Abstract
Objectives
Glycosylation or the modification of a cellular component with a carbohydrate moiety has been demonstrated in all three domains of life as a basic post-translational process important in a range of biological processes. This review will focus on the latest studies attempting to exploit bacterial N-linked protein glycosylation for glycobiotechnological applications including glycoconjugate vaccine and humanised glycoprotein production. The challenges that remain for these approaches to reach full biotechnological maturity will be discussed.
Key findings
Oligosaccharyltransferase-dependent N-linked glycosylation can be exploited to make glycoconjugate vaccines against bacterial pathogens. Few technical limitations remain, but it is likely that the technologies developed will soon be considered a cost-effective and flexible alternative to current chemical-based methods of vaccine production. Some highlights from current glycoconjugate vaccines developed using this in-vivo production system include a vaccine against Shigella dysenteriae O1 that has passed phase 1 clinical trials, a vaccine against the tier 1 pathogen Francisella tularensis that has shown efficacy in mice and a vaccine against Staphylococcus aureus serotypes 5 and 8. Generation of humanised glycoproteins within bacteria was considered impossible due to the distinct nature of glycan modification in eukaryotes and prokaryotes. We describe the method used to overcome this conundrum to allow engineering of a eukaryotic pentasaccharide core sugar modification within Escherichia coli. This core was assembled by combining the function of the initiating transferase WecA, several Alg genes from Saccharomyces cerevisiae and the oligosaccharyltransferase function of the Campylobacter jejuni PglB. Further exploitation of a cytoplasmic N-linked glycosylation system found in Actinobacillus pleuropneumoniae where the central enzyme is known as N-linking glycosyltransferase has overcome some of the limitations demonstrated by the oligosaccharyltransferase-dependent system.
Summary
Characterisation of the first bacterial N-linked glycosylation system in the human enteropathogen Campylobacter jejuni has led to substantial biotechnological applications. Alternative methods for glycoconjugate vaccine production have been developed using this N-linked system. Vaccines against both Gram-negative and Gram-positive organisms have been developed, and efficacy testing has thus far demonstrated that the vaccines are safe and that robust immune responses are being detected. These are likely to complement and reduce the cost of current technologies thus opening new avenues for glycoconjugate vaccines. These new markets could potentially include glycoconjugate vaccines tailored specifically for animal vaccination, which has until today thought to be non-viable due to the cost of current in-vitro chemical conjugation methods. Utilisation of N-linked glycosylation to generate humanised glycoproteins is also close to becoming reality. This ‘bottom up’ assembly mechanism removes the heterogeneity seen in current humanised products. The majority of developments reported in this review exploit a single N-linked glycosylation system from Campylobacter jejuni; however, alternative N-linked glycosylation systems have been discovered which should help to overcome current technical limitations and perhaps more systems remain to be discovered. The likelihood is that further glycosylation systems exist and are waiting to be exploited.
Keywords: glycoconjugates, humanised glycoproteins, PglB, vaccines
Introduction
Glycoconjugate vaccines
The majority of bacterial pathogens are coated with a carbohydrate moiety. Unsurprisingly, upon infecting a host, these carbohydrates are often the very first epitopes detected by the immune system. The idea that surface carbohydrates from bacterial pathogens may serve as vaccines has been around since the 1920s. However, in the majority of patients, this carbohydrate component alone is insufficient to generate a long lasting immune response. Most polysaccharides are considered T-cell independent antigens,[1] unable to induce IgM to IgG switching, and to sustain T-cell memory.
Particularly at-risk populations are children under the age of 5 and the elderly. In young children, the reason for this weakened immune response is thought to be due to immature B cells with reduced CD21 expression, and reduced concentrations of IgG2 which are required to react to polysaccharide antigens.[2] In the elderly population, there is a limited magnitude of antibody responses to polysaccharides; the reason for this weakened response is thought to be due to a low reservoir of IgM+ memory B cells and their weaker differentiation into antibody producing cells.[3]
The discovery by Oswald Avery and Walter Goebel in 1929 that a specific polysaccharide functioned only as a true antigen when attached with some other constituent of the cell was ground breaking. Just as insightful was the demonstration that asymmetry in the sugar component alone, a switch of glucose and galactose, could lead to distinct serological responses.[4]
The latter finding gave the scientific community a glimpse into how infection with two seemingly identical bacteria with distinct sugar structures could give rise to such different immunological responses.
These were the foundations for glycoconjugate vaccinology. Surprisingly, however, no glycoconjugate vaccine was licensed for almost 60 years. In 1987, a vaccine directed against Haemophilus influenzae type b (Hib) was licensed and was based on the attachment of the polyribosylribitol phosphate capsular polysaccharide to various immunogenic carrier proteins.[5,6]
The global vaccine market is projected to be worth approximately US $30 billion by 2015; of this total, glycoconjugate vaccines will make up more than US $7 billion. However, production is expensive and time consuming. The accepted technique to make glycoconjugate vaccines is chemical conjugation; polysaccharides are chemically activated at random sites, or alternatively at the reducing end.[7] There are a multitude of options to link the glycan to the carrier protein. However, they all require initial steps to strip the polysaccharide from the pathogenic organism against which the vaccine is targeted: removal of contaminating endotoxin, purification of the acceptor protein from the organism of choice, before protein and glycan are treated to link the two to make the final glycoconjugate. Preparation of the acceptor protein is often not a simple procedure, for example, the carrier protein CRM197 used to be isolated from the culture supernatant of Corynebacterium diphtheriae, which required dedicated biosafety level 2 facilities with appropriate containment.[8]
These methods work; however, production is a multistep process and requires several rounds of purification to ensure that the glycoconjugate has been assembled correctly, therefore rendering glycoconjugate vaccine production an expensive process and prone to batch-to-batch variation (Figure 1).
Figure 1.
Example of chemical conjugation procedure used to generate glycoconjugate vaccines. In this example, production requires purification of lipopolisaccharide (LPS) from Francisella tularensis subs. tularensis, endotoxin cleavage, further purification, chemical activation and linkage to carrier protein CRM197.
Hib vaccine
H. influenzae is a non-motile Gram-negative coccobacillus; it can cause considerable morbidity and mortality with disease presentations such as meningitis, pneumonia, osteomyelitis, epiglottis and otitis. The bacterium is carried in the upper respiratory tract and is transmissible from person to person via aerosols.
