Abstract
Helicobacter pylori is associated with inflammatory diseases and can cause gastric cancer and mucosa-associated lymphoma. One of the bacterium’s key proteins is high temperature requirement A (HpHtrA) protein, an extracellular serine protease that cleaves E-cadherin of gastric epithelial cells, which leads to loss of cell-cell adhesion. Inhibition of HpHtrA may constitute an intervention strategy against H. pylori infection. Guided by the computational prediction of hypothetical ligand binding sites on the surface of HpHtrA, we performed residue mutation experiments that confirmed the functional relevance of an allosteric region. We virtually screened for potential ligands addressing this surface cleft located between the catalytic and PDZ1 domains. Our receptor-based computational method represents protein surface pockets in terms of graph frameworks and retrieves small molecules that satisfy the constraints given by the pocket framework. A new chemical entity was identified that blocked E-cadherin cleavage in vitro by direct binding to HpHtrA, and efficiently blocked pathogen transmigration across the gastric epithelial barrier. A preliminary crystal structure of HpHtrA confirms the validity of a comparative “homology” model of the enzyme, which we used for the computational study. The results of this study demonstrate that addressing orphan protein surface cavities of target macromolecules can lead to new bioactive ligands.
More than 50% of the human population is infected by the gram-negative bacterium Helicobacter pylori (H. pylori).1 It is associated with several inflammatory diseases such as ulceration and gastritis.2,3 H. pylori colonizes the host’s gastric epithelium where it is able to destroy mucosal integrity and therefore can pass the epithelial barrier.4 In severe cases, an infection can lead to gastric cancer and mucosa-associated lymphoid tissue lymphoma, which is why H. pylori has been rated as a class 1 carcinogen by the World Health Organization.5,6 One of the key factors for migration across the epithelial barrier is the bacterium’s high temperature requirement A (HpHtrA) protein that functions as a secreted protease cleaving E-cadherin.7 This essential serine protease cleaves the ectodomain of E-cadherin and consequently impairs cell–cell adhesion of the epithelial cells. It has previously been shown that inhibition of HtrA-mediated cleavage of E-cadherin significantly reduces migration of bacteria through polarized epithelial monolayers and might therefore be an effective strategy to treat H. pylori infections.7 Only a few HpHtrA inhibitors have been published so far.8–10 In 2010, Hoy et al.7 discovered the first inhibitor HHI, which was obtained by virtual screening with a comparative (“homology”) model of HpHtrA,9 focusing on the active site around catalytic Ser221. HHI also served as the starting point for a ligand-based virtual screen that led to another inhibitor of HpHtrA (compound 1, Fig. 1).8b The actual binding site of this ligand is unknown. While there are many high-resolution crystallographic structures available of HtrA homologues, e.g. Escherichia coli DegP, HpHtrA has eluded full structural characterization to date. Here, we propose that several ligand binding sites might exist on the surface of H. pylori enzyme, and disclose inhibitors supposed to bind to an allosteric surface cavity.
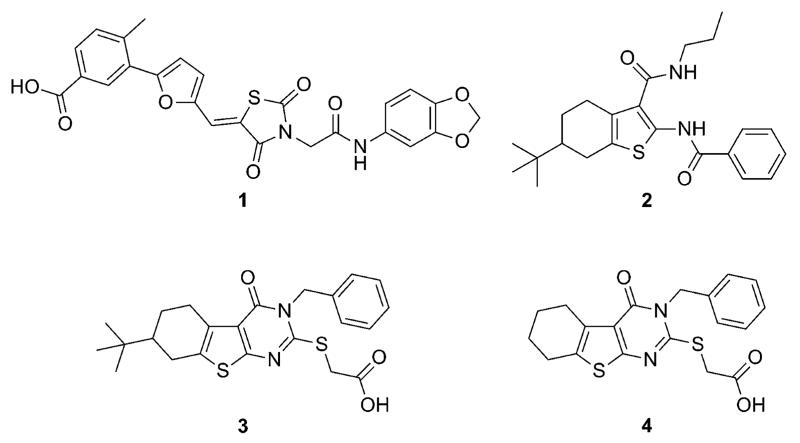
Fig. 1.
Reference compound 1 (ref. 8b) and three tested compounds that were suggested by virtual screening with the PoLiMorph21 software.
As demonstrated for the mentioned example of the HpHtrA active site but also for several other targets,11–13 receptor-based virtual screening provides a starting point to obtain new chemical entities with desired biological activity.14 One such method for receptor-based virtual screening is automated ligand docking, where potential ligands are placed into the respective binding site and scored according to their interactions with the target protein.15 Alternatives to docking are pharmacophore-based methods that compare the pharmacophoric feature distribution of candidate compounds and their potential binding site.16 There are several tools available for receptor-based pharmacophore modelling and compound screening, for example Catalyst (Accelrys, Inc., http://accelrys.com, San Diego, California), FLAP,17 PseudoLigand,18 and VirtualLigand,9 just to mention some prominent representatives. These methods implement grid-based approaches like GRID19 or LUDI20 to determine pharmacophoric features in the protein pockets. Such grid representations are usually transformed into three-dimensional (3D) pharmacophore models and encoded by pocket-fingerprints (FLAP) or correlation vectors (VirtualLigand), which allow for the rapid comparison of pocket features and screening compounds.
