Abstract
IMPORTANCE
Most evidence to date highlights the importance of genetic influences on the liability to autism and related traits. However, most of these findings are derived from clinically ascertained samples, possibly missing individuals with subtler manifestations, and obtained estimates may not be representative of the population.
OBJECTIVES
To establish the relative contributions of genetic and environmental factors in liability to autism spectrum disorder (ASD) and a broader autism phenotype in a large population-based twin sample and to ascertain the genetic/environmental relationship between dimensional trait measures and categorical diagnostic constructs of ASD.
DESIGN, SETTING, AND PARTICIPANTS
We used data from the population-based cohort Twins Early Development Study, which included all twin pairs born in England and Wales from January 1, 1994, through December 31, 1996. We performed joint continuous-ordinal liability threshold model fitting using the full information maximum likelihood method to estimate genetic and environmental parameters of covariance. Twin pairs underwent the following assessments: the Childhood Autism Spectrum Test (CAST) (6423 pairs; mean age, 7.9 years), the Development and Well-being Assessment (DAWBA) (359 pairs; mean age, 10.3 years), the Autism Diagnostic Observation Schedule (ADOS) (203 pairs; mean age, 13.2 years), the Autism Diagnostic Interview–Revised (ADI-R) (205 pairs; mean age, 13.2 years), and a best-estimate diagnosis (207 pairs).
MAIN OUTCOMES AND MEASURES
Participants underwent screening using a population-based measure of autistic traits (CAST assessment), structured diagnostic assessments (DAWBA, ADI-R, and ADOS), and a best-estimate diagnosis.
RESULTS
On all ASD measures, correlations among monozygotic twins (range, 0.77-0.99) were significantly higher than those for dizygotic twins (range, 0.22-0.65), giving heritability estimates of 56% to 95%. The covariance of CAST and ASD diagnostic status (DAWBA, ADOS and best-estimate diagnosis) was largely explained by additive genetic factors (76%-95%). For the ADI-R only, shared environmental influences were significant (30% [95% CI, 8%-47%]) but smaller than genetic influences (56% [95% CI, 37%-82%]).
CONCLUSIONS AND RELEVANCE
The liability to ASD and a more broadly defined high-level autism trait phenotype in this large population-based twin sample derives primarily from additive genetic and, to a lesser extent, nonshared environmental effects. The largely consistent results across different diagnostic tools suggest that the results are generalizable across multiple measures and assessment methods. Genetic factors underpinning individual differences in autismlike traits show considerable overlap with genetic influences on diagnosed ASD.
Twin studies of autism,1-6 conducted from 1977 onward, provided the first clear evidence that genetic factors were etiologically important. Recent reviews of this literature5,7-9 show general agreement across studies that concordance for autism in monozygotic (MZ) twin pairs is typically at least double that in dizygotic (DZ) twin pairs, resulting in high heritability estimates (60%-90%)10-14 and suggesting little influence of shared environmental factors. Two twin studies15,16 stand in contrast and reported only moderate heritability (21%-38%), with a substantial shared environmental component explaining 58% to 78% of the variance in liability to autism spectrum disorder (ASD). In comparison, 1 recent twin study did not confirm significant shared environmental effects and reported heritability of 95%.17 In addition, a large population study of extended families (approximately 2 million individuals)18 reported estimates of 50% for heritability and nonshared environmental factors. Most recently, in the same population, molecular genetic analysis19 indicated that 95% of variance in ASD is accounted for by common allelic variants, supporting a polygenic model. This finding contrasts markedly with heritability estimates of around 0 derived from single-nucleotide polymorphism data (GCTA) in an arguably underpowered sample.20 Given the interest in possible environmental factors in the etiology of autism, these contradictory findings have reopened the discussion of high heritability and the possibility that findings may be biased by sample selection and screening. The first aim of the present study was to examine the relative importance of genetic and environmental factors in liability to ASD in a large systematically screened, population-based twin sample.
Twin and family studies21,22 have also shown that the genetic liability to autism confers a risk for a broader range of impairments in social communication, restricted and repetitive behaviors, and behaviors that extend beyond the traditional diagnostic boundaries for autism.9,23,24 These pioneering studies contributed to the revision and broadening of diagnostic criteria and to the conceptualization of autism as a spectrum encompassing subtypes of pervasive developmental disorders, such as Asperger syndrome, atypical autism, and subtler presentations.25,26 Research27-31 has explored autismlike traits in community samples and provided evidence of a genetic correlation between autismlike traits at the extremes and in the rest of the population. Our second aim was therefore to quantify the genetic and environmental relationship between dimensional trait measures and the categorical diagnostic constructs of ASD (from criterion-standard instruments), which, to our knowledge, has not been tried before.
To provide a more definitive picture that addresses these 2 aims and incorporates current diagnostic concepts, we used rigorous approaches and screened an age-specific epidemiologic sample of twins to ascertain all twins with possible ASD. We then undertook independent, in-depth evaluations using an additional screening instrument and well-established diagnostic assessment tools. The purpose was to minimize methodological artifacts and provide results that can be used as a benchmark for comparison in future research.
Our approach is novel and contrasts with those of other recent twin and family studies in sample ascertainment and analytic methods. Previous studies15,16 have identified their twin samples through clinical services. Such a strategy could result in sampling bias; if registration or participation is influenced by concordance, probandwise concordance rates in DZ twin pairs might be increased, resulting in inflated estimates of common environmental influence. In addition, sole reliance on clinical ascertainment could result in underidentification of cases with high levels of functioning.32 Investigators should include these cases and define the genetic liability as a continuous distribution that extends beyond stringent diagnostic categories. Our study is novel in using criterion-standard, in-person, clinical diagnostic tools with a population-based (vs a clinic-based) sample in which ascertainment was good (62.1% response rate from the eligible sample compared with 17% in the study by Hallmayer et al15).
