Abstract
Background:
As a DNA repair protein, flap endonuclease 1 is a key enzyme in maintaining genomic instability and preventing carcinogenesis. Two single nucleotide polymorphisms (SNPs), -69G>A and 4150G>T are associated with DNA damage. This meta-analysis is to evaluate the genetic effects of FEN1 gene SNPs (-69G/A and 4150G/T) and the susceptibility to diseases, including glioma risk, breast cancer, lung cancer, keratoconus (KC) and fuchs’ endothelial corneal dystrophy (FECD).
Methods:
A literature search of PubMed and Embase was conducted to identify all eligible published studies. Five case-control studies were included with a total of 5612 cases and 6703 controls in this meta-analysis. Crude odds ratios (ORs) with their corresponding confidence intervals (95%CI) were used to assess the strength of the association.
Results:
The FEN1 -69G/A and 4150G/T polymorphisms were significantly associated with the disease risk. Our meta-analysis showed the FEN1 -69GG genotype was correlated to increase risk for the contained diseases compared with the -69AG genotype (OR=0.77, 95%CI=0.71∼0.83). Moreover, the FEN1 4150GG genotype could increase diseases risk compared with the 4150TG genotype (OR=0.81, 95%CI=0.75∼0.87).
Conclusion:
The variant genotypes of the FEN1 -69G/A and FEN1 4150G/T polymorphisms may be associated with diseases susceptibility. However, more studies are needed to detect the disease risk in different ethnic populations.
Keywords: Flap endonuclease 1, Genetic variants, SNPs, Susceptibility, Disease
Introduction
Genome stability is critical to cell survival and normal cell growth such as genome instability caused numerous human cancers (1). It ensures that replication, repair and recombination occur with high fidelity (2). There are still many unknown details of DNA metabolic, because of the multiple functions of proteins that cause diseases. Flap endonuclease1 (FEN1) is one of the DNA metabolism enzyme that possesses multiple functions. FEN1 is a member of the RAD2 structure-specific nuclease family (3). It is best known for its essential roles in the penultimate steps of Okazaki fragment maturation and 5′-flap removal during long-patch base excision repair (BER) (4, 5). Besides, FEN1 has also been implicated in other major DNA metabolic pathways, including rescue of stalled DNA replication forks, maintenance of telomere stability and apoptotic fragmentation of DNA (6, 7). In addition, FEN1 is also a 5′ exonuclease (EXO) in eliminating heterologous sequences at DNA damage site and facilitating DNA repair by homologous recombination (8). The third activity of FEN1 has been identified is gap-dependent endonuclease (GEN) activity, which is critical in resolving stalled replication fork (6, 7). FEN1 haploinsufficiency appears to lead to genome instability and carcinogenesis in mice, indicating that the FEN1 gene product is important for development and its expression might be necessary for cell survival or proliferation (9). The expression of its mRNA is up regulated in small cell lung cancer and the protein is high expressed in sorts of lung cells comparing with normal lung cells (10). FEN1 is highly expressed in gastric tumor cell lines compared with normal cell lines (11).
A group of mutations in human FEN1 is associated with several cancers and cause nuclease activity deficiency (12). The disease susceptibility may be increased due to the aberrant expression of FEN1. Therefore, we thought the gene polymorphisms also could affect the risk of diseases. Both -69G/A (rs174538, in the gene promoter region) and 4150G/T(rs4246215, in gene 3′-UTR) SNPs could influence gene expression in vivo (13, 14). Then we identified two SNPs, -69G/A and 4150G/T to analyze the association between these variant and disease risk. Our study included nine diseases in Asians and Caucasians: glioma risk, breast cancer, lung cancer, hepatocellular carcinoma (HCC), esophageal cancer (EC), gastric cancer (GC), colorectal cancer (CRC), keratoconus (KC) and fuchs’ endothelial corneal dystrophy (FECD). Cancer is one of serious disease that threatening human health.
Hence, we performed this meta-analysis from these studies and hope it might be potentially important for early identification of individuals with diseases.
Materials and Methods
Search strategy
An extensive literature search for relevant studies was conducted on PubMed and Embase by using the following terms: {“FEN1” or “flap endonuclease 1”} and “disease risk” and {“SNPs” or “polymorphisms” or “variation”}.