Here, we present a receptor-based virtual screening method as an extension to our software package PoLiMorph (Pocket Ligand Morphing).21 Protein surface pockets and small molecules are represented by fuzzy labelled graphs that store information about the shape and pharmacophoric features of the respective binding site. These graphs are conceptual models of ligands and their binding. Graph superimposition and comparison of the distributions of pharmacophoric features are used to predict potential receptor-ligand interaction. We demonstrate the software’s ability to support hit identification by retrieving an HpHtrA inhibitor from a large pool of screening compounds. The inhibitor was identified through application of the PoLiMorph receptor-based screening module.
Pocket frameworks
The software PoLiMorph was originally developed for the comparison of ligand binding sites on protein surfaces,21 and has recently been extended to allow ligand-based virtual screening.22 Taking the graph description of both binding sites and small molecules, we developed a matching scheme that evaluates complementarity of pockets and ligands without the need for explicit ligand docking into surface cavities. We expect lipophilic, hydrophilic and uncharged regions of the ligand graphs to be matched to regions of the protein pocket with similar properties. Specifically, positively or negatively charged ligand regions should face oppositely charged regions of the pocket. Hydrogen-bond donors should be located opposite to hydrogen-bond acceptors and vice versa. Accordingly, the fit between a pocket graph vertex vp and a ligand graph vertex vl is determined on the basis of complementary properties:
| (1) |
where F is the set of all ligand graph potentials, v(f) corresponds to the value of property f in vertex v, and comp(f) is the complementary graph potential to f. The fit between two vertices in feature f is weighted according to predetermined correlation values cf, which we obtained from observed property distributions in a set of known ligand-protein complexes (PDBbind core set).23 Briefly, graph representations of all binding sites and their bound ligands were calculated, and all vertices of the ligand graphs were assigned to their closest vertex of the respective pocket graph. The correlation values between the resulting paired property distributions were taken as weighting factors cf. To be consistent with the vertex-fit calculations of the other modules of PoLiMorph that employ z-scoring procedures, the vertex-fit distributions of all assigned vertices within the PDBbind core set were determined. Means and standard deviations of these distributions were used for rescoring of the matched graphs (for technical and mathematical details see ref. 21). eqn (1) allows for computing similarity values for property-labeled graph representations of protein pockets and potential ligands.
As a first evaluation of PoLiMorph’s receptor-based virtual screening module, ligands for six drug targets were exemplarily chosen from the COBRA (v10.3) collection of drugs and lead compounds,24 and corresponding protein structures were taken from the Protein Data Bank (PDB):25 human immunodeficiency virus 1 (HIV-1) protease-1 (PDB-ID: 1dmp),26 peroxisome proliferator activator receptor gamma (PPARγ, PDB-ID: 1zgy),27 cyclooxygenase 2 (COX2, PDB-ID: 6cox),28 retinoic acid receptor gamma (RARγ, PDB-ID: 1fcx),29 human vitamin D3 receptor (VDR, PDB-ID: 3b0t),30 and factor Xa (fXa, PDB-ID: 2bok).31 From each protein structure, the software PocketPicker32 extracted binding pockets, for which PoLiMorph constructed a graph representation. Then, graph representations of the ligands were created and ranked according to their calculated pocket matching score (eqn (1)). On average the matching of a pair of pocket and ligand graphs took 15 ms on a MacPro dual Quad-Core Intel Xeon machine (non-parallelized), rendering the method applicable to screening large compound databases.