In summary, our first aim was to estimate heritability of the liability to ASD using a population-based sample that was selected using several screening instruments sent to all twins in a 3-year birth cohort. The second aim was to study the genetic/environmental relationship between dimensional trait measures and categorical diagnostic constructs of ASD. In contrast to previous approaches,15,18 we assumed a continuous liability distribution underlying ASD and a more broadly defined phenotype with high-level autism traits that fell short of thresholds for an ASD diagnosis. We predicted a strong genetic overlap between dimensional and diagnostic measures in keeping with previous twin analyses based on extreme cases.21-23
Methods
Participants
The participants were recruited from the Twins Early Development Study,33 a longitudinal study of twin pairs ascertained from population records of twin births in England and Wales from January 1, 1994, through December 31, 1996. The Twins Early Development Study sample is considered representative of the population of the United Kingdom in terms of maternal ethnicity (92.8% white) and educational level (40.1% with A level qualifications or higher, the equivalent of some college education in the United States). Ethical authorization, including authorization to work with children, was given by the Institute of Psychiatry ethics committee. Parents were given a letter describing the general purpose of the study, and written parental consent was required. Participation was voluntary and participants could withdraw from the study whenever they wished.
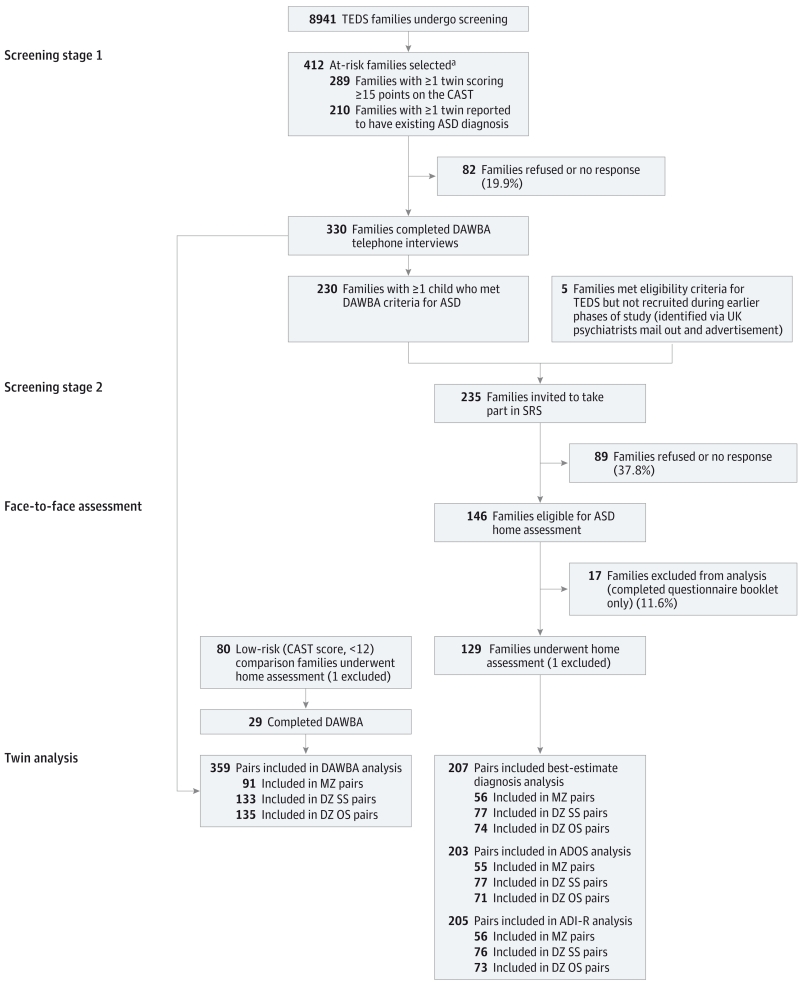
The ASD and co-twin sample were selected after a 2-stage screening process outlined in Figure 1 and section 1 of eAppendix 1 in the Supplement. Of the 412 eligible families for the Social Relationship Study at stage 1, 80.1% completed the Development and Well-being Assessment (DAWBA) interview.34,35 At stage 2, 62.1% of the 235 eligible families underwent diagnostic evaluations. Two researchers worked with each family. One researcher administered the Autism Diagnostic Interview–Revised (ADI-R)36 and the other administered the Autism Diagnostic Observation Schedule (ADOS)37 for the first twin; the reseachers then swapped assessments for the second twin. This design meant that different assessors administered the ADI-R and ADOS assessments within each pair to minimize any effects of rater bias.
Figure 1. Social Relationship Study (SRS) Sample Selection Stages and the Overall Number of Participants Included in the Analysis.
ADI-R indicates Autism Diagnostic Interview–Revised; ADOS, Autism Diagnostic Observation Schedule; ASD, autism spectrum disorder; DAWBA, Development and Well-being Assessment; DZ, dizygotic; MZ, monozygotic; OS, opposite-sex; SS, same-sex; and TEDS, Twins Early Development Study.
aSome twins met both criteria, so numbers total more than 412.
Within the final sample, participants with ASD were broadly comparable to those eligible for participation (score of ≥15 on the Childhood Autism Spectrum Test [CAST]25 or with suspected ASD) but who did not take part (zygosity, [P = .23]; socioeconomic status, t397 = −1.2 [P = .25, independent t test with 2-tailed significance]; and CAST result, t420 = −1.5 [P = .14, independent t test with 2-tailed significance]), with the exception of sex ( [P < .001]). Among the group with high CAST scores or suspected ASD, 36.4% were female compared with 16.6% of the final sample.
Measures
Childhood Autism Spectrum Test
The CAST is an informant-completed questionnaire based on behavioral descriptions of ASD as delineated in the International Statistical Classification of Diseases, Tenth Revision (ICD-10) and the DSM-IV. The 31 items are scored yes or no and summed; a cutoff score of 15 or greater is reported to have 100% sensitivity, 97% specificity, and a positive predictive value of 50% for a diagnosis of ASD.28 The CAST data from at least 1 twin were available from 6423 pairs (MZ pairs, 2261; DZ same-sex pairs, 2097; and DZ opposite-sex pairs, 2065), with a mean (SD) age of 7.9 (0.5) years. Of these, 289 pairs (4.5% of all pairs; 317 individuals) had scores greater than the cutoff value.