The impact of Hib glycoconjugate vaccination has been impressive; in 2000, Hib caused an estimated 8–13 million serious illnesses and 371 000 deaths worldwide, most of these in developing countries.[9] In the absence of vaccination, the global incidence of Hib pneumonia was 1304 per 100 000 children younger than 5 years.[9] In countries where Hib glycoconjugate vaccination has been introduced such as the United Kingdom, this rate has declined to approximately 0.6 per 100 000 in children younger than 5 years.[10]
Streptococcus pneumoniae vaccine
S. pneumoniae is a Gram-positive coccus and a major cause of bacterial meningitis, sepsis and pneumonia. The organism is carried in the upper respiratory tract of approximately 5% of the world’s population and is transmitted via droplet spread. It is estimated that up to one million children die each year from S. pneumoniae infection. The greatest challenge for vaccine manufacture is that there are over >90 serotypes, based on capsular polysaccharide structure.[11] In 2000, a glycoconjugate vaccine targeting seven of the most prevalent serotypes (4, 6B, 9V, 14, 18C, 19F and 23F) conjugated to CRM197 (a detoxified version of diphtheria toxin, generated by a single amino acid substitution from glycine 52 to glutamic acid) was introduced in the United States. This resulted in a 73% decrease in hospital admissions for pneumonia in children aged below 5 years old, and a 39% decrease in children below the age of 2.[12] Despite this excellent efficacy and the subsequent generation of 10 and 13 valent glycoconjugate vaccine formulations, the number of children dying each year remains high, particularly in developing countries. This is partly due to the cost of manufacturing the vaccine, the complexity of such a varied number of serotypes and the fact that there is different serotype prevalence in different parts of the world.
Neisseria meningitidis vaccine
N. meningitidis is a Gram-negative diplococcus with 13 distinct capsule types (A, B, C, D, 29E, H, I, K, L, W135, X, Y and Z). The highest incidence rates in the world occur within an area known as the ‘meningitis belt’ in Sub-Saharan Africa that spans from Senegal to Ethiopia. The organism is a major cause of meningitis with considerable morbidity and mortality worldwide. The organism is carried in the nasopharynx of at least 10% of the general population and is spread through person to person contact, such as inhalation of respiratory droplets from the nose and throats of infected persons. Within the meningitis belt region of Africa, disease rates have been reported to reach 1000 per 100 000 population.[13] In Africa, the majority of cases are caused by serotype A while in Europe, N. meningitidis serotype B appears to be the major causative agent of disease. A serogroup A polysaccharide (a homopolymer of 2-acetamido-2-deoxy-D-mannopyranosyl phosphate)[7] conjugated to tetanus toxoid known as MenAfriVac was licensed in India in 2009 and prequalified by the World Health Organisation in 2010. Early tests in one of the endemic countries, Chad, indicates that meningitis rates in unvaccinated regions reached 43.8 per 100 000 while in vaccinated regions, it dropped to 2.48 per 100 000. Moreover, none of the reported cases in vaccinated regions appear to be due to serotype A.[14]
How glycoconjugate vaccines generate the desired immune response
Glycoconjugate vaccines provide enormous health benefits and superior immunity compared with polysaccharides alone, especially in the <5-year-old age group, the elderly and the immunocompromised. By attaching the sugar moiety to a typical T-cell dependent antigen such as a protein, the glycoconjugate becomes capable of inducing formation of polysaccharide-specific IgM to IgG switching, memory T-cell proliferation and memory B cell development. Despite this knowledge, a working model explaining why a glycoconjugate could provide such a response has been difficult to elucidate.
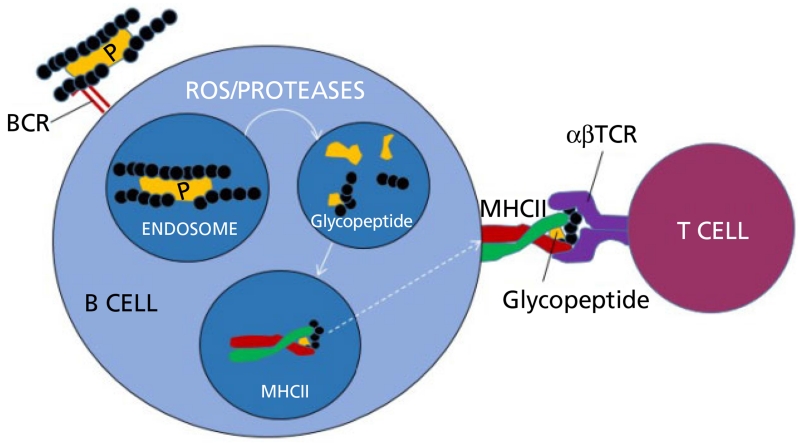
In 2011, Avci et al.[15] presented a plausible working model; one that appears to indicate that the glycoconjugate bound to a T-cell receptor was internalised and, in the endosome, was broken down into smaller peptides. Crucially, both the peptide and polysaccharide are trimmed allowing for binding of a glycopeptide to major histocompatibility complex (MHC) class II and presentation of this moiety to the T cell. This model appears to answer the conundrum as to why the protein and the sugar must be covalently bound to induce the optimal immune response (Figure 2).
Figure 2.
Simplified working model of T-cell activation by glycoconjugate vaccines. The glycan portion of the glycoconjugate binds to the B cell receptor (BCR), the glycoprotein is internalised into an endosome where the action of reactive oxygen species (ROS) and proteases process the glycoprotein into smaller glycopeptides, with shorter polysaccharide repeat units. The peptide portion of the newly formed glycopeptide binds to major histocompatibility complex (MHC) class II and enables presentation of the hydrophilic carbohydrate to the αβ T-cell receptor (αβTCR). Downstream processes enable maturation of a B cell to become a memory B cell and produce carbohydrate specific IgG antibodies (adapted from Avci et al.[15]). P, peptide; carbohydrate component shown as black circles.
The model also provides important clues as to how current glycoconjugate vaccines could be improved by rationally designing the protein carrier and the glycan attached to it. It may be possible that, by first identifying which shortened peptides are presented to the T-cell receptor by MHC class II, one may be able to reduce the size of the carrier, making protein expression and purification much easier and cheaper. It also indicates that the location of the sugar-peptide glycosidic bond is absolutely critical and that the next generation of glycoconjugate vaccines may be designed based on this location.
Glycoproteins in bacteria
The post-translational modification of proteins with glycan structures was for many years thought to be a feature unique to eukaryotic organisms. In 1976, the first study was published indicating that glycoproteins were present also in bacteria. Sleytr and Thorne analysed the surface layers of Clostridium thermasaccharolyticum and C. thermohydrosulfuricum and revealed that the proteins making up these surface layers were glycosylated.[16] We now understand that protein glycosylation systems are abundant in bacteria and that they can be separated into separate classes. For example, glycans can covalently attach to asparagine (N-linked) or serine/threonine residues (O-linked); they can modify single, or multiple proteins; glycans can be added sequentially or en bloc; and glycosylation can occur in different compartments of the cell. Additionally, they often involve oligosaccharyltransferases (OTases), a family of glycosyltransferases that catalyse the transfer of an oligosaccharide from a lipid donor to an acceptor molecule, usually a protein. Recent studies have demonstrated at least four classes of protein glycosylation systems in bacterial pathogens. These include two evolutionary distinct N-linked general glycosylation systems: one typified by the Campylobacter jejuni PglB system, where en bloc glycosylation operates in the periplasm through an N-OTase[17] and the other by the H. influenzae N-glycosyltransferase system where sequential glycosylation operates in the cytoplasm.[18,19] Similarly, there are en bloc and sequential O-glycosylation pathways typified by the Neisseria PglL periplasmic system, which requires an O-OTase[20] and the surface glycosylation of Escherichia coli AIDA-1 adhesins that is OTase independent.[21]
The expanding bacterial sequence dataset coupled with developments in mass spectrometry and analytical glycosciences has identified protein glycosylation systems in most bacterial pathogens. Indeed, many bacterial species have multiple glycosylation systems. However, despite their potential therapeutic importance, bacterial glycosylation systems remain largely uncharacterised, partly due to the complexity of the glycostructures. Thus, bacterial glycosylation represents an important but largely unexplored frontier in microbial science.