With ROC-AUC values (in brackets: BEDROC scores, α = 0.05)33 of 0.87 (0.37), 0.65 (0.11), 0.97 (0.612), and 0.99 (0.94) for the target classes HIV-1 protease, COX-2, RARγ, and VDR respectively, PoLiMorph performed well for four out of the six selected targets. ROC-AUC values of 0.49 (0.03) and 0.43 (0.05) for fXa and PPARγ indicate that for these target classes PoLiMorph imperfectly captured essential ligand properties. This preliminary retrospective evaluation indicates that without further methodological tuning the applicability of virtual screening with PoLiMorph might be restricted to certain targets or target classes. Improved performance might be achievable by considering excluded pocket volumes to avoid clashes between ligands and proteins during the matching process. Also, the incorporation of target-specific information by up- and down-weighting of binding site regions or pharmacophoric features could result in better screening performance, which was shown by Hähnke et al.34
Pocket identification and structure-based virtual screening for HtrA inhibitors
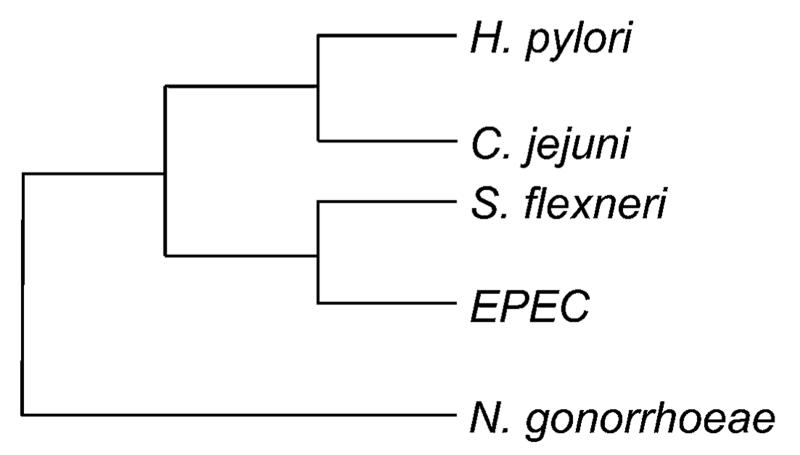
Keeping these caveats in mind, our search for new HpHtrA inhibitors started with the analysis of potential binding sites on the surface of a comparative model of HpHtrA.7 For model construction, we retrieved HtrA protein sequences from H. pylori (UniProt-ID:35 G2J5T2), Campylobacter jejuni (UniProt-ID: A1W0L1), enteropathogenic E. coli (EPEC; UniProt-ID: 6GXK7), Shigella flexneri (UniProt-ID: E3YA49) and Neisseria gonorrhoeae (UniProt-ID: E8SRH2) via PubMed from the National Center for Biotechnology Information (U.S. National Library of Medicine, Bethesda MD, USA). HtrAs from these species, except N. gonorrhoeae, efficiently cleave E-cadherin.36 We computed BLAST37 sequence alignments for each of the HtrA homologues to identify the best structures available from the PDB (Fig. 2). In the alignment, PDB entry 3mh6 (ref. 38) showed the highest sequence identity to all queries and ranked first based on the calculated E-values. Its identity to S. flexneri and EPEC HtrAs was 99% (E-value = 0). C. jejuni HtrA shared 41% sequence identity (E-value = 2e−72), and N. gonorrhoeae HtrA exhibited the lowest sequence pairwise identity of 36% (E-value = 2e−88) compared to 3mh6. The sequence in crystal structure 3mh6 was aligned to all sequences using ClustalW.39 We constructed the comparative HpHtrA model based on this alignment with the software Modeller 9v3 (ref. 40) (Fig. 3).
Fig. 2.
Sequence-based neighbour-joining tree of HtrA homologues.
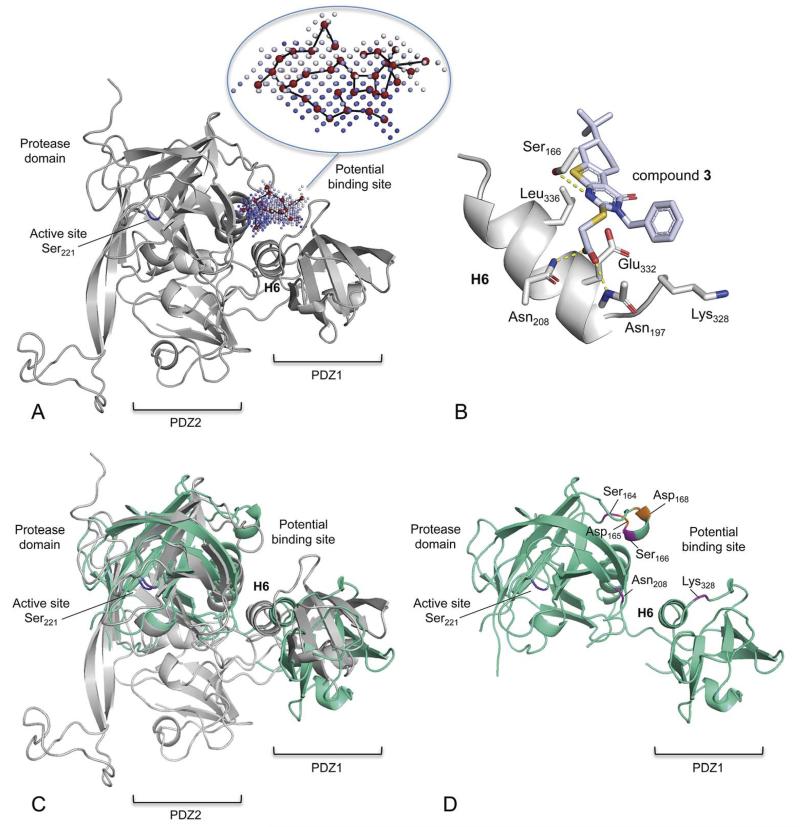
Fig. 3.
Structural model of H. pylori HtrA and a potential binding mode of compound 3. (A) Comparative (“homology”) protein model of HpHtrA. The enzyme contains a protease domain and two PDZ domains (PDZ1, PDZ2). The potential allosteric pocket that was used for virtual screening is located between the protease domain and the PDZ1 domain. This pocket lies distant to the active site (catalytic Ser221) and is flanked by helix 6 (H6) and two loops. The pocket graph (red vertices) computed by the PoLiMorph software is shown inside the allosteric pocket that was extracted by the PocketPicker tool. The intensity of the blue colour of the pocket grid points correlates with their buriedness. (B) Computed docking pose of compound 3. Potential hydrogen-bond interactions are indicated by yellow dotted lines. (C) Structural alignment of the homology model (gray) and a preliminary X-ray structure of HpHtrA (green). (D) X-ray structural model of HpHtrA with the mutated residues highlighted.