Development and Well-being Assessment
Telephone interviews using the ASD module of the DAWBA were performed at the second stage35 and included 15 questions about social difficulties; 14 questions about repetitive, restricted behaviors and interests; and 3 questions about developmental language milestones. The same parent rated both twins during a telephone call with a single interviewer. A child received a DAWBA diagnosis of autism when the operational criteria in the DSM-IV and ICD-10 were met. A diagnosis of Asperger syndrome was given when parent reports indicated that all autism criteria were met but the child’s early language development was not delayed and the child’s intellectual ability was in the normal range. A diagnosis of ASD (other) was assigned if the parents reported a minimum of 3 probable or 2 definite symptoms from the social difficulties domain, 2 probable or 1 definite symptom from the communication domain, and 2 probable or 1 definite symptom from the repetitive, restricted behaviors and interests domain. The measure used in analysis was a 3-category diagnosis of ASD, where 0 indicates no ASD/controls; 1, ASD (other); and 2, Asperger syndrome or autism.
Autism Diagnostic Interview-Revised
The ADI-R is a well-established diagnostic tool for the assessment of autism.36 It consists of a semistructured caregiver interview inquiring about current function and developmental history (93 items) and is administered by a trained investigator. We used criteria from the Autism Genetics Resource Exchange (http://agre.autismspeaks.org/site/c.lwLZKnN1LtH/b.5332889/k.B473/AGRE.htm) to assign cases to 1 of the following 3 categories: ASD (consisting of Autism Genetics Resource Exchange categories autism and not quite autism), broad-spectrum disorder, and unaffected (operational definitions are given in eAppendix 2 in the Supplement). The measure used in the analysis was a 3-category diagnosis of ASD, where 0 indicates no ASD/controls; 1, broad-spectrum disorder; and 2, ASD.
Autism Diagnostic Observation Schedule
The ADOS is a well-validated, semistructured observational assessment designed to accompany the ADI-R in the diagnosis of ASD.37 The present study used recent updates to the ADOS algorithm (Catherine Lord, PhD, written communication, 2008; described in eAppendix 2 in the Supplement) to yield scores for communication, social interaction, and restricted behaviors and interests. Clinical cutoffs were available for ASD and autism, and these diagnostic groups were combined to create a single ASD category. An additional broad-spectrum category included individuals who scored just below the cutoff (−2 points) to correspond to the broad-spectrum category on the ADI-R. The measure used in the analysis was a 3-category diagnosis of ASD, where 0 indicates no ASD/controls; 1, broad-spectrum disorder; and 2, ASD.
Best-Estimate Diagnosis
Diagnoses were assigned with investigators blinded to zygosity and co-twin diagnostic status after review of all available information (ADI-R, ADOS, and DAWBA assessments and clinical reports). When all available sources of information were in agreement, cases were assigned to that category. In 89 cases, the diagnostic classifications across instruments were inconsistent. In these cases, all available data were assessed by expert clinicians (E.C., S.R.C., and/or P.B.), and best-estimate diagnoses (BeDs) were assigned based on this review. Additional details are given in eAppendix 2 in the Supplement. Best-estimate diagnosis was used in the analysis as a 3-category measure of ASD, where 0 indicates no ASD/controls; 1, broad-spectrum disorder; and 2, ASD.
Data Analysis
Twin Correlations
Twin data analysis was performed in the structural equation modeling program OpenMx.38 We used the full information maximum likelihood estimation to jointly analyze the continuous (CAST) and ordinal (ASD) measures, assuming a liability threshold model to reflect the risk for ASD.39,40 To obtain unbiased estimates, thresholds were fixed to z values corresponding to the following known population prevalences because of the selection of individuals with ASD: first at 5%,41,42 discriminating between unaffected and broad-spectrum disorder, and second at 1%, discriminating between broad-spectrum disorder and ASD. The assumption of a joint multivariate normal distribution for CAST scores and the 3 ASD diagnostic categories (unaffected, broad-spectrum disorder, and ASD) allowed the estimation of correlations within and across MZ/DZ twin pairs. The MZ:DZ ratios of these correlations indicate the relative importance of genetic and environmental influences on variation within each measure and on the covariance between them; these correlations were formally tested in the bivariate genetic model.
The Bivariate Genetic Model
With the use of biometrical genetic theory, the covariance of the CAST score and each ASD diagnosis was modeled as the effects of additive genetic (A), shared environmental (C), and nonshared environmental and measurement error(E) factors.43 Because the order of traits is immaterial, we interpreted the standardized solution in which the paths from the A1 factor to the CAST score and the A2 factor to ASD are the square roots of their respective heritabilities, and the correlation between A1 and A2 is the genetic correlation between them (rA).44,45 The same logic applies to the nonshared environmental effects (Figure 2). Shared environmental factors were modeled on ASD only; they do not influence the variance of the CAST scores46 and therefore cannot explain the covariance with ASD. In addition to the standardized path estimates, we calculated the phenotypic correlation (rph) due to genetic effects (rph_A) as √h12 × rA × √h22 and the phenotypic correlation due to nonshared environmental and measurement error effects (rph_E) as √e12 × rE × √ e22, which can be expressed as proportions of phenotypic correlation.47,48
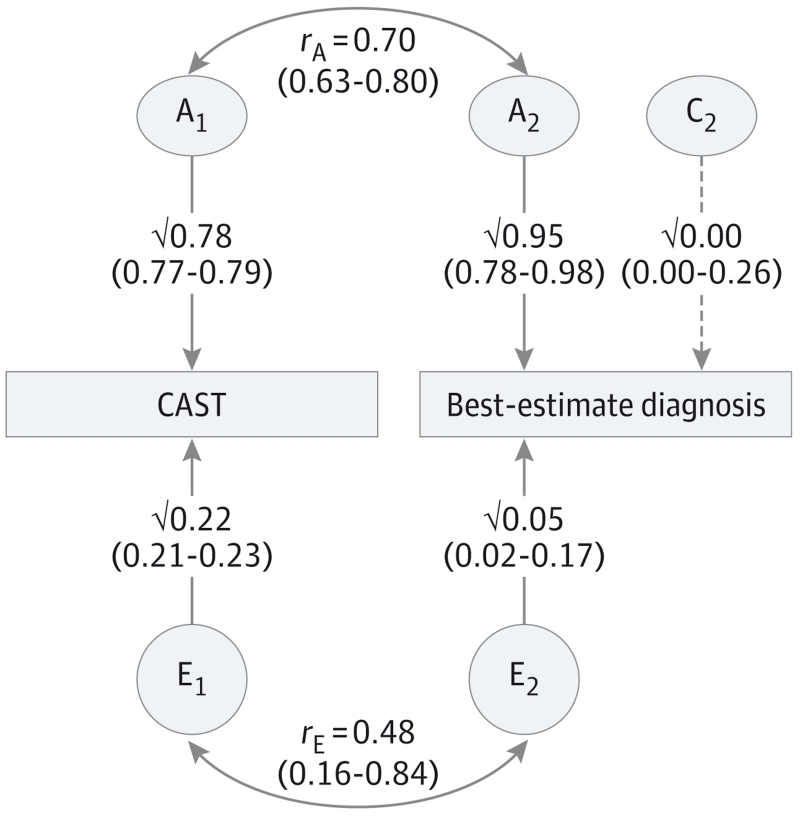
Figure 2. Diagram of the Correlated Factors Solution of the Joint Continuous-Ordinal Model of Effects on the Childhood Autism Spectrum Test (CAST) Results and Best-Estimate Diagnosis (BeD).