PglB the first bacterial N-linking OTase
Sequencing of the human gut pathogen C. jejuni NCTC 11168 in 2000 confirmed the presence of a genetic locus previously identified in C. jejuni 81-176 that looked like a polysaccharide coding region.[22-24] Within this polysaccharide coding region, an open reading frame that has significant similarity to an OTase subunit called STT3 involved in protein glycosylation in Saccharomyces cerevisiae was found. The region was tentatively named pgl for protein glycosylation locus and the OTase orthologue, PglB. Studies over the last 14 years have revealed the function of the locus in making a heptasaccharide with the structure GalNAc-α1,4-GalNAc-α1,4-[Glcβ1,3-]GalNAc-α1, 4-GalNAc-α1,4-GalNAc-α1,3-Bac-β1 where Bac is bacillosamine or 2,4-amino 2,4,6 trideoxy-D-glucose, GalNAc is N-acetylgalactosamine and Glc is glucose.[25,26] Crucially, these studies also revealed that PglB was an OTase and that the similarity in function meant that it was the first OTase identified in bacteria that could modify asparagines residues within a protein. The enzyme was found to accept diverse glycans when heterologously expressed in E. coli[27,28] and the amino acid sequence on an acceptor protein required for glycan modification was similar to that seen in the eukaryotic STT3 counterpart. The eukaryotic enzyme recognises an N-X-S/T sequon where is X is any amino acid except for proline, while substrate proteins for PglB require a negatively charged residue at the −2 position, extending the sequon to D/E-X1-N-X2-S/T where X is any amino acid except for proline.[28] The presence of an aspartic acid or a glutamic acid at the −2 position is an interesting difference between the eukaryotic and prokaryotic counterpart, since in eukaryotes, the presence of an acidic residue near the glycosylation site is disfavoured.[29] Currently, an absolute requirement of both eukaryotic and prokaryotic counterparts (including C. jejuni PglB) is the presence of an acetamido group at the C2 position of the reducing end sugar.
Experiments demonstrated that proteins that did not have native glycosylation sequons could become glycosylated with the addition of a glycosylation sequon. This was demonstrated elegantly in the glycosylation of cholera toxin[30] and green fluorescent protein, also indicating that the glycosylation motif must be present within an accessible domain such as a flexible loop within a nascent polypeptide, in order for it to be accessible to PglB.[31]
More recently, the structure of PglB was elucidated from Campylobacter lari. Crystallisation of the enzyme in complex with a peptide consisting of DQNATF (the optimal acceptor peptide for PglB)[32] revealed 13 transmembrane domains and provided some clues to indicate that the extended acceptor sequon may be necessary for tighter peptide binding. For the first time, a plausible working model of how PglB functions is possible and is explained in great detail in Lizak et al.[33]
The advent of next generation sequencing has revealed that PglB is not unique to the Campylobacter species but is present in a large number of epsilonproteobacteria, including deep sea vent organisms such as Nitratiruptor tergarcus. This finding makes it tempting to speculate that perhaps this type of protein modification evolved to enable proteins to have higher thermotolerance. The abundance of STT3 orthologues extends outside this subdivision to deltaproteobacteria and also archae,[34] and provides a source of enzymes that may have different specificities to the C. jejuni PglB currently being utilised.
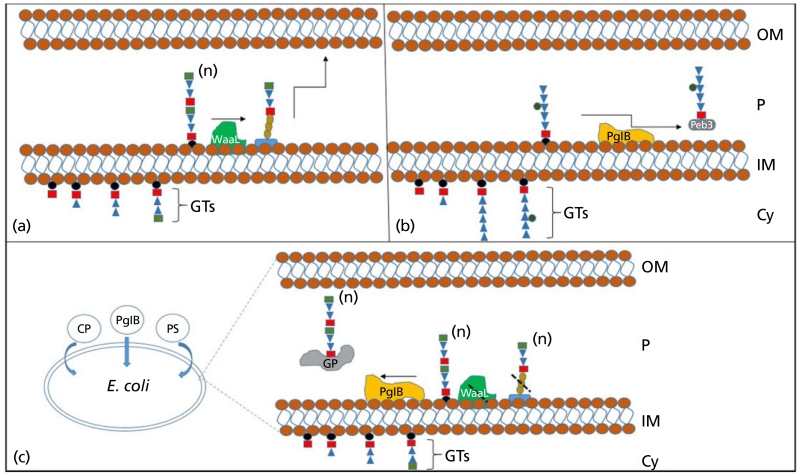
The discovery of the N-linked glycosylation system in C. jejuni is forerunner in glycoconjugate vaccine generation. To fully appreciate the biotechnological applications of OTases such as PglB, one needs only to compare the mechanism of heptasaccharide assembly in C. jejuni and typical O-antigen biosynthesis (Figure 3).
Figure 3.
Utilisation of PglB to make glycoconjugate vaccines. (a) Typical O-antigen assembly model; glycosyltransferases (GTs) sequentially build polysaccharide on undecaprenyl pyrophosphate (black circles), this is flipped into the periplasmic compartment of the cell where a ligase (WaaL) transfers the polysaccharide onto a lipid A core, before this is transported and presented on the surface of the bacterium. (b) Model of N-linked glycoprotein assembly in Campylobacter jejuni; GTs sequentially build a heptasaccharide on undecaprenyl pyrophosphate; this is flipped into the periplasmic compartment of the cell before PglB using the UndPP linked sugar as a substrate, transfers it onto an acceptor protein (Peb3) within the acceptor sequon D/E-X1-N-X2-S/T. (c) Protein glycan coupling technology; plasmids coding for a carrier protein (CP), the OTase (PglB) and a polysaccharide of choice (PS) are transformed into a laboratory strain of E. coli, expressing the genetic contents of the three plasmids within an E. coli strain lacking WaaL allows for glycoconjugate vaccine production (GP). Cy, cytoplasm; IM, inner membrane; P, periplasm; OM, outer membrane; (n) indicates polymerisation status of polysaccharide.
This is an in-vivo approach to generate glycoconjugate vaccines and has a number of advantages when compared against current chemically conjugated vaccines. Firstly, following subcloning of the required genetic content coding for the polysaccharide of choice, the entire process of glycoconjugate production can be transferred into a safe, laboratory strain of E. coli. This means that the glycoconjugate can conceivably be made without ever coming into contact with the pathogenic organism. The polysaccharide component and the carrier protein are linked in vivo, meaning that they do not have to undergo any purification steps before attachment, rendering the process much cheaper when compared with in-vitro conjugation. Once the three plasmid system is transformed into an E. coli strain, an inexhaustible supply of homogenous glycoconjugate is produced. Finally, the system has built-in flexibility. If the carrier protein or glycan need to be changed, then a simple plasmid swap is undertaken, meaning that multiple glycoconjugate combinations could be assembled and tested rapidly.