Selection criteria
1) The case-control studies focus on the relationship between FEN1 gene polymorphisms (including -69G/A and 4150G/T) and disease; 2) studies published from 2009 to 2014; (3) the method of genotype detection was PCR-RFLP; 4) the methods of statistics were appropriate and the data were reliable.
Exclusion criteria
1) Articles excluded based on screening of titles and abstracts, 2) The data in these studies were not exact; 3) studies that had no control group.
Statistical analysis
The ORs with their corresponding 95%CIs were calculated under five genetic models: the allele model, the homozygous model, the heterozygous model, the dominant model, and the recessive model were used to estimating the associations between FEN1 genotypes and risk of the included diseases. Genotype distributions of FEN1 polymorphisms between patients and controls were examined by chi-square test. The significance of the combined OR was determined by Z test and P<0.05 was considered as significant. We estimated the degree of heterogeneity among studies using the Q-statistic of Review-Manager software, which is considered significant at PH<0.05. In addition, the I2 test was used to quantify the heterogeneity (15, 16). When PH<0.05, we used random effects model, else we used fixed-effects model. After that, we constructed the funnel plot to investigate whether publication bias might affect the validity of the estimates. The symmetry of the funnel plot was further evaluated by Egger’s linear regression test. All the analyses were calculated using the Review-Manager 5.2 (Cochrane Library Software, Oxford, UK) and Stata 12.0 (Stata Corp, TX, USA) software.
Results
Included studies
Fifteen case-control studies were searched. Five studies in Asians and Caucasians with 5612 cases and 6703 controls were involved (13, 14, 17–19). Publication year ranged from 2009 to 2014. One study had four different types of diseases: HCC, EC, GC, and CRC. Another study with Caucasian population had two types of disease: KC and FECD, but it only had one control group, so we combined these two diseases for analyzing (Table 1).
Table 1:
Characteristics and methodological quality of studies included in meta-analysis
| Reference Number | Country/Ethnicity | Disease Type | Genotyping | -69G>A(rs174538) | 4150G>T(rs4246215) | ||
|---|---|---|---|---|---|---|---|
| case AA/GA/GG | control AA/GA/GG | case TT/GT/GG | control TT/GT/GG | ||||