The model represents the standardized effects of additive genetic (A1 and A2) and nonshared environmental factors (E1 and E2) on each trait separately and the A and E correlations between the 2 variables (rA and rE). We found no shared environmental influences at play for the CAST score; therefore, no covariance due to correlated shared environmental factors is possible with the autism spectrum disorder (ASD) measures. We modeled the shared environmental effects for the ASD measures, which were nonsignificant for most as shown for the BeD (striped line).
Results
Probandwise Concordance Rates
Probandwise concordance rates were calculated as [2 × (number of concordant pairs)]/[2 × (number of concordant pairs) + discordant pairs] (Table 1). These calculations express the probability that the co-twin of a proband (affected twin) is also affected and are commonly used as an index of twin resemblance. The high MZ (0.62-0.94) and low DZ (0.05-0.61) concordances suggest substantial genetic influence. For example, MZ concordances are 0.87 for ASD and 0.94 for BeD, in contrast to 0.22 and 0.46, respectively, for DZ concordances. However, concordance rates cannot be used to estimate genetic and environmental parameters because they do not take population prevalence rates into account.
Table 1. MZ and DZ Probandwise Concordance Rates Across MZ and DZ Affected Twinsa.
| MZ Twin Pairs |
DZ Twin Pairs |
|||
|---|---|---|---|---|
| Measure | No. of Discordant/ Concordant Pairs |
Probandwise Concordance Rate |
No. of Discordant/ Concordant Pairs |
Probandwise Concordance Rate |
| ASDb | ||||
|
| ||||
| DAWBA | 12/15 | 0.71 | 74/2 | 0.05 |
|
| ||||
| ADI-R | 15/12 | 0.62 | 80/8 | 0.17 |
|
| ||||
| ADOS | 8/12 | 0.75 | 57/9 | 0.40 |
|
| ||||
| Best- estimate diagnosis |
8/17 | 0.87 | 77/11 | 0.22 |
|
| ||||
| ASD and Broad-Spectrum Disorderc | ||||
|
| ||||
| DAWBA | 16/24 | 0.75 | 118/5 | 0.08 |
|
| ||||
| ADI-R | 4/24 | 0.92 | 54/43 | 0.61 |
|
| ||||
| ADOS | 7/16 | 0.82 | 56/18 | 0.39 |
|
| ||||
| Best- estimate diagnosis |
3/24 | 0.94 | 70/30 | 0.46 |
Abbreviations: ADI-R, Autism Diagnostic Interview-Revised; ADOS, Autism Diagnostic Observation Schedule; ASD, autism spectrum disorder; DAWBA, Development and Well-being Assessment; DZ, dizygotic; MZ, monozygotic.
Includes same-sex and opposite-sex twin pairs.
Rates reflect twins included in category 2 (ASD) only.
Rates reflect pairs in which a child was included in category 1 (broad-spectrum disorder) or 2.
Diagnostic Agreement
Agreement of classification of individuals into the 3 categories (unaffected, broad-spectrum disorder, and ASD) for different diagnostic measures was calculated by means of weighted κ coefficients (Stata software; StataCorp) with pre-defined weights used so that the 0-2 cells get a full weight of 1 and the 0-1 and 1-2 cells only a weight of 25% in calculating disagreement. These values (−1 to 1) represent the observed agreement between 2 diagnostic tests relative to the expected agreement between tests occurring by chance alone.49 We found moderate κ agreement for DAWBA and the ADOS assessment (0.58) and substantial agreement for DAWBA and the ADI-R assessment and for DAWBA and BeD (both 0.72), for the ADI-R and ADOS assessments (0.67), and for the ADOS assessment and BeD (0.79). Agreement for the ADI-R assessment and BeD was almost perfect (0.91).
Twin Correlations
The 2:1 MZ:DZ ratio of the cross-twin within-trait correlations for the ADOS assessment and BeD suggest a significant contribution of genetic effects, with the remainder explained by nonshared environmental effects (Table 2). This contribution is not the case for DAWBA, for which the DZ correlation is less than half that of the MZ pairs, pointing to nonadditive genetic effects. For the ADI-R assessment, the DZ correlation is more than half the MZ correlation, indicating genetic and shared environmental effects. The MZ:DZ ratio of the cross-twin cross-trait correlations for the CAST score and each diagnosis indicates mainly genetic and nonshared environmental influences on their overlap.
Table 2. MZ and DZ Within-Trait and Cross-Trait Twin Correlationsa.
| Within-Trait Correlation, r (95% CI)b |
Cross-Twin Correlation, r (95% CI)c |
|||
|---|---|---|---|---|
| Measure | MZ | DZ | MZ Cross-Trait | DZ Cross-Trait |
| CAST | 0.79 (0.77-0.80)d,e | 0.31 (0.28-0.33)d,e | NA | NA |
|
| ||||
| DAWBA | 0.82 (0.67-0.90)e | 0.22 (0.00-0.42) | 0.40 (0.34-0.45)e | −0.01 (0.00-0.07) |
|
| ||||
| ADI-R | 0.87 (0.77-0.93)e | 0.65 (0.55-0.73)e | 0.60 (0.54-0.65)e | 0.40 (0.35-0.45)e |
|
| ||||
| ADOS | 0.77 (0.62- 0.87)e | 0.40 (0.26-0.63)e | 0.56 (0.49-0.61)e | 0.30 (0.23-0.37)e |
|
| ||||
| Best-estimate diagnosis | 0.99 (0.98-0.99)e | 0.53 (0.41-0.63)e | 0.61 (0.57-0.66)e | 0.37 (0.31-0.42)e |
Abbreviations: See Table 1. CAST, Covariance of the Childhood Autism Spectrum Test; NA, not applicable.