The technology has been used to make a glycoconjugate using the Shigella dysenteriae serotype O1 O-antigen repeat unit and attaching it to a detoxified form of Pseudomonas aeruginosa exotoxin A (EPA).[35] In February 2010, GlycoVaxyn, a company based on the utilisation of PglB to make glycoconjugate vaccines, reported that phase 1 clinical trials had begun using the S. dysenteriae O1-EPA conjugate, and in October of 2010, they reported that the conjugate demonstrated excellent safety profiles and that robust immune responses were elicited. More recently, initiation of phase 1 clinical trials on a vaccine developed against extra-intestinal pathogenic E. coli was announced (www.GlycoVaxyn.com, Press release).
The technology has been exploited by the Wren laboratory to generate a glycoconjugate vaccine against Francisella tularensis subs. tularensis by linking the O-antigen repeat unit to EPA.[36] The most recent breakthrough has been the utilisation of this technique to make a capsular polysaccharide glycan from a Gram-positive organism, Staphylococcus aureus serotypes 5 and 8 within E. coli. The flexibility of this technology is best demonstrated in this latest study where the acceptor protein was switched from EPA to a S. aureus toxoid HlaH35L that resulted in superior vaccine efficacy.[37]
One of the difficulties related to the use of PglB, in glycoconjugate vaccine design, is the assignment of a glycosylation sequon to a particular site within a protein. This meant that to find a location accessible for modification, partial or predicted structural information will be required. This problem was addressed by Fisher et al. who demonstrated that a D/E-X1-N-X2-S/T acceptor sequon could be added to the C- or N-terminus of a protein to render it a target for glycosylation.[38] The malE gene coding for maltose-binding protein was cloned and expressed in E. coli with four glycosylation tags (GT). Expression of malE alongside the entire pgl locus from C. jejuni resulted in tetraglycosylated material, indicating that every GT was occupied.
This technique, also known as protein glycan coupling technology (PGCT), may for the first time enable the cheap manufacture of glycoconjugate vaccines aimed at bacterial animal pathogens. Very few attempts have been made to generate such vaccines due to the prohibitive cost of manufacture, but examples exist generated through chemical conjugation, that show efficacy. In the swine pathogen Actinobacillus pleuropneumoniae serotype 1, the capsular polysaccharide and the O-antigen were chemically conjugated to the haemolysin protein from the same organism. Following vaccination, pigs were found to have significantly increased IgG antibodies against capsular polysaccharide and lipopolisaccharide, and sera from vaccinated pigs were opsonic in phagocytic assays.[39,40]
A study by Iwashkiw et al. attempting to make a novel glycoconjugate vaccine against Brucella abortus, the causative agent of brucellosis, demonstrated that PglB could be transformed and expressed directly within a host organism other than E. coli. The authors ‘hijacked’ Yersinia enterocolitica O9 O-antigen production by introducing PglB into the organism because this bacterium shares identical O-antigen structure to B. abortus. Transformation of a second plasmid coding for a glycosylatable acceptor protein resulted in glycoprotein generation within Y. enterocolitica.[41] This work demonstrated the possibility to rapidly obtain glycoconjugate vaccine candidates by introducing PglB directly into host organisms and our group among others are attempting this approach.
In summary, glycoconjugate vaccine production using PGCT has enormous potential to make a significant impact on bacterial disease, both in humans and in animals. The proof of principle experiments have been completed, and a number of vaccines are now in phase I clinical trials. There are, of course, considerable hurdles to overcome before vaccines generated with this technique become widely available. New vaccines generated against human bacterial pathogens require enormous financial backing. Not least for the costs of carrying out larger scale, testing of vaccine efficacy and then moving to tens of thousands of study participants required for phase 4 trials. However, the evidence is mounting that the technology works, and further technical modification are being developed to improve yield further.
Humanised glycoprotein production
Bacterial N-linked glycosylation systems are also being exploited to attempt to tap into a second multibillion dollar industry, namely generation of humanised glycoproteins. Unlike glycoconjugate vaccine production where significant similarities can be observed between assembly of the heptasaccharide that modifies C. jejuni proteins and O-antigen biosynthesis, the eukaryotic glycoprotein assembly mechanism is quite distinct. N-linked glycosylation in eukaryotes begins by assembly on a dolichol phosphate (DolP) lipid carried within the cytosolic compartment of the endoplasmic reticulum (ER). A core glycan of 14 units is assembled by the addition of monosaccharides through the action of glycosyltransferases. This core takes the form of Glc3Man9GlcNAc2 where Glc is glucose, Man is mannose and GlcNAc is N-acetyl glucosamine. The dolichol pyrophosphate (DolPP) loaded core sugar is flipped to the luminal side of the ER before the OTase transfers it onto a growing polypeptide, within the recognition sequon N-X-S/T. This step is important in enabling correct protein folding. The core glycan is trimmed within the ER by glucosidases and in some cases mannosidases, then enters the Golgi and is trimmed by class I mannosidases to generate a smaller core made of a pentasaccharide consisting of two N-acetyl glucosamines and three mannose residues. This structure forms the core of eukaryotic glycans; however, it can be further altered by various glycosyltransferases to form three groupings of modifications known as high mannose, complex and hybrid (Figure 4).[42] High mannose types have mannose residues added to the pentasaccharide core, complex types have no further mannose residues but have multiple branches with additions of N-acetyl glucosamine, and hybrid types have a combination of both mannose and N-acetyl glucosamine. Complex and hybrid types can have further additions of fucose and N-acetyl neuraminic acid to increase complexity. It should be stressed that in nature, glycoproteins show a high degree of heterogeneity in terms of glycan content and structure.
Figure 4.
Major classes of N-linked glycans. N indicates the asparagine residues onto which the reducing end sugar is attached. GlcNAc is N-acetyl glucosamine; NeuAc is N-acetyl neuraminic acid. Boxed area indicates core pentasaccharide structure present in all eukaryotic glycans.
It is estimated that over half of all eukaryotic proteins are glycosylated and that 90% of these carry an N-linked modification. A study utilising mass spectrometry to investigate the heterogeneity of human serum N-linked glycosylation revealed that complex type glycans made up 96% of the total glycans identified while hybrid and high mannose types made up the remainder.[43]
In eukaryotes, N-linked glycosylation plays multiple roles, from correct protein folding, stability, intracellular and intercellular trafficking to cell–cell interactions.[44] N-linked glycosylation is so important in humans that removal of this type of glycan modification is lethal.[45] Glycosylation is so important that 1–2% of our genome is dedicated to encoding for enzymes involved in glycan formation[46] and defects in enzymes involved in N-linked glycosylation can lead to a host of genetic disorders. For example, mutation of DPM1 coding for a Dol-P-mannosyltransferase leads to severe mental retardation, epilepsy and hypotonia. A host of muscular dystrophies are caused by defects in glycosylation pathways, an example is seen in Walker–Warburg syndrome caused by a defect in the O-mannosyltransferase I enzyme encoded by POMT1/POMT2 which can lead to cerebellar malformations, eye abnormalities and death in infancy.
