y, Cheng Y, Yu Hsiang, Chuang LM. Molecular Genetics of Metabolic Syndrome. eLS John Wiley & Sons Online Library; 2013. [Google Scholar]
- 24.Al-Daghri NM, Al-Attas OS, Alkharfy KM, Khan N, Mohammed AK, Vinodson B, et al. Association of VDR-gene variants with factors related to the metabolic syndrome, type 2 diabetes and vitamin D deficiency. Gene. 2014;542:129–133. doi: 10.1016/j.gene.2014.03.044. [DOI] [PubMed] [Google Scholar]
- 25.Ntzani EE, Kavvoura FK. Genetic risk factors for type 2 diabetes: insights from the emerging genomic evidence. Curr Vasc Pharmacol. 2012;10:147–155. doi: 10.2174/157016112799305030. [DOI] [PubMed] [Google Scholar]
- 26.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Ann N Y Acad Sci. 2010;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alharbi KK, Khan IA, Munshi A, Alharbi FK, Al-Sheikh Y, Alnbaheen MS. Association of the genetic variants of insulin receptor substrate 1 (IRS-1) with type 2 diabetes mellitus in a Saudi population. Endocrine. 2014;47:472–477. doi: 10.1007/s12020-014-0177-2. [DOI] [PubMed] [Google Scholar]
- 28.Alharbi KK, Khan IA, Al-Daghri NM, Munshi A, Sharma V, Mohammed AK, et al. ABCA1 C69T gene polymorphism and risk of type 2 diabetes mellitus in a Saudi population. Journal of Biosciences. 2013;38:893–897. doi: 10.1007/s12038-013-9384-x. [DOI] [PubMed] [Google Scholar]
- 29.Al-Daghri NM, Alkharfy KM, Alokail MS, Alenad AM, Al-Attas OS, Mohammed AK, et al. Assessing the contribution of 38 genetic loci to the risk of type 2 diabetes in the Saudi Arabian Population. Clin Endocrinol (Oxf) 2014;80:532–537. doi: 10.1111/cen.12187. [DOI] [PubMed] [Google Scholar]
- 30.Bazzi MD, Nasr FA, Alanazi MS, Alamri A, Turjoman AA, Moustafa AS, et al. Association between FTO, MC4R, SLC30A8, and KCNQ1 gene variants and type 2 diabetes in Saudi population. Genet Mol Res. 2014;13:10194–10203. doi: 10.4238/2014.December.4.14. [DOI] [PubMed] [Google Scholar]
- 31.Filus A, Trzmiel A, Kuliczkowska-Plaksej J, Tworowska U, Jedrzejuk D, Milewicz A, et al. Relationship between vitamin D receptor BsmI and FokI polymorphisms and anthropometric and biochemical parameters describing metabolic syndrome. Aging Male. 2008;11:134–139. doi: 10.1080/13685530802273426. [DOI] [PubMed] [Google Scholar]
- 32.Bid HK, Konwar R, Aggarwal CG, Gautam S, Saxena M, Nayak VL, et al. Vitamin D receptor (FokI BsmI and TaqI) gene polymorphisms and type 2 diabetes mellitus: a North Indian study. Indian J Med Sci. 2009;63:187–194. [PubMed] [Google Scholar]
- 33.Cauchi S, El Achhab Y, Choquet H, Dina C, Krempler F, Weitgasser R, et al. TCF7L2 is reproducibly associated with type 2 diabetes in various ethnic groups: a global meta-analysis. J Mol Med (Ber) 2007;85:777–782. doi: 10.1007/s00109-007-0203-4. [DOI] [PubMed] [Google Scholar]
- 34.Vimaleswaran KS, Loos RJ. Progress in the genetics of common obesity and type 2 diabetes. Expert Rev Mol Med. 2010;12:e7. doi: 10.1017/S1462399410001389. [DOI] [PubMed] [Google Scholar]
- 35.Grarup N, Andersen G, Krarup NT, Albrechtsen A, Schmitz O, Jørgensen T, et al. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes. 2008;57:2534–2540. doi: 10.2337/db08-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingelsson E, Langenberg C, Hivert M-F, Prokopenko I, Lyssenko V, Dupuis J, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59:1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatanaka M, Tanabe K, Yanai A, Ohta Y, Kondo M, Akiyama M, et al. Wolfram syndrome 1 gene (WFS1) product localizes to secretory granules and determines granule acidification in pancreatic beta-cells. Hum Mol Genet. 2011;20:1274–1284. doi: 10.1093/hmg/ddq568. [DOI] [PubMed] [Google Scholar]