Based on 4 bivariate analyses of the CAST score with each diagnostic ASD measure.
Maximum likelihood correlations for MZ and DZ twins (including same-sex and opposite-sex DZ pairs) are estimated in a model with the 2 thresholds on the liability to ASD set to population values of broad-spectrum disorder (5%) and ASD (1%) prevalence.
For the CAST score, 4 sets of correlations are available because 4 bivariate analyses were performed; here, only 1 is given (the other 3 were of similar value and with overlapping 95% CIs).
Maximum likelihood cross-twin, cross-CAST correlation was obtained for each diagnostic variable and the CAST score separately.
Indicates significant estimates (ie, 95% CIs not spanning 0).
Bivariate Genetic Model
Table 3 reports the standardized results of the bivariate ACE models. Variance in the CAST score (age and sex regressed) resulted from genetic influences (78% [95% CI, 77%-79%]) and nonshared environmental effects (22% [95% CI, 21%-23%]), as reported previously.30 Genetic influences were significant for all clinical measures with the highest heritability reported for BeD (95% [95% CI, 74%-98%]) and shared environments significantly explaining the variance of the ADI-R assessment only (30% [95% CI, 8%-47%]). The correlations between the CAST score and each of the ASD variables (rph) is the sum of the paths via the additive genetic and nonshared environmental factors (A and E) connecting the 2 variables (rph_A and rph_E). The phenotypic correlations were moderate to high (0.52-0.65), and genetic factors accounted for 77% to 100% of the covariance. The genetic correlations, that is, the extent to which the same genetic factors influence the CAST score and clinical measures independent of their heritabilities, are substantial (0.52-0.89). The remainder of the covariance was explained by non-shared environmental factors (rph_E), although nonsignificant for the overlap between the CAST score and the ADI-R or the ADOS assessment. Figure 2 depicts the path diagram with standardized estimates of the reduced bivariate AE model (in which no shared environmental influences are found) for the CAST score and BeD, BeD being the best diagnostic estimate of ASD in our study. The findings indicate strong and overlapping genetic influences on dimensional and categorical measures.
Table 3. Standardized Estimates of the Reduced A(C)E Bivariate Models of CAST Scores and Each ofthe 4 Clinical Measures of ASDa.
| Standardized Estimates (95% CI) |
Part of Phenotypic Correlation (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Measure | h 2 | c 2 | e 2 | r A b | r E b | r ph c | r ph_A d | r ph_E d |
| CAST | 0.78 (0.77-0.79)e |
0.00 (0.00-0.00) |
0.22 (0.21-0.23)e |
NA | NA | NA | NA | NA |
|
| ||||||||
| DAWBA | 0.78 (0.48-0.87)e |
0.00 (0.00-0.00) |
0.22 (0.13-0.36)e |
0.52 (0.47-0.67)e |
0.48 (0.32-0.64)e |
0.52 (0.48-0.55)e |
0.40 (77)e | 0.12 (23)e |
|
| ||||||||
| ADI-R | 0.56 (0.37-0.82)e |
0.30 (0.08-0.47)e |
0.14 (0.07-0.47)e |
0.89 (0.70-0.99)e |
0.19 (0.00-0.41) |
0.61 (0.56-0.66)e |
0.58 (≈100)e | 0.03 (≈0) |
|
| ||||||||
| ADOS | 0.76 (0.41-0.86)e |
0.00 (0.00-0.30) |
0.24 (0.14-0.39)e |
0.73 (0.63-0.99)e |
−0.02 (0.00-0.15) |
0.54 (0.51-0.60)e |
0.56 (≈100)e | −0.02 (≈0) |
|
| ||||||||
| Best-estimate diagnosis |
0.95 (0.74-0.98)e |
0.00 (0.00-0.26) |
0.05 (0.02-0.17)e |
0.70 (0.63-0.80)e |
0.48 (0.16-0.84)e |
0.65 (0.24-0.67)e |
0.60 (92)e | 0.05 (8)e |
Abbreviations: See Table 1. c2, shared environmental factors estimate; CAST, Covariance of the Childhood Autism Spectrum Test; e2, nonshared environmental factors estimate; h2, heritability estimate; NA, not applicable.
Thresholds on the ASD liability were fixed at 5% (broad spectrum) and1% (ASD); the estimates for the CAST score across the 4 models were of similar value and with overlapping 95% CIs.
Indicates additive genetic (A) and nonshared environmental (E) correlations between the CAST score and ASD measure.
Indicates phenotypic (ph) correlation between the CAST score and ASD measures.
Indicates part of the phenotypic correlation due to additive genetic (A) and unique environmental (E) influences (percentage of phenotypic correlation). Proportions cannot be calculated for the ADOS assessment owing to the opposite signs of rph_A and rph_E; however, if we disregard the nonsignificant contributions of rph_E for the CAST-ADOS and CAST-ADI-R relationships, shared genetic effects explain nearly all of the observed correlations.
Indicates significant estimates (ie, 95% CIs not spanning 0).
Discussion
The present study examines the genetic and environmental contributions to ASD in a large systematically screened population-based twin sample and the genetic/environmental overlap between a continuous measure of autismlike traits and categorical diagnostic assessments. Our study was novel in including twins with high subclinical levels of traits and selected low-risk twins as well as those with diagnosed ASD to capture the full range of liability. The probandwise concordance rates and liability threshold model analyses reassert the importance of genetic factors in the etiology of ASD. Analyses partitioning liability into genetic and shared and nonshared environmental components indicate that most liability could be attributed to additive genetic influences and a smaller proportion attributed to non-shared environmental influences. This finding held across a number of different measures. We found very little evidence of shared environmental effects overall, which is contrary to the findings of Hallmayer et al,15 although the wide CIs in their results for additive genetic and shared environmental effects overlap with some of the present estimates. In our study, only the ADI-R parent-reported developmental history measure showed significant shared environment effects. Because the ADI-R assessment was completed by the same parent for both twins, the estimated influence of shared environment may be inflated by rater bias. However, the wide CIs (0.08-0.47) warrant caution in interpretation.