Current manufacture
Sales of therapeutic proteins by pharmaceuticals companies reached over $63 billion in 2012; the highest selling class of biologics was made up of monoclonal antibodies (mAb), with US sales reaching $24.6 billion. The vast majority of these mAb are humanised by the addition of glycan modifications achieved through the expression of the protein of interest in mammalian cell lines; examples of such products are Actemra (Genentech, San Francisco, CA, USA), an anti IL-2 humanised IgG1k, prescribed for the treatment of arthritis. Xgeva (Amgen Inc., Thousand Oaks, CA, USA), the anti-RANK ligand human IgG2k mAb, prescribed to patients with bone metastases from solid tumours. The pharmaceutical company Roche alone has 11 monoclonals on the market,[47] demonstrating that the biotech sector is growing and that much of this growth is fuelled by glyco-protein production. The proven benefits of adding human-like glycan modifications to proteins include enhancing stability, reducing immunogenicity and increasing circulating half-life.[48]
When preparing for humanised glycoprotein production, a number of factors must be considered: the first is to determine if glycosylation is necessary for the therapeutic profile of the target glycoprotein. If not, then one must ascertain if this protein, without glycan modification, could become more immunogenic due to peptide unmasking.
If glycan modification is necessary, then one must understand if the glycan epitope can be simplified and still achieve full therapeutic activity. Finally, there is the added complexity of ensuring that glycosylation can be reproduced faithfully without batch-to-batch variation.[49]
To date, a number of alternative systems are employed to generate proteins with ‘human-like’ glycan modification. The industry standard is to use mammalian cell line expression systems such as Chinese hamster ovary (CHO) cells. These build the glycan on the same DolPP precursor and generate identical core sugar structures to human proteins. However, there are considerable difficulties with current approaches that leave the door open for alternative, cheaper, efficient, production strategies. Animals and humans differ in their glycan modifications when the glycans extend beyond the core pentasaccharide. Humans do not have the ability to synthesise N-glycolyneuraminic acid (Neu5Gc) due to an inactivation of cmah, coding for CMP-N-acetylneuraminic acid hydroxylase, utilising instead N-acetyl neuraminic acid (NeuAc).[50] Humans are also unable to produce N-linked glycans with terminal galactose 1–3 galactose residues (α Gal), in contrast to CHO cell lines. These modifications are actually immunogenic and high levels of contaminating Neu5Gc in proteins have been linked with a reduction in the circulating glycoprotein half-life.[51] Correct terminal sialic acid decoration on therapeutic glycoproteins is known to play a role in reducing clearance rate, increasing protein stability, reducing immunogenicity and increasing enzymatic activity.[52]
There are a number of further complications related to the use of mammalian cell lines. Difficulties in scalability mean that the process is expensive, mammalian cells require bovine serum albumin for culture of which there is a global shortage and there is also a concern of viral disease spread through the use of products generated by this approach.
Humanised glycoprotein production in bacteria
The discovery of PglB and the subsequent characterisation of its enzyme capabilities led researchers to speculate that humanised glycoprotein production might be a possibility within a prokaryotic host. Generation of a humanised glycoprotein within a bacterium is considered a ‘bottom-up’ approach because prokaryotic and eukaryotic N-linked glycans are structurally distinct. This is an attractive proposition because the method should produce a homogenous glycan modification. However, to succeed, a number of technical hurdles must be overcome. Transfer of a complete glycan biosynthetic pathway is not plausible primarily because in eukaryotes sugars are assembled on DolP while in bacteria polysaccharides are assembled on UndP. The specificity of C. jejuni PglB for a longer acceptor sequon (D/E-X-N-X-S/T) and the discovery that eukaryotic sites disfavour acidic amino acids near the glycosylated asparagines would also mean that the human acceptor protein would likely have to be modified, potentially altering protein characteristics. Finally, sugar biosynthetic pathways in the prokaryotic host may need to be altered to drive sugar precursors towards the required product.
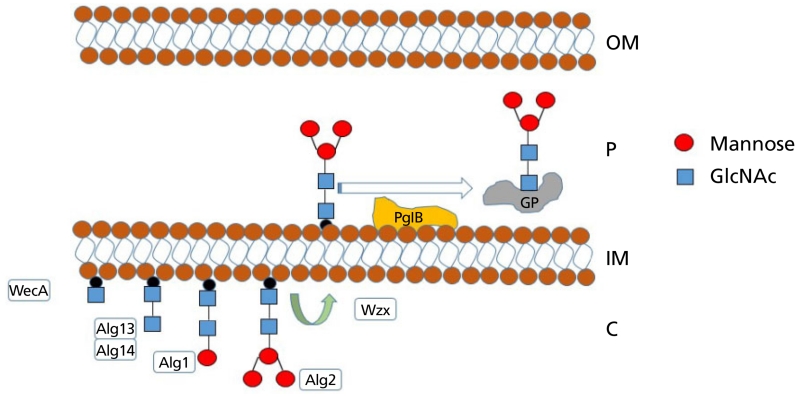
To resolve these limitations, Valderrama-Rincon et al. developed a hybrid approach where sugar biosynthesis was initiated using WecA (an initiating transferase used by E. coli to generate enterobacterial common antigen) to attach N-acetylglucosamine-1-phosphate (GlcNAc-1-phosphate) to UndP. Crucially, this is the same initial sugar seen in eukaryotic N-linked glycans and provides a ‘priming’ site onto which glycan biosynthesis may progress. The addition of Alg14/13/1/2 from Saccharomyces cerevisiae continued the process to make an intact eukaryotic pentasaccharide core (Figure 5). To increase the availability of guanosine diphosphate-mannose (GDP-mannose) and aid in the assembly of the mannose-containing core, the authors also deleted a GDP-mannose dehydratase, therefore preventing GDP-mannose from continuing its normal conversion to GDP-L-fucose.
Figure 5.
Schematic demonstrating prokaryotic assembly of the eukaryotic glycan pentasaccharide core and transfer to an acceptor protein. WecA attaches GlcNAc to UndP, Alg13/14 add a second GlcNAc residue, before Alg1 and Alg2 attach three mannose residues to generate GlcNAc2Man3 core. The Escherichia coli flippase, Wzx, then moves the UndPP pentasaccharide to the periplasmic compartment of the cell where upon recognition by PglB, the glycan moiety is transferred to GP within the acceptor sequon D-X-N-X-S/T. GP, glycoprotein.
Valderrama-Rincon et al. demonstrate glycosylation of a number of human glycoprotein targets, including the Fc domain of IgG1, by adding a native periplasmic signal sequence to the human protein and introducing C. jejuni PglB into this E. coli strain. As expected, the Fc domain only became glycosylated with the pentasaccharide once the acceptor sequon had been modified and extended to DQNAT.