Then we searched cavities on the surface of our HpHtrA model using the software PocketPicker. In addition to the active site cavity located around the catalytic Ser221, a second large pocket was found. This ‘orphan’ allosteric pocket is placed in the interface between PSD-95/Discs-Large/ZO domain 1 (PDZ1) and the serine protease domain of HpHtrA (Fig. 3A). Two residues (Asp165, Asp168; Fig. 3D) flanking the potential binding site were previously shown to be important for protease activity of HpHtrA oligomers.41 We expected a potential loss of enzymatic activity upon binding of small molecules to this hypothetical allosteric pocket. Of note, Krojer et al. showed for the E.coli homolog of HpHtrA, DegP, that the large cavity on the opposite side of helix H6 to our hypothetical pocket, located between the catalytic and the PDZ1 domains, is in fact an allosteric regulation site for the proteolytic chaperone activity of DegP.42,43
We calculated the PoLiMorph graph description of the suggested new binding site (Fig. 3A) and used it as query for virtually screening a database containing 127,138 ligand graphs. The compound pool contained Specs Natural Products 08/2010 and the Specs Screening Collection 08/2010 (Specs, Delft, The Netherlands). All compounds were pre-processed using the ‘wash’ function of the Molecular Operating Environment (MOE) software (2009.10, Chemical Computing Group, Montreal, Canada). For each compound, we generated a single heuristic conformation with CORINA (v3.26, Molecular Networks, Erlangen, Germany).
Among the top ranked compounds (1250 molecules, ~1% of the screening pool) 10.4% contained a tetrahydrobenzothiophene scaffold, which represents a four-fold enrichment of this substructure compared to its overall prevalence in the compound pool (Table 1). Moreover, 63% of the top-ranking compounds contained a tertiary-butyl group (17-fold enrichment). We selected the best-ranked tetrahydrobenzothiophene derivative (compound 2, rank 75) for testing in vitro binding to HtrA. In addition, we performed a follow-up ligand-based similarity searching experiment using PoLiMorph to detect analogues of compound 2, which retrieved compounds 3 and 4. In these compounds the tetrahydrobenzothiophene scaffold is replaced by thienopyrimidinone (Fig. 1).
Table 1.
Scaffold frequency among the virtual hit list
| Scaffold | Occurrence in top-ranked compounds | Overall occurrence in database | Fold enrichment in top-ranked compounds |
|---|---|---|---|
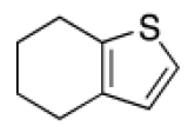
|
10.4% | 2.6% | 4 |
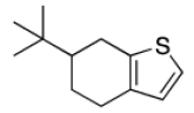
|
6.6% | 0.4% | 17 |
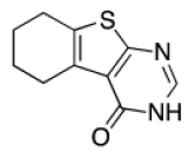
|
7.9% | 0.05% | 17 |
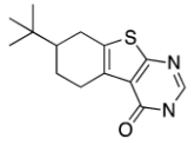
|
2.2% | 0.005% | 45 |
A subset of retrieved compounds block HpHtrA enzymatic activity
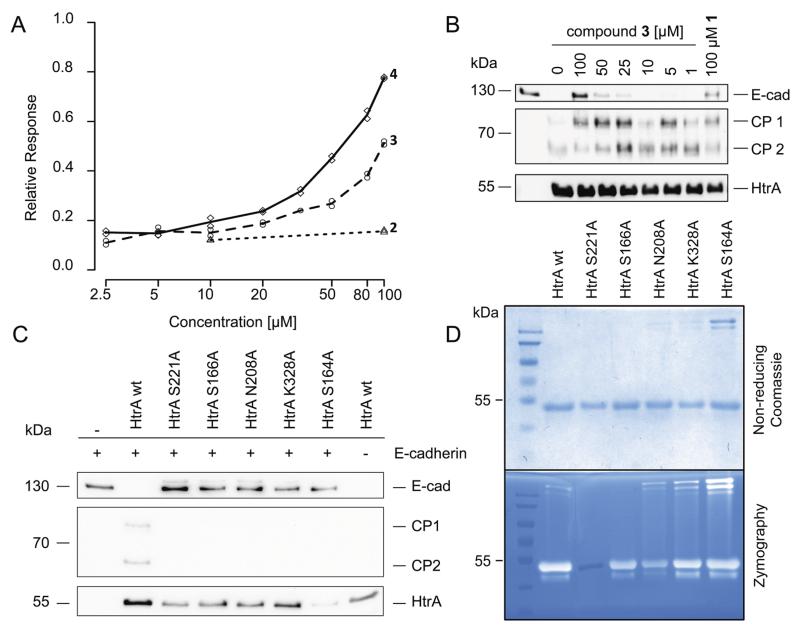
Surface plasmon resonance (SPR) experiments revealed concentration-dependent binding of compounds 3 and 4 to HpHtrA, while for compound 2 no binding was observed (Fig. 4A). In an in vitro E-cadherin cleavage assay we tested if 3 and 4 also inhibit the enzymatic activity of HpHtrA. Compound 3 showed concentration-dependent inhibition with an estimated IC50 of 30–40 μM (n = 4) (Fig. 4B), while compound 4 had no effect on E-cadherin processing by HpHtrA.†
Fig. 4.