Our findings also confirm that the heritability of the liability to ASD is high when incorporating subclinical cases with high trait scores into the model, extending support for the notion that the genetic liability to autism confers a risk for a broader autism phenotype. Indeed, the relationship between the CAST score and diagnostic assessments indicated a substantial genetic correlation and a significant correlation in the nonshared environmental factors that influence variations in both traits. This result indicates common etiologic underpinnings for individual differences in autistic traits across the whole spectrum and in our 3 clinically meaningful categories (ASD, high subclinical trait level, and low risk/trait level). This result provides support for examining broader autistic traits in the general population as a complementary strategy for identifying the genetic risk factors for ASD.50-52 Our findings are broadly in line with those of recent twin and family studies and point toward strong genetic effects in ASD and no strong influence from shared environmental factors. The strengths of the present study add validity to these conclusions because previous research has often lacked the rigor and systematic approach to the sample selection used herein. The population-based sampling in the present study, the 2-stage systematic screening methods used, and the inclusion of individuals with subclinical disorders ensured the capture of a more complete picture of genetic risk (additive and nonadditive) for ASD than in previous studies. A novel contribution is the strong evidence that the same genetic influences are largely responsible for the overlap between dimensional trait measures and categorical diagnostic constructs of ASD. In addition, to the best of our knowledge, this study is one of the largest screened population-based twin studies yet reported.
The limitations of this study include the fact that few of the potentially eligible twin pairs did not enroll in the study. Second, although one of the largest twin studies, the sample size was insufficient to allow any meaningful analyses of the basis for sex differences in ASD. In addition, twin study methods assume that the environments of MZ and DZ53 twins are equal and that twins are not at especially high risk for the disorder under investigation. The available evidence indicates that both assumptions are justified in this study.54 Another issue is that genetic modeling assumes that no gene-environment interactions or correlations exist; if they exist, the estimates of environmental and genetic effects may be inflated.55 Heritability estimates are also population specific and depend on the dynamic interaction with the current environment. Our analysis took a liability threshold approach, but other types of analyses (eg, continuous data modeling, DeFries-Fulker quantile regression56) are possible and may be warranted by future developments in the molecular genetics of ASD. Recent findings lend support to a polygenic trait approach.19
Conclusions
The present study combines the strengths of previous studies and provides a more complete picture than any of them individually by being nationally representative and incorporating dimensional and categorical measures using a systematic repeated screening method. We conclude that liability to ASD and a more broadly defined high-level autism trait phenotype in UK twins 8 years or older derives from substantial genetic and moderate nonshared environmental influences. Genetic influences on diagnosed ASD are shared with those on autistic traits in the general population.
Supplementary Material
Acknowledgments
Funding/Support: The Twins Early Development Study (TEDS) is supported by program grant G0901245 (previously G0500079) from the UK Medical Research Council (MRC). The Social Relationship Study was supported by grant G0500870 from the MRC. This study is also supported by 1+3 PhD studentship MR/J500380/1 from the MRC (Ms Tick), by a senior investigator award from the National Institute for Health Research (Dr Bolton), by the Biomedical Research Centre in Mental Health at the South London and Maudsley NHS Trust (Dr Bolton), and by an Autism Speaks grant (Dr Bolton).
Role of the Funder/Sponsor: The funding source had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Author Contributions
Dr Colvert and Ms Tick contributed equally to this article, as did Drs Happé and Bolton. Ms Tick and Dr Rijsdijk had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Colvert, Tick, Ronald, Plomin, Rijsdijk, Happé, Bolton.
Acquisition, analysis, or interpretation of data: Colvert, Tick, McEwen, Stewart, Curran, Woodhouse, Gillan, Hallett, Lietz, Garnett, Ronald, Happé, Bolton.
Drafting of the manuscript: Colvert, Tick, McEwen, Curran, Rijsdijk, Happé, Bolton.
Critical revision of the manuscript for important intellectual content: Colvert, Tick, McEwen, Stewart, Woodhouse, Gillan, Hallett, Lietz, Garnett, Ronald, Plomin, Rijsdijk, Happé, Bolton.
Statistical analysis: Colvert, Tick, Ronald, Rijsdijk, Bolton.
Obtained funding: Plomin, Happé, Bolton.
Administrative, technical, or material support: Colvert, McEwen, Stewart, Woodhouse, Gillan, Garnett, Ronald.
Study supervision: Curran, Rijsdijk, Happé, Bolton.
Footnotes
Conflict of Interest Disclosures: None reported.
Additional Contributions: We thank the TEDS participants and their families for their ongoing contribution.