The work demonstrated proof of principle that humanised glycoproteins could soon be assembled in E. coli. The elegance of this approach is that by using S. cerevisiae enzymes, the linkages between individual sugars were confirmed to be identical to the human N-linked glycan core.[53]
The glycan would, most likely still, require a number of additions to generate a complex class of modification but this is an impressive starting point. A recent study by Meuris et al. indicates that if the therapeutic goal is antigen neutralisation, the epitope could be shortened to GlcNAc-Gal–NeuAc,[54] making in-vivo glycoprotein assembly in bacteria a strong possibility.
Next generation sequencing technologies have led to the identification of a plethora of PglB orthologues, even outside of the epsilonproteobacteria subdivision. Discovery and characterisation of a PglB orthologues in the deltaproteobacterium Desulfovibrio desulfuricans (PglBDD) revealed that this enzyme could transfer mono- and polysaccharides when expressed heterologously in E. coli. Studies also revealed that the enzyme had an altered amino acid specificity, requiring only the eukaryotic sequon (N-X-S/T) to be present for glycosylation to occur.[34]
A PglB capable of efficiently glycosylating proteins using shortened eukaryotic sequons would have clear advantages in making humanised glycoproteins when compared with C. jejuni PglB. Unfortunately, PglBDD does not appear to be the answer, as glycosylation efficiency is much reduced compared with C. jejuni PglB. In addition, it appears as though PglBDD was unable to modify the wild-type CH2 domain of human IgG1.[53] The characterisation of other PglB orthologues is expected to eventually resolve this sequon dilemma. Alternatively, the structure of C. lari PglB has revealed important clues regarding the amino acid residues responsible for extended sequon recognition; this means that there is a possibility that C. jejuni PglB could be mutated to make a variant capable of N-X-S/T recognition, although it is unknown what influence this may have to glycoprotein yield.[33]
The discovery and characterisation of a cytoplasmic N-linking glycosyltransferase (NGT), in the respiratory swine pathogen A. pleuropneumoniae, has also provided an alternative approach to in-vivo expression systems. NGT is distinct from PglB because it uses UDP-Glu as the sugar substrate and catalyses formation of the glycosidic bond directly onto N-X-S/T acceptor sequons within a protein.[55] Lomino et al. demonstrated that using a chemoenzymatic approach, the Glc-peptide linkage could be used as a priming site for transglycosylation by endoglycosidases. This allowed build up of high mannose, asialylated complex and fully sialylated complex bi-antennary glycans, even demonstrating modification of C34, a potent human immunodeficiency virus (HIV) fusion inhibitor derived from HIV-1 gp41 envelope glycoprotein.[56] Further work is required to demonstrate the efficacy of this approach on a range of human proteins, and the scalability of the process.
Finally, Hug et al. report the generation of a Lewis × (Lex) glycan epitope consisting of Galβ1-4[Fucα1-3]GlcNAc (where Fuc is fucose) on a carrier protein to generate a potential immunosuppressed protein. The authors utilised the E. coli WecA protein to attach a priming GlcNAc onto which the Lex epitope could be completed. This was achieved by using glycosyltransferases from the H. influenzae lipooligosaccharide biosynthetic cluster. The fucose modification was added in vitro to complete the sugar structure.[57]
Conclusions and future perspectives
This review focused on N-linked glycosylation and Table 1 summarises key publications demonstrating utilisation of this modification system; included in these highlights are a number of studies that have overcome current technical limitations of humanised glycoprotein expression within E. coli by combining in-vivo and in-vitro enzymatic steps. Bacteria also carry several O-linked glycosylation systems, and there is also potential for these systems to be exploited in the manufacture of glycoconjugate vaccines. O-linked OTases are related to WaaL ligase enzymes; the biggest advantage of PglL-based enzymes is that they do not have the current C. jejuni OTase limitation of an absolute requirement of an acetamido group at the C2 position of the reducing end sugar. Currently however, a peptide consensus sequon has yet to be identified meaning that acceptor proteins are strictly limited.[20,59]
Table 1.
Publications related to utilisation of N-linked glycosylation for glycoconjugate vaccine and humanised glycoprotein production
| Study | Key findings |
|---|---|
| Production of glycoprotein vaccines in Escherichia coli. By Ihssen et al.[35] | First report of a glycoconjugate vaccine generated using PGCT. Shigella dysenteriae O1 antigen coupled to EPA. |
| Exploitation of bacterial N-linked glycosylation to develop a novel recombinant glycoconjugate vaccine against Francisella tularensis. By Cuccui et al.[36] | Stepwise demonstration of glycoconjugate vaccine development and efficacy testing in animal models. Francisella tularensis subs. tularensis O-antigen coupled to EPA. |
| Prevention of Staphylococcus aureus infections by glycoprotein vaccines synthesized in Escherichia coli. By Wacker et al.[37] | First demonstration of glycoconjugate vaccines developed against a Gram-positive organisms. Demonstration of efficacy using Staphylococcus aureus serotype 5 and 8 capsular polysaccharide attached to EPA as the carrier protein and demonstration of increased efficacy when the carrier protein is switched to the S. aureus-specific protein Hla. |
| An engineered eukaryotic protein glycosylation pathway in Escherichia coli. By Valderrama-Rincon et al.[53] | Demonstration of humanised glycoprotein production within Escherichia coli by combining E. coli, Saccharomyces cerevisiae and Campylobacter jejuni enzymes. Eukaryotic core pentasaccharide Man3GlcNAc2 assembled and transferred onto the eukaryotic proteins IgG1, bovine RNase A and human growth hormone. |
| A two-step enzymatic glycosylation of polypeptides with complex N-glycans. By Lomino et al.[56] | Utilised a novel N-linking glycosyltransferase from Actinobacillus pleuropneumoniae to form a priming sugar onto which complex N-glycans could be assembled in vitro through the use of an endoglycosidase (ENGase) catalysed transglycosylation. This method was used to generate a complex glycoform on an HIV inhibitor. |
| Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. By Hug et al.[57] | By combining WecA from Escherichia coli to attach GlcNAc onto the lipid carrier UndPP and the Haemophilus influenzae lipooligosaccharide (LOS) biosynthesis cluster a precursor glycolipid GalGlcNAcGalGlcNAc-UndPP was formed and transferred to the acceptor protein AcrA. Addition of the fucose residue to the exterior GlcNAc residue by Helicobacter pylori FucT in vitro completed the synthesis of the Lewis x-glycoprotein. |
| A combined method for producing homogeneous glycoproteins with eukaryotic N-glycosylation. By Schwarz et al.[58] | Humanised glycoprotein were generated by altering the Pgl locus from Campylobacter jejuni. A (GalNAc)5GlcNAc-PP-undecaprenyl scaffold was assembled and presenting this hexasaccharide to PglB within an Escherichia coli cell resulted in transfer to an acceptor protein. Finally, in-vitro endoglycosidase-catalysed transglycosylation using pre-assembled N-glycan species as substrates gave rise to a homogenous pool of glycoproteins. |
Bacterial glycosylation systems are also attractive targets for new class of antimicrobials that target glycosyltransferases[60] because these systems are often critical for full virulence.