Binding potential and inhibition of HpHtrA by compounds 2, 3, and 4, and the activity of HpHtrA mutants. (A) SPR response for binding of compounds 2, 3, and 4 to immobilized HpHtrA. (B) Recombinant E-cadherin was incubated with HpHtrA and several concentrations of compounds 1 (reference inhibitor) and 3. Reduction of full-length E-cadherin (E-Cad), cleavage products (CP1, CP2) and enzyme loading (HtrA) are shown. (C) HpHtrA wt cleaves E-cadherin. HpHtrA S221A, S166A, N208A, K328A and S164A show no or very limited proteolytic activity against E-cadherin. (D) HpHtrA wt, HpHtrA S166A, N208A, K328A and S164A are proteolytically active against casein, only the active-site mutation S221A loses all activity.
It is of note that we did not observe sigmoidal response curves in the SPR experiments, which might indicate unspecific binding or compound aggregation. Such a binding behaviour is not uncommon for drug-like compounds, as reported by Browner and coworkers,44 and points to potentially promiscuous or allosteric ligands.45
To obtain a preliminary idea about the relevant pharmacophore points and underlying structure-activity relationship, we docked compound 3 into the predicted allosteric site of HpHtrA using PoLiMorph. For this procedure, we used the vertex assignments of the matched pocket and ligand graphs to construct a rotation matrix with the Kabsch algorithm46 that served to place the ligand in the presumed binding site. The resulting complex was energy minimized in MOE. The result suggested that compound 3 might fit into the presumable binding site and form interactions with the protein (Fig. 3B). The carboxamide group of Asn208 and the backbone amide nitrogen of Asn197 might act as hydrogen-bond donors for the carboxyl group of the ligand. Also, a hydrogen bridge between Ser166 and the sp2-hybridized nitrogen atom in the thienopyrimidinone scaffold could be formed. Hydrophobic interactions between Lys328 and the phenyl ring of compound 3 might also contribute to ligand binding. Given the fact that compound 4 does not inhibit the catalytic activity of HpHtrA, the interaction of Leu336 and the tertiary butyl group of 3 seems to be important for enzyme activity inhibition. One might speculate that this group is essential for stabilizing the protein in an inactive conformation by blocking the relative movement of the protease and the adjacent PDZ1 domain.
In order to test this hypothesis, we generated three single mutations: S166A, N208A, K328A. Caseinolytic activity of these single residue mutations mutants was retained, as determined by casein zymography (Fig. 4D). Additionally, we generated the S164A mutation, which also showed casein digestion. The catalytic site mutation S221A served as negative control in the assays. However, the mutations within the potential allosteric site affected E-cadherin cleavage by HpHtrA. Despite their ability to cleave casein, these mutants lost their ability to cleave the natural substrate E-cadherin (Fig. 4C). This observation supports our hypothesis of the ligand binding site being relevant for the functional regulation of HpHtrA.
As a main outcome of the virtual screening study, we identified compound 3 as a new direct inhibitor of H. pylori HtrA protease activity by addressing a computationally identified allosteric pocket without the need to explicitly dock all screening compounds into the presumed binding cavity.
Crystal structure of HpHtrA
In an attempt to validate experimentally the comparative structural model of the complex between compound 3 and HpHtrA, we pursued crystallization experiments. While no co-complex was to date producible (crystallization trials ongoing), we were able to solve the apo-structure of HpHtrA at 2.6 Å. The species captured in the crystal is a hexameric form of the protein. PDZ2 was not visible in the electron density maps, so protease and PDZ1 domains alone (amino acids 48 to 364) from refined chain A only were used for a comparison with the homology model (amino acids 48 to 475). (Note, the final, fully refined structure will be reported elsewhere.) Superposition of the experimentally determined and homology models shows a good agreement in the relative positions of protease and PDZ1 domains (Fig. 3C). Furthermore, when comparing individual domains the structural alignments of both protease domains together, or both PDZ1 domains have root-mean-square deviation values of 0.68Å and 0.69 Å, respectively. Thus, the comparative model produced is a reasonably accurate representation of the HpHtrA structure in terms of tertiary structure and domain arrangement, specifically in the region around the presumed allosteric binding site, which is located between the PDZ1 and protease domains.
Conclusions
Structure-based virtual screening by a new graph-based method based on the PoLiMorph algorithm has resulted in the identification of functional ligands that efficiently block E-cadherin processing by the serine protease HtrA from the human pathogen H. pylori. We validated the applicability of the comparative protein model used in the virtual screen by a new crystal structure of apo-HpHtrA. Our results suggest an allosteric mode of action for the most potent inhibitor 3, although there is no direct experimental proof of the presumed binding site. Although cocrystallization experiments failed, we were able to solve the first apo-structure of HpHtrA at a resolution of 2.6 Å. The experimentally obtained structure corroborates the suitability of an advanced comparative (“homology”) protein model for computational hit and lead finding and motivates ligand screening for presumed allosteric surface cavities. The inhibitors identified in our study can now serve as starting points for hit-to-lead optimization and scaffold hopping to other chemotypes.
Acknowledgements
This research was supported by a grant from the OPO-Foundation Zurich to GS. SF acknowledges funding from the EU FP7 consortium TARGETBINDER and the State of Saxony-Anhaltine (Research Focus Molecular Biosciences). SW was supported by grant P-24074 from the Austrian Science Fund (FWF).