- 38.Wang H, Antinozzi PA, Hagenfeldt KA, Maechler P, Wollheim CB. Molecular targets of a human HNF1 alpha mutation responsible for pancreatic beta-cell dysfunction. EMBO J. 2000;19:4257–4264. doi: 10.1093/emboj/19.16.4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu H, Dembski M, Yang Q, Yang D, Moriarty A, Tayber O, et al. Dual specificity mitogen-activated protein (MAP) kinase phosphatase-4 plays a potential role in insulin resistance. J Biol Chem. 2003;278:30187–30192. doi: 10.1074/jbc.M302010200. [DOI] [PubMed] [Google Scholar]
- 40.Brunham LR, Kruit JK, Pape TD, Timmins JM, Reuwer AQ, Vasanji Z, et al. Beta-cell ABCA1 influences insulin secretion, glucose homeostasis and response to thiazolidinedione treatment. Nat Med. 2007;13:340–347. doi: 10.1038/nm1546. [DOI] [PubMed] [Google Scholar]
- 41.Grarup N, Rose CS, Andersson EA, Andersen G, Nielsen AL, Albrechtsen A, et al. Studies of association of variants near the HHEX CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes. 2007;56:3105–3111. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 42.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 43.Yamagata K, Senokuchi T, Lu M, Takemoto M, Fazlul Karim M, Go C, et al. Voltage-gated K(+) channel KCNQ1 regulates insulin secretion in MIN6 beta-cell line. Biochem Biophys Res Commun. 2011;407:620–625. doi: 10.1016/j.bbrc.2011.03.083. [DOI] [PubMed] [Google Scholar]
- 44.Nelson DL, Cox MM. Biosignaling. In: Lehninger Principles of Biochemistry. 2nd ed. New York (NY): WH Freeman & Company; 2008. [Google Scholar]
- 45.Ehret GB. Genome-Wide Association Studies: Contribution of Genomics to Understanding Blood Pressure and Essential Hypertension. Curr Hypertens Rep. 2010;12:17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ji LD, Zhang LN, Xu J. Genome-wide association studies of hypertension: achievements, difficulties and strategies. World J Hypertens. 2011;1:10–14. [Google Scholar]
- 47.Alghasham A, Settin AA, Ali A, Dowaidar M, Ismail H. Association of MTHFR C677T and A1298C gene polymorphisms with hypertension. Int J Health Sci (Qassim) 2012;6:3–11. doi: 10.12816/0005968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali A, Alghasham A, Ismail H, Dowaidar M, Settin A. ACE I/D and eNOS E298D gene polymorphisms in Saudi subjects with hypertension. J Renin Angiotensin Aldosterone Syst. 2013;14:348–353. doi: 10.1177/1470320312459976. [DOI] [PubMed] [Google Scholar]
- 49.Alghasham A, Ali A, Ismail H, Dowaidar M, Settin AA. CYP2J2-50 G/T and ADRB2 G46A gene polymorphisms in Saudi subjects with hypertension. Genet Test Mol Biomarkers. 2012;16:1027–1031. doi: 10.1089/gtmb.2012.0006. [DOI] [PubMed] [Google Scholar]
- 50.Srivastava K, Sundriyal R, Meena PC, Bhatia J, Narang R, Saluja D. Association of angiotensin converting enzyme (insertion/deletion) gene polymorphism with essential hypertension in northern Indian subjects. Genet Test Mol Biomarkers. 2012;16:174–177. doi: 10.1089/gtmb.2011.0155. [DOI] [PubMed] [Google Scholar]
- 51.Xi B, Shen Y, Zhao X, Chandak GR, Cheng H, Hou D, et al. Association of common variants in/near six genes (ATP2B1, CSK, MTHFR, CYP17A1, STK39 and FGF5) with blood pressure/hypertension risk in Chinese children. J Hum Hypertens. 2014;28:32–36. doi: 10.1038/jhh.2013.50. [DOI] [PubMed] [Google Scholar]
- 52.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 53.Stehouwer CD, van Guldener C. Does homocysteine cause hypertension? Clin Chem Lab Med. 2003;41:1408–1411. doi: 10.1515/CCLM.2003.216. [DOI] [PubMed] [Google Scholar]
- 54.Baudin B. New aspects on angiotensin-converting enzyme: from gene to disease. Clin Chem Lab Med. 2002;40:256–265. doi: 10.1515/CCLM.2002.042. [DOI] [PubMed] [Google Scholar]