REFERENCES
- 1.Folstein S, Rutter M. Genetic influences and infantile autism. Nature. 1977;265(5596):726–728. doi: 10.1038/265726a0. [DOI] [PubMed] [Google Scholar]
- 2.Folstein S, Rutter M. Infantile autism: a genetic study of 21 twin pairs. J Child Psychol Psychiatry. 1977;18(4):297–321. doi: 10.1111/j.1469-7610.1977.tb00443.x. [DOI] [PubMed] [Google Scholar]
- 3.Ritvo ER, Freeman BJ, Mason-Brothers A, Mo A, Ritvo AM. Concordance for the syndrome of autism in 40 pairs of afflicted twins. Am J Psychiatry. 1985;142(1):74–77. doi: 10.1176/ajp.142.1.74. [DOI] [PubMed] [Google Scholar]
- 4.Steffenburg S, Gillberg C, Hellgren L, et al. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30(3):405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 5.Bailey A, Le Couteur A, Gottesman I, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25(1):63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenstein P, Carlström E, Råstam M, Gillberg C, Anckarsäter H. The genetics of autism spectrum disorders and related neuropsychiatric disorders in childhood. Am J Psychiatry. 2010;167(11):1357–1363. doi: 10.1176/appi.ajp.2010.10020223. [DOI] [PubMed] [Google Scholar]
- 7.Taniai H, Nishiyama T, Miyachi T, Imaeda M, Sumi S. Genetic influences on the broad spectrum of autism: study of proband-ascertained twins. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):844–849. doi: 10.1002/ajmg.b.30740. [DOI] [PubMed] [Google Scholar]
- 8.Rosenberg RE, Daniels AM, Law JK, Law PA, Kaufmann WE. Trends in autism spectrum disorder diagnoses: 1994-2007. J Autism Dev Disord. 2009;39(8):1099–1111. doi: 10.1007/s10803-009-0723-6. [DOI] [PubMed] [Google Scholar]
- 9.Ronald A, Hoekstra RA. Autism spectrum disorders and autistic traits: a decade of new twin studies. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(3):255–274. doi: 10.1002/ajmg.b.31159. [DOI] [PubMed] [Google Scholar]
- 10.Ronald A, Happé F, Plomin R. The genetic relationship between individual differences in social and nonsocial behaviours characteristic of autism. Dev Sci. 2005;8(5):444–458. doi: 10.1111/j.1467-7687.2005.00433.x. [DOI] [PubMed] [Google Scholar]
- 11.Ronald A, Happé F, Price TS, Baron-Cohen S, Plomin R. Phenotypic and genetic overlap between autistic traits at the extremes of the general population. J Am Acad Child Adolesc Psychiatry. 2006;45(10):1206–1214. doi: 10.1097/01.chi.0000230165.54117.41. [DOI] [PubMed] [Google Scholar]
- 12.Ronald A, Happé F, Plomin R. A twin study investigating the genetic and environmental aetiologies of parent, teacher and child ratings of autistic-like traits and their overlap. Eur Child Adolesc Psychiatry. 2008;17(8):473–483. doi: 10.1007/s00787-008-0689-5. [DOI] [PubMed] [Google Scholar]
- 13.Skuse DH, Mandy WPL, Scourfield J. Measuring autistic traits: heritability, reliability and validity of the Social and Communication Disorders Checklist. Br J Psychiatry. 2005;187:568–572. doi: 10.1192/bjp.187.6.568. [DOI] [PubMed] [Google Scholar]
- 14.Hoekstra RA, Bartels M, Verweij CJ, Boomsma DI. Heritability of autistic traits in the general population. Arch Pediatr Adolesc Med. 2007;161(4):372–377. doi: 10.1001/archpedi.161.4.372. [DOI] [PubMed] [Google Scholar]
- 15.Hallmayer J, Cleveland S, Torres A, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frazier TW, Thompson L, Youngstrom EA, et al. A twin study of heritable and shared environmental contributions to autism. J Autism Dev Disord. 2014;44(8):2013–2025. doi: 10.1007/s10803-014-2081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordenbæk C, Jørgensen M, Kyvik KO, Bilenberg N. A Danish population-based twin study on autism spectrum disorders. Eur Child Adolesc Psychiatry. 2014;23(1):35–43. doi: 10.1007/s00787-013-0419-5. [DOI] [PubMed] [Google Scholar]
- 18.Sandin S, Lichtenstein P, Kuja-Halkola R, Larsson H, Hultman CM, Reichenberg A. The familial risk of autism. JAMA. 2014;311(17):1770–1777. doi: 10.1001/jama.2014.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaugler T, Klei L, Sanders SJ, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46(8):881–885. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trzaskowski M, Dale PS, Plomin R. No genetic influence for childhood behavior problems from DNA analysis. J Am Acad Child Adolesc Psychiatry. 2013;52(10):1048–1056.e3. doi: 10.1016/j.jaac.2013.07.016. doi:10.1016/j.jaac.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishop DVM, Maybery M, Wong D, Maley A, Hallmayer J. Characteristics of the broader phenotype in autism: a study of siblings using the Children’s Communication Checklist-2. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(2):117–122. doi: 10.1002/ajmg.b.30267. [DOI] [PubMed] [Google Scholar]
- 22.Constantino JN, Todorov A, Hilton C, et al. Autism recurrence in half siblings: strong support for genetic mechanisms of transmission in ASD. Mol Psychiatry. 2013;18(2):137–138. doi: 10.1038/mp.2012.9. [DOI] [PubMed] [Google Scholar]
- 23.Sucksmith E, Roth I, Hoekstra RA. Autistic traits below the clinical threshold: re-examining the broader autism phenotype in the 21st century. Neuropsychol Rev. 2011;21(4):360–389. doi: 10.1007/s11065-011-9183-9. [DOI] [PubMed] [Google Scholar]
- 24.Pickles A, Starr E, Kazak S, et al. Variable expression of the autism broader phenotype: findings from extended pedigrees. J Child Psychol Psychiatry. 2000;41(4):491–502. [PubMed] [Google Scholar]
- 25.Williams JG, Allison C, Scott FJ, et al. The Childhood Autism Spectrum Test (CAST): sex differences. J Autism Dev Disord. 2008;38(9):1731–1739. doi: 10.1007/s10803-008-0558-6. [DOI] [PubMed] [Google Scholar]
- 26.Baron-Cohen S, Wheelwright S, Skinner R, Martin J, Clubley E. The Autism-Spectrum Quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. J Autism Dev Disord. 2001;31(1):5–17. doi: 10.1023/a:1005653411471. [DOI] [PubMed] [Google Scholar]
- 27.Constantino JN. The quantitative nature of autistic social impairment. Pediatr Res. 2011;69(5, pt 2):55R–62R. doi: 10.1203/PDR.0b013e318212ec6e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams J, Scott F, Stott C, et al. The CAST (Childhood Asperger Syndrome Test): test accuracy. Autism. 2005;9(1):45–68. doi: 10.1177/1362361305049029. [DOI] [PubMed] [Google Scholar]