Footnotes
Surface Plasmon Resonance. The SPR binding study was performed at 25 °C on a SPR-2 instrument from Sierra Sensors GmbH (Hamburg, Germany) with a temperature-stabilized light source and a flow rate of 25 μl min−1. Freshly prepared 10 mM HEPES buffered saline (HBS-P), 150 mM NaCl and 0.05% Tween was used as immobilization running buffer. For immobilization on sensor chips with a carboxymethyl dextran matrix (SPR-2 Affinity Sensor Amine, batch 10-BB-02-349-A, Sierra Sensors) an amino coupling method with activation solution [200 mM N-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) and 50 mM N-hydroxysuccinimide (NHS)] was performed for 5 min. 1 mg HpHtrA and 1 mg trypsin (TPP, T0303, lot #089K7358, Sigma) were dissolved in 1 ml of 10 mM HEPES (HEPES buffer 1 M Solution, pH 7.3, Fisher Scientific). Protein injection over 7 min and chip inactivation with 1 M ethanolamine at pH 8.5 for 6 min led to an HpHtrA response of 2569 RU on spot2 and a trypsin response of 895 RU on spot1. After immobilization the system was primed with the assay running buffer, 10 mM HEPES buffered saline containing 3% DMSO (HBS-PD). 20 mM DMSO stock solutions of compounds 2, 3, and 4 were prepared and diluted to 600 μM in 1.02 × HBS buffer without DMSO. These 600 μM solutions were diluted to yield eight concentrations ranging from 100 μM to 2.5 μM.
E-cadherin cleavage. For in vitro E-cadherin cleavage studies, 100 ng recombinant E-cadherin (R&D Systems) were incubated with 100 ng recombinant HpHtrA in 50 mM HEPES (pH 7.4) at 37 °C for 16 h. HtrA inhibitors were dissolved in DMSO and added to obtain a final concentration of 100 μM unless otherwise stated. We stopped the reaction by adding SDS loading buffer. Proteins were separated by SDS-PAGE followed by Western Blot analysis. Recombinant E-cadherin was visualized using anti E-cadherin antibody H108 (Santa Cruz) and HtrA antiserum was raised against the N-terminal peptide (DKIKVTIPGSNKEY) of HpHtrA (Biogenes GmbH).
Non-reducing SDS PAGE and zymography. HpHtrA variants (3 μg) were separated by SDS PAGE under non-reducing conditions. For zymography, 0.1% casein (Roth) was copolymerized in the gel. After electrophoresis, zymography gels were incubated in a 2.5% Triton X-100 solution at room temperature for 1 h under gentle agitation, equilibrated in developing buffer (50 mM Tris pH 7.5, 200 mM NaCl, 5 mM CaCl2, 0.02% Brij-35) for 30 min at room temperature and then incubated in fresh developing buffer for 16 h at 37 °C. The gels were stained either with Coomassie G250 (Roth) for the regular SDS-PAGE, or with Coomassie R250 (Roth) for zymography. The gels were imaged with the ChemiDoc XRS+ using the ImageLab software (Biorad).
HtrA mutagenesis. The cloning of HpHtrA wt and HpHtrA S221A has been described previously.8a The HpHtrA mutants (S164A, S166A, N208A, K328A) were generated using the QuickChange Lightning Site directed mutagenesis Kit (Stratagene). The following primers were used: S164Af: 5′-CCC ACG ATC AAA TTC GCT GAT TCT AAT GAT ATT; S164Ar: 5′-AAT ATC ATT AGA ATC AGC GAA TTT GAT CGT GGG; S166Af: 5′-CCC ACG ATC AAA TTC TCT GAT GCT AAT GAT ATT; S166Ar: 5′-AAT ATC ATT AGC ATC AGA GAA TTT GAT CGT GGG; N208Af: 5′-ATC AAC AGC TAT GAG GCT TTC ATT CAA ACA GAC; N208Ar: 5′-GTC TGT TTG AAT GAA AGC CTC ATA GCT GTT GAT; K328Af: 5′- AAT GGG AAA AAG GTT GCA AAC ACG AAT GAG TTA, K328Ar: 5′-TAA CTC ATT CGT GTT TGC AAC CTT TTT CCC ATT.
HpHtrA preparation. For overexpression and purification of the recombinant HpHtrA transformed E. coli was grown in 1500 ml TB medium at 37 °C. At an OD550 of 0.5 the expression was induced by the addition of 100 μM isopropylthiogalactoside (IPTG). After incubation for 4 h at 25 °C the culture was pelleted for 20 min (4000 × g) at 4 °C and lysed in PBS by sonication. The lysate was cleared by centrifugation (10 000 × g) and the supernatant was incubated with glutathione sepharose (GE Healthcare) at 4 °C overnight. After washing the sepharose, the bound GST-fusion proteins were cleaved by PreScission Protease (GE Healthcare) at 4 °C overnight.
Computational docking. Ligand docking was done with GOLD v5.0.2.47 We performed ligand placement 50 times in a sphere with a radius of 10Å around residue N208 in the HpHtrA homology model. Ligand poses were evaluated by the ChemPLP scoring function.
Crystallization and structure determination. Protein crystals of HpHtrA were produced by the sitting-drop method after mixing protein solution in a 2 : 1 ratio with reservoir solution containing: 0.2 M NaF, 0.1 M bis–tris propane pH 6.5, 20% PEG 3350 and 10% ethylene glycol. Data were collected on beamline I24 at Diamond Light Source (Harwell, UK). A preliminary model only was available for this study, however the backbone α-carbon atoms from chain A were well refined with acceptable geometry according to Ramachandran dihedrals as quality indicators (97.6% in preferred or allowed regions), and thus were usable as a reference in validating the comparative homology model. Full details of crystallization, data collection and structural refinement of the completed oligomeric model will be published in a separate work.