Recent research is providing compelling evidence for protein glycosylation being central to the survival and pathogenesis of many bacteria[61-64] and thus represents an ideal target to disable them. We consider bacterial glycosylation systems a largely untapped resource to counteract the continued threat posed by bacterial pathogens, either through the development of novel antimicrobials or via glycoconjugate vaccines. However, to fully realise this benefit, and in the longer term to produce recombinant human glycoproteins of considerable therapeutic value, a better understanding of the basic mechanisms and pathways of bacterial glycosylation is essential. Glycoconjugate vaccine production stalled for a number of years, primarily due to the discovery of antibiotics. We are living in an era where the rise of multidrug resistant bacteria mean that alternative vaccine formulations are desperately needed. This is especially true in susceptible young children within the developing world.
The N-linked glycosylation system from C. jejuni is a powerful tool for glycoconjugate vaccine development, the most advanced vaccine generated using PGCT, targeting S. dysenteriae has passed phase 1 clinical trials, and a glycoconjugate vaccine against extra-intestinal pathogenic E. coli has commenced phase 1 testing.
It should be noted that there is potential for this technology to be exploited to develop glycoconjugate vaccines against animal pathogens; to achieve this, novel genetic tools are being developed and tested by our labs and others.
Alternative uses for bacterial N-linked glycosylation systems are being explored and some successes have been reported in the generation of ‘human-like’ glycans. A eukaryotic pentasaccharide core can now be assembled using the initiating transferase WecA as the starting sugar and combining this with S. cerevisiae glycosyltransferases to build the pentasaccharide. However, this technical application is still in its infancy and further modifications are required to make full-length humanised glycoprotein. The complexity of human glycoproteins is such that the type of modification will likely need to be different depending on the target protein being modified and the therapeutic effect aimed for.
Finally, it should be stressed that glycosylation systems are abundant in all kingdoms of life and await biotechnological exploitation.
Acknowledgements
We thank Dr Emily Kay, Dr Vanessa Terra and Mr Sherif Abouelhadid for editing and for their critical reading of this manuscript.
References
- 1.Coutinho A, Moller G. B cell mitogenic properties of thymus-independent antigens. Nat New Biol. 1973;245:12–14. doi: 10.1038/newbio245012a0. [DOI] [PubMed] [Google Scholar]
- 2.Timens W, et al. Immaturity of the human splenic marginal zone in infancy. Possible contribution to the deficient infant immune response. J Immunol. 1989;143:3200–3206. [PubMed] [Google Scholar]
- 3.Shi Y, et al. Regulation of aged humoral immune defense against pneumococcal bacteria by IgM memory B cell. J Immunol. 2005;175:3262–3267. doi: 10.4049/jimmunol.175.5.3262. [DOI] [PubMed] [Google Scholar]
- 4.Avery OT, Goebel WF. Chemo-immunological studies on conjugated carbohydrate-proteins: II. Immunological specificity of synthetic sugar-protein antigens. J Exp Med. 1929;50:533–550. doi: 10.1084/jem.50.4.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schneerson R, et al. Preparation, characterization, and immunogenicity of Haemophilus influenzae type b polysaccharide-protein conjugates. J Exp Med. 1980;152:361–376. doi: 10.1084/jem.152.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schneerson R, et al. Haemophilus influenzae type B polysaccharide-protein conjugates: model for a new generation of capsular polysaccharide vaccines. Prog Clin Biol Res. 1980;47:77–94. [PubMed] [Google Scholar]
- 7.Kuberan B, Linhardt RJ. Carbohydrate based vaccine. Curr Org Chem. 2000;4:653–677. [Google Scholar]
- 8.Brady C, et al. A new paradigm for rapidly translating novel conjugate vaccines into the clinic. Bioprocess Tech. 2012;10:50–55. [Google Scholar]
- 9.Watt JP, et al. Burden of disease caused by Haemophilus influenzae type b in children younger than 5 years: global estimates. Lancet. 2009;374:903–911. doi: 10.1016/S0140-6736(09)61203-4. [DOI] [PubMed] [Google Scholar]
- 10.Ladhani SN. Two decades of experience with the Haemophilus influenzae serotype b conjugate vaccine in the United Kingdom. Clin Ther. 2012;34:385–399. doi: 10.1016/j.clinthera.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 11.Bentley SD, et al. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet. 2006;2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grijalva CG, et al. Decline in pneumonia admissions after routine childhood immunisation with pneumococcal conjugate vaccine in the USA: a time-series analysis. Lancet. 2007;369:1179–1186. doi: 10.1016/S0140-6736(07)60564-9. [DOI] [PubMed] [Google Scholar]
- 13.Yazdankhah SP, Caugant DA. Neisseria meningitidis: an overview of the carriage state. J Med Microbiol. 2004;53(Pt 9):821–832. doi: 10.1099/jmm.0.45529-0. [DOI] [PubMed] [Google Scholar]
- 14.Daugla DM, et al. Effect of a serogroup A meningococcal conjugate vaccine (PsA-TT) on serogroup A meningococcal meningitis and carriage in Chad: a community study [corrected] Lancet. 2014;383:40–47. doi: 10.1016/S0140-6736(13)61612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Avci FY, et al. A mechanism for glycoconjugate vaccine activation of the adaptive immune system and its implications for vaccine design. Nat Med. 2011;17:1602–1609. doi: 10.1038/nm.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sleytr UB, Thorne KJ. Chemical characterization of the regularly arranged surface layers of Clostridium thermosaccharolyticum and Clostridium thermohydrosulfuricum. J Bacteriol. 1976;126:377–383. doi: 10.1128/jb.126.1.377-383.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott NE, et al. Simultaneous glycanpeptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics. 2011;10:M000031–MCP201. doi: 10.1074/mcp.M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grass S, et al. The Haemophilus influenzae HMW1 adhesin is glycosylated in a process that requires HMW1C and phosphoglucomutase, an enzyme involved in lipooligosaccharide biosynthesis. Mol Microbiol. 2003;48:737–751. doi: 10.1046/j.1365-2958.2003.03450.x. [DOI] [PubMed] [Google Scholar]
- 19.McCann JR, St Geme JW., III The HMW1C-like glycosyltransferases-an enzyme family with a sweet tooth for simple sugars. PLoS Pathog. 2014;10:e1003977. doi: 10.1371/journal.ppat.1003977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faridmoayer A, et al. Functional characterization of bacterial oligosaccharyltransferases involved in O-linked protein glycosylation. J Bacteriol. 2007;189:8088–8098. doi: 10.1128/JB.01318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benz I, Schmidt MA. Glycosylation with heptose residues mediated by the aah gene product is essential for adherence of the AIDA-I adhesin. Mol Microbiol. 2001;40:1403–1413. doi: 10.1046/j.1365-2958.2001.02487.x. [DOI] [PubMed] [Google Scholar]
- 22.Fry BN, et al. The lipopolysaccharide biosynthesis locus of Campylobacter jejuni 81116. Microbiology. 1998;144(Pt 8):2049–2061. doi: 10.1099/00221287-144-8-2049. [DOI] [PubMed] [Google Scholar]