- 29.Robinson EB, Koenen KC, McCormick MC, et al. Evidence that autistic traits show the same etiology in the general population and at the quantitative extremes (5%, 2.5%, and 1%) Arch Gen Psychiatry. 2011;68(11):1113–1121. doi: 10.1001/archgenpsychiatry.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ronald A, Happé F, Bolton P, et al. Genetic heterogeneity between the three components of the autism spectrum: a twin study. J Am Acad Child Adolesc Psychiatry. 2006;45(6):691–699. doi: 10.1097/01.chi.0000215325.13058.9d. [DOI] [PubMed] [Google Scholar]
- 31.Lundström S, Chang Z, Råstam M, et al. Autism spectrum disorders and autistic like traits: similar etiology in the extreme end and the normal variation. Arch Gen Psychiatry. 2012;69(1):46–52. doi: 10.1001/archgenpsychiatry.2011.144. [DOI] [PubMed] [Google Scholar]
- 32.Baron-Cohen S, Scott FJ, Allison C, et al. Prevalence of autism-spectrum conditions: UK school-based population study. Br J Psychiatry. 2009;194(6):500–509. doi: 10.1192/bjp.bp.108.059345. [DOI] [PubMed] [Google Scholar]
- 33.Haworth CMA, Davis OSP, Plomin R. Twins Early Development Study (TEDS): a genetically sensitive investigation of cognitive and behavioral development from childhood to young adulthood. Twin Res Hum Genet. 2013;16(1):117–125. doi: 10.1017/thg.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dworzynski K, Happé F, Bolton P, Ronald A. Relationship between symptom domains in autism spectrum disorders: a population based twin study. J Autism Dev Disord. 2009;39(8):1197–1210. doi: 10.1007/s10803-009-0736-1. [DOI] [PubMed] [Google Scholar]
- 35.Goodman R, Ford T, Richards H, Gatward R, Meltzer H. The Development and Well-Being Assessment: description and initial validation of an integrated assessment of child and adolescent psychopathology. J Child Psychol Psychiatry. 2000;41(5):645–655. [PubMed] [Google Scholar]
- 36.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24(5):659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 37.Lord C, Rutter M, Goode S, et al. Autism Diagnostic Observation Schedule: a standardized observation of communicative and social behavior. J Autism Dev Disord. 1989;19(2):185–212. doi: 10.1007/BF02211841. [DOI] [PubMed] [Google Scholar]
- 38.Boker S, Neale M, Maes H, et al. OpenMx: an open source extended structural equation modeling framework. Psychometrika. 2011;76(2):306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Falconer DS. The inheritance of liability to certain diseases, estimated from the incidence among relatives. Ann Hum Genet. 1965;29(1):51–76. [Google Scholar]
- 40.Pearson K. On the laws of inheritance in man, II: on the inheritance of the mental and moral characters in man, and its comparison with the inheritance of the physical characters. Biometrika. 1904;3(2/3):131–190. [Google Scholar]
- 41.Brugha TS, McManus S, Bankart J, et al. Epidemiology of autism spectrum disorders in adults in the community in England. Arch Gen Psychiatry. 2011;68(5):459–465. doi: 10.1001/archgenpsychiatry.2011.38. [DOI] [PubMed] [Google Scholar]
- 42.Baird G, Simonoff E, Pickles A, et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368(9531):210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- 43.Neale MC, Cardon LR. Methodology for Genetic Studies of Twins and Families. Kluwer Academic Publishers; Dordrecht, the Netherlands: 1992. [Google Scholar]
- 44.Loehlin JC. The Cholesky approach: a cautionary note. Behav Genet. 1996;26(1):65–69. [Google Scholar]
- 45.Rijsdijk FV, van Haren NE, Picchioni MM, et al. Brain MRI abnormalities in schizophrenia: same genes or same environment? Psychol Med. 2005;35(10):1399–1409. doi: 10.1017/S0033291705005167. [DOI] [PubMed] [Google Scholar]
- 46.Holmboe K, Rijsdijk FV, Hallett V, Happé F, Plomin R, Ronald A. Strong genetic influences on the stability of autistic traits in childhood. J AmAcad Child Adolesc Psychiatry. 2014;53(2):221–230. doi: 10.1016/j.jaac.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owens SF, Picchioni MM, Ettinger U, et al. Prefrontal deviations in function but not volume are putative endophenotypes for schizophrenia. Brain. 2012;135(pt 7):2231–2244. doi: 10.1093/brain/aws138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Plomin R, DeFries JC, Knopik VS, Neiderhiser JM. Behavioural Genetics. 6th ed. Worth Publishers; New York, NY: 2013. [Google Scholar]
- 49.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med. 2005;37(5):360–363. [PubMed] [Google Scholar]
- 50.Ronald A, Butcher LM, Docherty S, et al. A genome-wide association study of social and non-social autistic-like traits in the general population using pooled DNA, 500 K SNP microarrays and both community and diagnosed autism replication samples. Behav Genet. 2010;40(1):31–45. doi: 10.1007/s10519-009-9308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steer CD, Golding J, Bolton PF. Traits contributing to the autistic spectrum. PLoS One. 2010;5(9):e12633. doi: 10.1371/journal.pone.0012633. doi:10.1371/journal.pone.0012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.St Pourcain B, Skuse DH, Mandy WP, et al. Variability in the common genetic architecture of social-communication spectrum phenotypes during childhood and adolescence. Mol Autism. 2014;5(1):18. doi: 10.1186/2040-2392-5-18. doi:10.1186/2040-2392-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rijsdijk FV, Sham PC. Analytic approaches to twin data using structural equation models. Brief Bioinform. 2002;3(2):119–133. doi: 10.1093/bib/3.2.119. [DOI] [PubMed] [Google Scholar]
- 54.Curran S, Dworzynski K, Happé F, et al. No major effect of twinning on autistic traits. Autism Res. 2011;4(5):377–382. doi: 10.1002/aur.207. [DOI] [PubMed] [Google Scholar]
- 55.Kendler KS, Eaves LJ. Models for the joint effect of genotype and environment on liability to psychiatric illness. Am J Psychiatry. 1986;143(3):279–289. doi: 10.1176/ajp.143.3.279. [DOI] [PubMed] [Google Scholar]
- 56.Logan JA, Petrill SA, Hart SA, et al. Heritability across the distribution: an application of quantile regression. Behav Genet. 2012;42(2):256–267. doi: 10.1007/s10519-011-9497-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.