Notes and references
- 1.Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat. Rev. Microbiol. 2007;5:441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 2.Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 2005;3:320–332. doi: 10.1038/nrmicro1095. [DOI] [PubMed] [Google Scholar]
- 3.Ishijima N, Suzuki M, Ashida H, Ichikawa Y, Kanegae Y, Saito I, Boren T, Haas R, Sasakawa C, Mimuro H. BabA-mediated adherence is a potentiator of the Helicobacter pylori type IV secretion system activity. J. Biol. Chem. 2011;286:25256–25264. doi: 10.1074/jbc.M111.233601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amieva MR, Vogelmann R, Covacci A, Tompkins LS, Nelson WJ, Falkow S. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300:1430–1434. doi: 10.1126/science.1081919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peek JRM, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 6. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans Schistosomes, liver flukes and Helicobacter pylori. World Health Organization; 61th edn. 1994. [PMC free article] [PubMed] [Google Scholar]
- 7.Hoy B, Löwer M, Weydig C, Carra G, Tegtmeyer N, Geppert T, Schröder P, Sewald N, Backert S, Schneider G, Wessler S. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010;11:798–804. doi: 10.1038/embor.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8 (a).Löwer M, Weydig C, Metzler D, Reuter A, Starzinski-Powitz A, Wessler S, Schneider G. Prediction of extracellular proteases of the human pathogen Helicobacter pylori reveals proteolytic activity of the Hp1018/19 protein HtrA. PLoS One. 2008;3:e3510. doi: 10.1371/journal.pone.0003510. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Klenner A, Hähnke V, Geppert T, Schneider P, Zettl H, Haller S, Rodrigues T, Reisen F, Hoy B, Schaible AM, Werz O, Wessler S, Schneider G. From virtual screening to bioactive compounds by visualizing and clustering of chemical space. Mol. Inf. 2012;31:21–26. doi: 10.1002/minf.201100147. [DOI] [PubMed] [Google Scholar]
- 9.Löwer M, Geppert T, Schneider P, Hoy B, Wessler S, Schneider G. Inhibitors of Helicobacter pylori protease HtrA found by ‘virtual ligand’ screening combat bacterial invasion of epithelia. PLoS One. 2011;6:e17986. doi: 10.1371/journal.pone.0017986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauske P, Meltzer M, Ottmann C, Krojer T, Clausen T, Ehrmann M, Kaiser M. Selectivity profiling of DegP substrates and inhibitors. Bioorg. Med. Chem. 2009;17:2920–2924. doi: 10.1016/j.bmc.2009.01.073. [DOI] [PubMed] [Google Scholar]
- 11.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat. Chem. Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Shoichet BK. Molecular docking and ligand specificity in fragment-based inhibitor discovery. Nat. Chem. Biol. 2009;5:358–364. doi: 10.1038/nchembio.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang H, Shen Y, Chen J, Jiang Q, Leng Y, Shen J. Structure-based virtual screening for identification of novel 11beta-HSD1 inhibitors. Eur. J. Med. Chem. 2009;44:1167–1171. doi: 10.1016/j.ejmech.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Löwer M, Proschak E. Structure-based pharmacophores for virtual screening. Mol. Inf. 2011;30:398–404. doi: 10.1002/minf.201100007. [DOI] [PubMed] [Google Scholar]
- 15.Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discovery. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 16.Horvath D, Bajorath J. Chemoinformatics and Computational Chemical Biology. Springer; New York: 2011. Pharmacophore-based virtual screening; pp. 261–298. [DOI] [PubMed] [Google Scholar]
- 17.Baroni M, Cruciani G, Sciabola S, Perruccio F, Mason JS. A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for Ligands and Proteins (FLAP): theory and application. J. Chem. Inf. Model. 2007;47:279–294. doi: 10.1021/ci600253e. [DOI] [PubMed] [Google Scholar]
- 18.Schüller A, Fechner U, Renner S, Franke L, Weber L, Schneider G. A pseudo-ligand approach to virtual screening. Comb. Chem. High Throughput Screening. 2006;9:359–364. doi: 10.2174/138620706777452375. [DOI] [PubMed] [Google Scholar]
- 19.Goodford PJ. A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J. Med. Chem. 1985;28:849–857. doi: 10.1021/jm00145a002. [DOI] [PubMed] [Google Scholar]
- 20.Böhm H-J. The computer program LUDI: a new method for the de novo design of enzyme inhibitors. J. Comput.-Aided Mol. Des. 1992;6:61–78. doi: 10.1007/BF00124387. [DOI] [PubMed] [Google Scholar]
- 21 (a).Weisel M, Kriegl JM, Schneider G. Architectural repertoire of ligand-binding pockets on protein surfaces. ChemBioChem. 2010;11:556–563. doi: 10.1002/cbic.200900604. [DOI] [PubMed] [Google Scholar]; (b) Reisen F, Weisel M, Kriegl JM, Schneider G. Self-organizing fuzzy graphs for structure-based comparison of protein pockets. J. Proteome Res. 2010;9:6498–6510. doi: 10.1021/pr100719n. [DOI] [PubMed] [Google Scholar]