| 13 | China/ Asian | glioma risk | PCR-RFLP | 35/122/160 | 130/356/316 | 34/120/160 | 130/363/309 |
| 14 | China/ Asian | HCC | PCR-RFLP | 68/290/290 | 134/334/270 | 73/295/280 | 129/335/274 |
| 17 | China/ Asian | EC | PCR-RFLP | 62/249/244 | 128/331/263 | 58/255/225 | 125/336/262 |
| GC | 41/182/189 | 76/209/169 | 42/177/183 | 76/212/166 | |||
| CRC | 28/104/104 | 54/144/109 | 27/102/100 | 52/145/109 | |||
| Breast cancer | 116/437/547 | 246/639/515 | 134/449/517 | 246/651/503 | |||
| 18 | China/ Asian | Lung cancer | PCR-RFLP TaqMan | 253/796/791 | 354/880/724 | 271/815/754 | 358/883/717 |
| 19 | Poland/Caucasian | KC, FECD | PCR-RFLP | 29/262/213 | 17/178/127 | 53/239/212 | 16/157/149 |
Association between FEN1 gene polymorphisms (-69G/A and 4150G/T) and susceptibility to disease
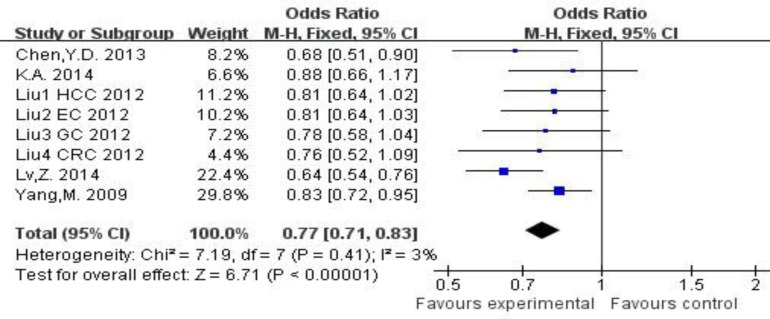
The studies on Table 2 were combined to show the result of FEN1 -69G/A and 4150G/T polymorphisms with disease risk, in which diseases were including breast cancer, KC, FECD, glioma risk, HCC, EC, GC, CRC and lung cancer. For the -69G/A polymorphisms, A vs. G: OR=0.74, 95%CI=0.70∼0.78, PH=0.05; AA vs. GG: OR=0.60, 95%CI=0.43∼0.83, PH<0.001; AG vs. GG: OR=0.77, 95%CI=0.71∼0.83, PH=0.41; (AA+AG) vs. GG: OR=0.71, 95%CI=0.66∼0.76, PH=0.17; AA vs. (AG+GG): OR=0.63, 95%CI=0.57∼0.70, PH=0.29. The result from the forest plot showed that FEN1 -69GG was significant associated with diseases (P<0.0001). (PH: P value for the heterogeneity test, Fig. 1)
Fig. 1:
Forest plot for FEN1 -69G/A polymorphisms and disease risk in the heterozygous model
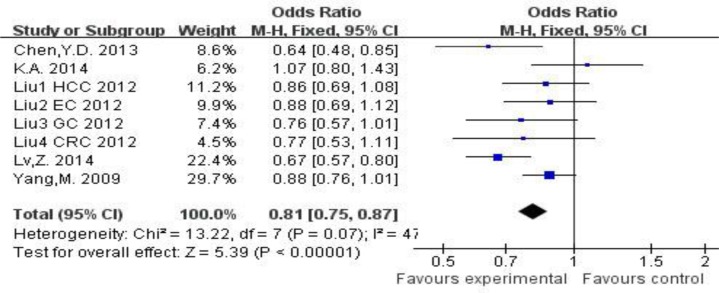
For the association between the 4150G/T polymorphisms and disease risk, T vs. G: OR=0.79, 95%CI=0.71∼0.89, PH<0.001; TT vs. GG: OR=0.63, 95%CI=0.50∼0.80, PH<0.001; TG vs. GG: OR=0.81, 95%CI=0.75∼0.87, PH=0.07; (TT+TG) vs. GG: OR=0.76, 95%CI=0.67∼0.87, PH=0.005; TT vs. (TG+GG): OR=0.71, 95%CI=0.58∼0.86, PH=0.004. The results revealed that the GG of 4150G/T polymorphisms may be associated with an increasing risk of the glioma risk, breast cancer, lung cancer, KC and FECD (P<0.00001, Fig. 2).
Fig. 2:
Forest plot for FEN1 4150G/T polymorphisms and disease risk in the heterozygous model
Evaluation of heterogeneity and publication bias
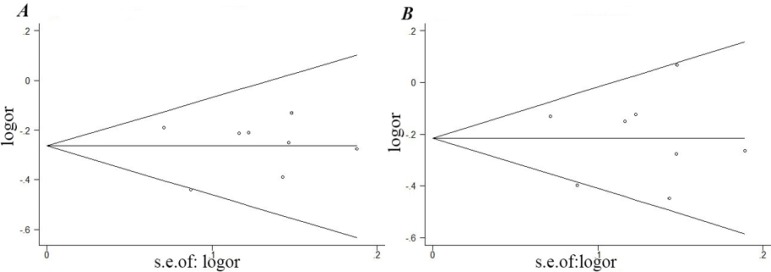
Funnel plot and Egger’s test were performed to access the publication bias of literature. The results did not show any evidence of publication bias for FEN1 -69G/A (t=0.03, P=0.975; Fig. 3A) or FEN1 4150G/T (t=−0.08, P=0.937; Fig. 3B).