- 55.Roman RJ. P-450 metabolites of arachidonic acid in the control of cardiovascular function. Physiol Rev. 2002;82:131–185. doi: 10.1152/physrev.00021.2001. [DOI] [PubMed] [Google Scholar]
- 56.Alharbi KK, Richardson TG, Khan IA, Syed R, Mohammed AK, Boustred CR, et al. Influence of adiposity-related genetic markers in a population of Saudi Arabians where other variables influencing obesity may be reduced. Dis Markers. 2014;2014:758232. doi: 10.1155/2014/758232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al-Daghri NM, Alkharfy KM, Al-Attas OS, Krishnaswamy S, Mohammed AK, Albagha OM, et al. Association between type 2 diabetes mellitus-related SNP variants and obesity traits in a Saudi population. Mol Biol Rep. 2014;41:1731–1740. doi: 10.1007/s11033-014-3022-z. [DOI] [PubMed] [Google Scholar]
- 58.Daghestani MH, Warsy A, Al-Odaib AN, Eldali A, Al-Eisa NA, Omer SA, et al. Arginine 16 Glycine Polymorphism in β2-Adrenergic Receptor Gene is Associated with Obesity, Hyperlipidemia, Hyperleptinemia and Insulin Resistance in Saudis. Int J Endocrinol. 2012;2012:945608. doi: 10.1155/2012/945608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dell’Angelica EC, Ohno H, Ooi CE, Rabinovich E, Roche KW, Bonifacino JS. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO J. 1997;16:917–928. doi: 10.1093/emboj/16.5.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCaffery JM, Papandonatos GD, Peter I, Huggins GS, Raynor HA, Delahanty LM, et al. Obesity susceptibility loci and dietary intake in the Look AHEAD Trial. Am J Clin Nutr. 2012;95:1477–1486. doi: 10.3945/ajcn.111.026955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boes E, Coassin S, Kollerits B, Heid IM, Kronenberg F. Genetic-epidemiological evidence on genes associated with HDL cholesterol levels: a systematic in-depth review. Exp Gerontol. 2009;44:136–160. doi: 10.1016/j.exger.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Falconer DS, Mackay TFC. Introduction to quantitative genetics. San Francisco (SF): Benjamin Cummings; 1996. [Google Scholar]
- 66.Neel JV. Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? American Journal of Human Genetics. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- 67.el-Hazmi MA, al-Swailem AR, Warsy AS, al-Swailem AM, Sulaimani R, al-Meshari AA. Consanguinity among the Saudi Arabian population. J Med Genet. 1995;32:623–626. doi: 10.1136/jmg.32.8.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.El Mouzan MI, Al Salloum AA, Al Herbish AS, Qurachi MM, Al Omar AA. Consanguinity and major genetic disorders in Saudi children: a community-based cross-sectional study. Ann Saudi Med. 2008;28:169–173. doi: 10.5144/0256-4947.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.El-Mouzan MI, Al-Salloum AA, Al-Herbish AS, Qurachi MM, Al-Omar AA. Regional variations in the prevalence of consanguinity in Saudi Arabia. Saudi Med J. 2007;28:1881–1884. [PubMed] [Google Scholar]
- 70.Bener A, Zirie M, Al-Rikabi A. Genetics, obesity, and environmental risk factors associated with type 2 diabetes. Croat Med J. 2005;46:302–307. [PubMed] [Google Scholar]
- 71.Anokute CC. Suspected synergism between consanguinity and familial aggregation in type 2 diabetes mellitus in Saudi Arabia. J R Soc Health. 1992;112:167–169. doi: 10.1177/146642409211200403. [DOI] [PubMed] [Google Scholar]
- 72.Gosadi IM, Goyder EC, Teare MD. Investigating the potential effect of consanguinity on type 2 diabetes susceptibility in Saudi population. Hum Hered. 2014;77:197–206. doi: 10.1159/000362447. [DOI] [PubMed] [Google Scholar]