- 22.Geppert T, Reisen F, Pillong M, Hähnke V, Tanrikulu Y, Perna AM, Perez TB, Schneider P, Schneider G. Virtual screening for compounds that mimic protein-protein interface epitopes. J. Comput. Chem. 2012;33:573–579. doi: 10.1002/jcc.22894. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Fang X, Lu Y, Yang CY, Wang S. The PDBbind database: methodologies and updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 24.Schneider P, Schneider G. Collection of bioactive reference compounds for focused library design. QSAR Comb. Sci. 2003;22:713–718. [Google Scholar]
- 25.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodge CN, Aldrich PE, Bacheler LT, Chang CH, Eyermann CJ, Garber S, Grubb M, Jackson DA, Jadhav PK, Korant B. Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450. Chem. Biol. 1996;3:301–314. doi: 10.1016/s1074-5521(96)90110-6. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9505–9510. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurumbail RG, Stevens AM, Gierse JK, McDonald JJ, Stegeman RA, Pak JY, Gildehaus D, Miyashiro JM, Penning TD, Seibert K, Isakson PC, Stallings WC. Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Nature. 1996;384:644–648. doi: 10.1038/384644a0. [DOI] [PubMed] [Google Scholar]
- 29.Klaholz BP, Mitschler A, Moras D. Structural basis for isotype selectivity of the human retinoic acid nuclear receptor. J. Mol. Biol. 2000;302:155–170. doi: 10.1006/jmbi.2000.4032. [DOI] [PubMed] [Google Scholar]
- 30.Hishiki A, Hashimoto H, Sato M, Shimizu T. PDB-ID: 3b0t. to be published.
- 31.Schärer K, Morgenthaler M, Paulini R, Obst-Sander U, Banner DW, Schlatter D, Benz J, Stihle M, Diederich F. Quantification of cation–π interactions in protein–ligand complexes: crystal-structure analysis of factor Xa bound to a quaternary ammonium ion ligand. Angew. Chem., Int. Ed. 2005;44:4400–4404. doi: 10.1002/anie.200500883. [DOI] [PubMed] [Google Scholar]
- 32.Weisel M, Proschak E, Schneider G. PocketPicker: analysis of ligand binding-sites with shape descriptors. Chem. Cent. J. 2007;1:1–7. doi: 10.1186/1752-153X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Truchon JF, Bayly CI. Evaluating virtual screening methods: good and bad metrics for the ‘early recognition’ problem. J. Chem. Inf. Model. 2007;47:488–508. doi: 10.1021/ci600426e. [DOI] [PubMed] [Google Scholar]
- 34.Hähnke V, Schneider G. Pharmacophore alignment search tool: influence of scoring systems on text-based similarity searching. J. Comput. Chem. 2011;32:1635–1647. doi: 10.1002/jcc.21741. [DOI] [PubMed] [Google Scholar]
- 35.The UniProt Consortium Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoy B, Geppert T, Boehm M, Reisen F, Plattner P, Gadermaier G, Sewald N, Ferreira F, Briza P, Schneider G, Backert S, Wessler S. Distinct roles of secreted HtrA proteases from gram-negative pathogens in cleaving the junctional protein and tumor suppressor E-cadherin. J. Biol. Chem. 2012;287:10115–10120. doi: 10.1074/jbc.C111.333419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 38.Krojer T, Sawa J, Huber R, Clausen T. HtrA proteases have a conserved activation mechanism that can be triggered by distinct molecular cues. Nat. Struct. Mol. Biol. 2010;17:844–852. doi: 10.1038/nsmb.1840. [DOI] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A. Comparative protein structure modeling with MODELLER. Curr. Protocols Bioinf. 2006;(suppl. 15):5.6.1–5.6.30. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geppert T, Hoy B, Wessler S, Schneider G. Context-based identification of protein–protein interfaces and ‘hot-spot’ residues. Chem. Biol. 2011;18:344–353. doi: 10.1016/j.chembiol.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Krojer T, Sawa J, Schäfer E, Saibil HR, Ehrmann M, Clausen T. Structural basis for the regulated protease and chaperone function of DegP. Nature. 2008;453:885–890. doi: 10.1038/nature07004. [DOI] [PubMed] [Google Scholar]
- 43.Krojer T, Pangerl K, Kurt J, Sawa J, Stingl C, Mechtler K, Huber R, Ehrmann M, Clausen T. Interplay of PDZ and protease domain of DegP ensures efficient elimination of misfolded proteins. Proc. Natl. Acad. Sci. U. S. A. 2008;105:7702–7707. doi: 10.1073/pnas.0803392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giannetti AM, Koch BD, Browner MF. Surface plasmon resonance based assay for the detection and characterization of promiscuous inhibitors. J. Med. Chem. 2008;51:574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 45.van Westen GJ, Gaulton A, Overington JP. Chemical, target, and bioactive properties of allosteric modulation. PLoS Comput. Biol. 2014;10:e1003559. doi: 10.1371/journal.pcbi.1003559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallogr., Sect. A: Cryst. Phys., Diffr., Theor. Gen. Crystallogr. 1976;32:922–923. [Google Scholar]
- 47.Jones G, Willett P, Glen RC. Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. J. Mol. Biol. 1995;245:43–53. doi: 10.1016/s0022-2836(95)80037-9. [DOI] [PubMed] [Google Scholar]