- 23.Szymanski CM, et al. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill J, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 25.Young NM, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–42539. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 26.Linton D, et al. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol. 2005;55:1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- 27.Feldman MF, et al. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci U S A. 2005;102:3016–3021. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wacker M, et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci U S A. 2006;103:7088–7093. doi: 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petrescu AJ, et al. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- 30.Kowarik M, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowarik M, et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 2006;314:1148–1150. doi: 10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- 32.Chen MM, et al. Polyisoprenol specificity in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2007;46:14342–14348. doi: 10.1021/bi701956x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lizak C, et al. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474:350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- 34.Ielmini MV, Feldman MF. Desulfovibrio desulfuricans PglB homolog possesses oligosaccharyl-transferase activity with relaxed glycan specificity and distinct protein acceptor sequence requirements. Glycobiology. 2011;21:734–742. doi: 10.1093/glycob/cwq192. [DOI] [PubMed] [Google Scholar]
- 35.Ihssen J, et al. Production of glycoprotein vaccines in Escherichia coli. Microb Cell Fact. 2010;9:61. doi: 10.1186/1475-2859-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuccui J, et al. Exploitation of bacterial N-linked glycosylation to develop a novel recombinant glycoconjugate vaccine against Francisella tularensis. Open Biol. 2013;3:130002. doi: 10.1098/rsob.130002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wacker M, et al. Prevention of Staphylococcus aureus infections by glycoprotein vaccines synthesized in Escherichia coli. J Infect Dis. 2014;209:1551–1561. doi: 10.1093/infdis/jit800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fisher AC, et al. Production of secretory and extracellular N-linked glycoproteins in Escherichia coli. Appl Environ Microbiol. 2011;77:871–881. doi: 10.1128/AEM.01901-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byrd W, et al. Protective efficacy of conjugate vaccines against experimental challenge with porcine Actinobacillus pleuropneumoniae. Vet Immunol Immunopathol. 1992;34:307–324. doi: 10.1016/0165-2427(92)90172-m. [DOI] [PubMed] [Google Scholar]
- 40.Byrd W, Kadis S. Preparation, characterization, and immunogenicity of conjugate vaccines directed against Actinobacillus pleuropneumoniae virulence determinants. Infect Immun. 1992;60:3042–3051. doi: 10.1128/iai.60.8.3042-3051.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iwashkiw JA, et al. Exploiting the Campylobacter jejuni protein glycosylation system for glycoengineering vaccines and diagnostic tools directed against brucellosis. Microb Cell Fact. 2012;11:13. doi: 10.1186/1475-2859-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zielinska DF, et al. Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol Cell. 2012;46:542–548. doi: 10.1016/j.molcel.2012.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Chu CS, et al. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varki A, Lowe JB. Biological roles of glycans. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd edn. Cold Spring Harbor; Huntington, NY: 2009. pp. 1–14. [Google Scholar]
- 45.Varki A, et al. Glycans in development and systemic physiology. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd edn. Cold Spring Harbor; Huntington, NY: 2009. pp. 1–6. [PubMed] [Google Scholar]
- 46.Bertozzi CR, et al. Glycans in biotechnology and the pharmaceutical industry. In: Varki A, et al., editors. Essentials of Glycobiology. 2nd edn. Cold Spring Harbor; Huntington, NY: 2009. pp. 1–14. [PubMed] [Google Scholar]
- 47.Aggarwal RS. What’s fueling the biotech engine-2012 to 2013. Nat Biotechnol. 2014;32:32–39. doi: 10.1038/nbt.2794. [DOI] [PubMed] [Google Scholar]
- 48.Ghaderi D, et al. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnol Genet Eng Rev. 2012;28:147–175. doi: 10.5661/bger-28-147. [DOI] [PubMed] [Google Scholar]
- 49.Jenkins N, et al. Getting the glycosylation right: implications for the biotechnology industry. Nat Biotechnol. 1996;14:975–981. doi: 10.1038/nbt0896-975. [DOI] [PubMed] [Google Scholar]
- 50.Nystedt J, et al. Human CMP-N-acetylneuraminic acid hydroxylase is a novel stem cell marker linked to stem cell-specific mechanisms. Stem Cells. 2010;28:258–267. doi: 10.1002/stem.250. [DOI] [PubMed] [Google Scholar]
- 51.Flesher AR, et al. Fluorophore-labeled carbohydrate analysis of immunoglobulin fusion proteins: correlation of oligosaccharide content with in vivo clearance profile. Biotechnol Bioeng. 1995;46:399–407. doi: 10.1002/bit.260460502. [DOI] [PubMed] [Google Scholar]
- 52.Son Y-D, et al. Enhanced sialylation of recombinant human erythropoietin in Chinese hamster ovary cells by combinatorial engineering of selected genes. Glycobiology. 2011;21:1019–1028. doi: 10.1093/glycob/cwr034. [DOI] [PubMed] [Google Scholar]
- 53.Valderrama-Rincon JD, et al. An engineered eukaryotic protein glycosylation pathway in Escherichia coli. Nat Chem Biol. 2012;8:434–436. doi: 10.1038/nchembio.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meuris L, et al. GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat Biotechnol. 2014;32:485–489. doi: 10.1038/nbt.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naegeli A, et al. Molecular analysis of an alternative N-glycosylation machinery by functional transfer from Actinobacillus pleuropneumoniae to Escherichia coli. J Biol Chem. 2014;289:2170–2179. doi: 10.1074/jbc.M113.524462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lomino JV, et al. A two-step enzymatic glycosylation of polypeptides with complex N-glycans. Bioorg Med Chem. 2013;21:2262–2270. doi: 10.1016/j.bmc.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hug I, et al. Exploiting bacterial glycosylation machineries for the synthesis of a Lewis antigen-containing glycoprotein. J Biol Chem. 2011;286:37887–37894. doi: 10.1074/jbc.M111.287755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwarz F, et al. A combined method for producing homogenous glycoproteins with eukaryotic N-glycosylation. Nat Chem Biol. 2010;6:264–266. doi: 10.1038/nchembio.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Faridmoayer A, et al. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J Biol Chem. 2008;283:34596–34604. doi: 10.1074/jbc.M807113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Descroix K, et al. Inhibition of galactosyltransferases by a novel class of donor analogues. J Med Chem. 2012;55:2015–2024. doi: 10.1021/jm201154p. [DOI] [PubMed] [Google Scholar]
- 61.Iwashkiw JA, et al. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog. 2012;8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerry P, et al. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60:299–311. doi: 10.1111/j.1365-2958.2006.05100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karlyshev AV, et al. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology. 2004;150(Pt 6):1957–1964. doi: 10.1099/mic.0.26721-0. [DOI] [PubMed] [Google Scholar]
- 64.Lithgow KV, et al. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol Microbiol. 2014;92:116–137. doi: 10.1111/mmi.12540. [DOI] [PubMed] [Google Scholar]