Fig. 3:
Begg’s funnel plots with pseudo 95% confidence limits of FEN1 -68G/A and 4150G/T polymorphisms in the heterozygous model. (A: -69G/A polymorphisms; B: 4150G/T polymorphisms)
Discussion
Our study is the first meta-analysis that reveals the effects of FEN1 -69G/A and 4150G/T polymorphisms to the diseases susceptibility which including the glioma risk, breast cancer, lung cancer, KC and FECD. FEN1 could increase the susceptibility of cancers (20–23). By analyzing we got that for the -69G/A, the G allele could increase the risk compared with A allele in Asians and Caucasians, and for the 4150G/T, the G allele could increase the risk. About 90% of human DNA sequence variants are single nucleotide polymorphisms (SNPs) (24). Testing the SNPs is helpful to know complex diseases such as we can get the relationship between the SNPs and relative diseases by using the case-control method (25). As a DNA structure-specific endonuclease, FEN1 plays pivotal roles in maturation of Okazaki fragments, long-patch base excision repair, restarting of stalled replication forks and telomere maintenance, and FEN1 is required for normal cell growth and proliferation (26, 27). Absence of FEN1 expression causes genome instability, for the homozygous, it causes embryonic lethality in mice, and for the heterozygous, it results in decreased survival rates (28). Involved studies confirmed that the -69G>A and 4150G>T have been associated with an increased frequency of serious diseases (13∼, 14, 17∼19). The -69G>A polymorphism happens in the promoter region, and the 4150G>T occurs in the 3′ untranslated region. Both of these polymorphisms can decrease the mRNA expression of FEN1 (16). These SNPs may contribute to disease risk, and could be used as markers for detection of those involved diseases. A lack of FEN1 in haploin sufficient cells could cause the rapid tumor progression (9).
Some limitations of this meta-analysis should be acknowledged. First, the number of the involved studies and populations were limited, and because this is a meta-analysis, so we did not know actual information at the patient levels, it has limited our statistical power to the relationship between gene and environment when analyzing. Secondly, though there were nine different diseases were involved, it is also not enough. The language we selected only in English, so we lacked kinds of other studies.
Although some parts were limited, our results did not show any publication bias by testing using Begg’s funnel plot and Egger’s testing (19), so this meta-analysis still had some strength.
Conclusion
Our results do support the hypothesis that the SNPs in FEN1 -69G/A and 4150G/T may serve as biological markers of some diseases. Larger and more kinds of populations are needed to confirm our finding.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgements
This work was supported by grants from National Natural Science Foundation of China (31401667), Natural Science Foundation of Zhejiang Province (LY16C050003), and Scientific Research Project of Zhejiang Education Department (Y201327362). The authors declare that there is no conflict of interests.
References
- 1. Shen BH, Singh P, Liu R, Qiu JZ, Zheng L, Finger DL, Alas S. (2005). Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays, 27( 7): 717–729. [DOI] [PubMed] [Google Scholar]
- 2. Kolodner RD, Putnam CD, Kyungjae M. (2002). Maintenance of genome stability in Saccharomyces cerevisiae. Science, 297( 5581): 552–557. [DOI] [PubMed] [Google Scholar]
- 3. Lieber MR. (1997). The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays, 19( 3): 233–240. [DOI] [PubMed] [Google Scholar]
- 4. Tomlinson CG, Atack JM, Brian C, Tainer JA, Grasby JA. (2010). Substrate recognition and catalysis by flap endonucleases and related enzymes. BiochemSoc T, 38( 2): 433–437. [DOI] [PubMed] [Google Scholar]
- 5. Bambara RA, Murante RS, Henricksen LA. (1997). Enzymes and reactions at the eukaryotic DNA replication fork. J BiolChem, 272( 8): 4647–4650. [DOI] [PubMed] [Google Scholar]
- 6. Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen BH. (2005). Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep, 6( 1): 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parrish JZ, Yang C, Shen BH, Xue D. (2003). CRN-1, a Caenorhabditiselegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO J, 22( 13): 3451–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kikuchi K, Taniguchi Y, Hatanaka A, Sonoda E, Hochegger H, Adachi N. (2005). Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol Cell Biol, 25( 16): 6948–6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kucherlapati M, Yang K, Kuraguchi M, et al. (2002). Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. P NatlAcadSci USA, 99( 15): 9924–9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sato M, Girard L, Sekine I, Sunaga N, Ramirez RD, Kamibayashi C, Minna JD. (2003). Increased expression and no mutation of the Flap endonuclease (FEN1) gene in human lung cancer. Oncogene, 22( 46): 7243–7246. [DOI] [PubMed] [Google Scholar]
- 11. Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO, Kim JH, Song KS, Rho SM, Yoo HS, Kim YS, Kim JG, Kim NS. (2005). Identification of gastric cancer–related genes using a cDNA microarray containing novel expressed sequence tags expressed in gastric cancer cells. Clin Cancer Res, 11( 2): 473–482. [PubMed] [Google Scholar]
- 12. Zheng L, Dai HF, Zhou M, Li M, Singh P, Qiu JZ, Tsark W, Huang Q, Kernstine K, Zhang XM, Lin DX, Shen BH. (2007). Fen1 mutations result in autoimmunity, chronic inflammation and cancers. Nat Med, 13( 7): 812–819. [DOI] [PubMed] [Google Scholar]
- 13. Chen YD, Zhang XJ, Qiu XG, Li JP, Yuan QP, Jiang T, Yang M. (2013). Functional FEN1 genetic variants and haplotypes are associated with glioma risk. J Neurooncol, 111( 2): 145–151. [DOI] [PubMed] [Google Scholar]
- 14. Liu L, Zhou C, Zhou L, Peng L, Li D, Zhang X, Zhou M, Kuang P, Yuan Q, Song X, Yang M. (2012). Functional FEN1 genetic variants contribute to risk of hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer. Carcinogenesis, 33( 1): 119–123. [DOI] [PubMed] [Google Scholar]
- 15. Jackson D, White IR, Riley RD. (2012). Quantifying the impact of between-study heterogeneity in multivariate meta-analyses. Stat Med, 31( 29): 3805–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peters JL, Sutton AJ, Jones DR, Abrams KR, Lesley R. (2006). Comparison of two methods to detect publication bias in meta-analysis. Jama-J Am Med Assoc, 295( 6): 676–680. [DOI] [PubMed] [Google Scholar]
- 17. Lv Z, Liu WL, Li DM, Liu LS, Wei JY, Zhang JF, Ge YX, Wang ZQ, Chen HW, Zhou CC, Yuan QP, Zhou LQ, Yang M. (2014). Association of functional FEN1 genetic variants and haplotypes and breast cancer risk. Gene, 538( 1): 42–45. [DOI] [PubMed] [Google Scholar]
- 18. Yang M, Guo H, Wu C, He Y, Yu D, Zhou L, Wang F, Xu J, Tan W, Wang G, Shen BH, Yuan J, Wu T, Lin D. (2009). Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Hum Mutat, 30( 9): 1320–1328. [DOI] [PubMed] [Google Scholar]
- 19. Wojcik KA, Synowiec E, Polakowski P, Glowacki S, Izdebska J, Lloyd S, Galea D, Blasiak J, Szaflik J, Szaflik JP. (2014). Polymorphism of the Flap Endonuclease 1 Gene in Keratoconus and Fuchs Endothelial Corneal Dystrophy. Int J MolSci, 15( 8): 14786–14802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. (1997). A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88( 2): 253–263. [DOI] [PubMed] [Google Scholar]
- 21. Xu H, Zheng L, Dai H, Zhou M, Hua Y, Shen BH. (2011). Chemical-induced cancer incidence and underlying mechanisms in Fen1 mutant mice. Oncogene, 30( 9): 1072–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zheng L, Jia J, Finger LD, Guo ZG, Zer C, Shen BH. (2010). Functional regulation of Fen1 nuclease and its link to cancer. Nucleic Acids Res, 39 (3): 781–794(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teasley DC, Parajuli S, Nguyen M, Moore HR, Alspach E, Lock YJ, Honaker Y, Saharia A, Piwnica-Worms H, Stewart SA. (2015). Flap endonuclease 1 limits telomere fragility on the leading strand. J BiolChem, 290( 24): 15133–15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collins FS, Brooks LD, Chakravarti A. (1998). A DNA polymorphism discovery resource for research on human genetic variation. Genome Res, 8( 12): 1229–1231. [DOI] [PubMed] [Google Scholar]
- 25. Longmate JA. (2001). Complexity and power in case-control association studies. Am J Hum Genet, 68( 5): 1229–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harrington JJ, Lieber MR. (1994). The characterization of a mammalian DNA structure-specific endonuclease. EMBO J, 13( 5): 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang KJ, Xie CH, Chen DR. (2014). Flap endonuclease 1 is a promising candidate biomarker in gastric cancer and is involved in cell proliferation and apoptosis. Int J Mol Med, 33( 5), 1268–1274. [DOI] [PubMed] [Google Scholar]
- 28. Larsen E, Gran C, Saether BE, Seeberg E, Klung-land A. (2003). Proliferation failure and gamma radiation sensitivity of Fen1 null mutant mice at the blastocyst stage. Mol Cell Biol, 23( 15): 5346–5353. [DOI] [PMC free article] [PubMed] [Google Scholar]